bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
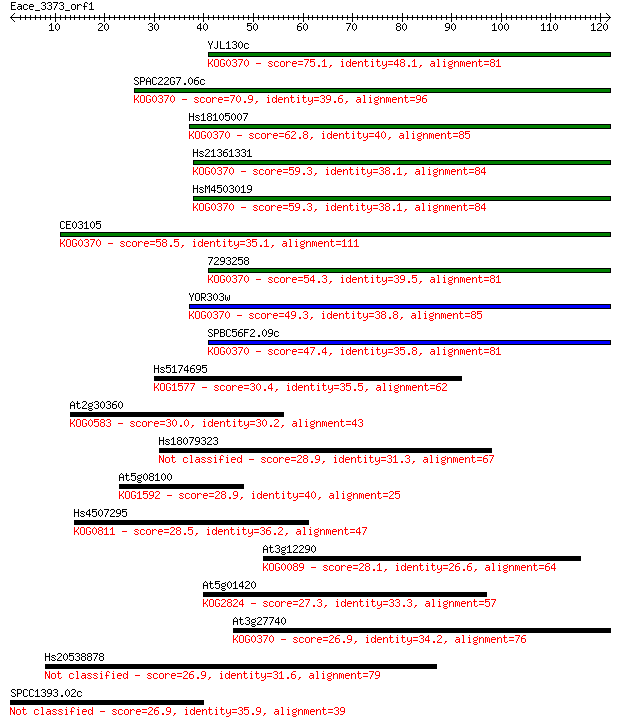
Query= Eace_3373_orf1
Length=121
Score E
Sequences producing significant alignments: (Bits) Value
YJL130c 75.1 3e-14
SPAC22G7.06c 70.9 5e-13
Hs18105007 62.8 2e-10
Hs21361331 59.3 2e-09
HsM4503019 59.3 2e-09
CE03105 58.5 3e-09
7293258 54.3 5e-08
YOR303w 49.3 2e-06
SPBC56F2.09c 47.4 8e-06
Hs5174695 30.4 0.86
At2g30360 30.0 1.2
Hs18079323 28.9 2.3
At5g08100 28.9 2.6
Hs4507295 28.5 2.8
At3g12290 28.1 3.9
At5g01420 27.3 6.8
At3g27740 26.9 8.6
Hs20538878 26.9 8.9
SPCC1393.02c 26.9 9.9
> YJL130c
Length=2214
Score = 75.1 bits (183), Expect = 3e-14, Method: Composition-based stats.
Identities = 39/90 (43%), Positives = 54/90 (60%), Gaps = 14/90 (15%)
Query 41 DPNSRNLVAEVSRKEPQVFVSPVN---------PSCHQVKVLAIDCGMKTNIYRQFLYSN 91
DPN +NLV++VS EP+++V P + P +++LAID GMK N R F+
Sbjct 189 DPNVQNLVSKVSINEPKLYVPPADNKHIELQTGPDGKVLRILAIDVGMKYNQIRCFIKRG 248
Query 92 FPDCNVVLKAVPWDYDISNDSFDGLFISNG 121
V LK VPW+YD + + +DGLFISNG
Sbjct 249 -----VELKVVPWNYDFTKEDYDGLFISNG 273
> SPAC22G7.06c
Length=2244
Score = 70.9 bits (172), Expect = 5e-13, Method: Composition-based stats.
Identities = 38/101 (37%), Positives = 58/101 (57%), Gaps = 14/101 (13%)
Query 26 WAAALQAAAGRLPLDDPNSRNLVAEVSRKEPQVFVSPVNPSCHQ-----VKVLAIDCGMK 80
W+ A + +P +PN+ NL ++VS KEP+++ + + +++L ID GMK
Sbjct 218 WSTAFE----DIPWKNPNTENLTSQVSIKEPKLYEPHPTTAIKKADGKIIRILVIDVGMK 273
Query 81 TNIYRQFLYSNFPDCNVVLKAVPWDYDISNDSFDGLFISNG 121
N R FL + V L VPWDYD + +++DGLFISNG
Sbjct 274 YNQIRCFL-----NRGVELLVVPWDYDFTKETYDGLFISNG 309
> Hs18105007
Length=2225
Score = 62.8 bits (151), Expect = 2e-10, Method: Composition-based stats.
Identities = 34/85 (40%), Positives = 48/85 (56%), Gaps = 10/85 (11%)
Query 37 LPLDDPNSRNLVAEVSRKEPQVFVSPVNPSCHQVKVLAIDCGMKTNIYRQFLYSNFPDCN 96
LP DPN+R LV EVS K P+VF + P ++LA+DCG+K N R
Sbjct 148 LPFLDPNARPLVPEVSIKTPRVFNTGGAP-----RILALDCGLKYNQIRCLCQRG----- 197
Query 97 VVLKAVPWDYDISNDSFDGLFISNG 121
+ VPWD+ + + ++GLF+SNG
Sbjct 198 AEVTVVPWDHALDSQEYEGLFLSNG 222
> Hs21361331
Length=1500
Score = 59.3 bits (142), Expect = 2e-09, Method: Composition-based stats.
Identities = 32/86 (37%), Positives = 51/86 (59%), Gaps = 12/86 (13%)
Query 38 PLD--DPNSRNLVAEVSRKEPQVFVSPVNPSCHQVKVLAIDCGMKTNIYRQFLYSNFPDC 95
P+D DPN +NL+AEVS K+ +V+ NP+ KV+A+DCG+K N+ R +
Sbjct 189 PVDFVDPNKQNLIAEVSTKDVKVY-GKGNPT----KVVAVDCGIKNNVIRLLVKR----- 238
Query 96 NVVLKAVPWDYDISNDSFDGLFISNG 121
+ VPW++D + +DG+ I+ G
Sbjct 239 GAEVHLVPWNHDFTKMEYDGILIAGG 264
> HsM4503019
Length=1500
Score = 59.3 bits (142), Expect = 2e-09, Method: Composition-based stats.
Identities = 32/86 (37%), Positives = 51/86 (59%), Gaps = 12/86 (13%)
Query 38 PLD--DPNSRNLVAEVSRKEPQVFVSPVNPSCHQVKVLAIDCGMKTNIYRQFLYSNFPDC 95
P+D DPN +NL+AEVS K+ +V+ NP+ KV+A+DCG+K N+ R +
Sbjct 189 PVDFVDPNKQNLIAEVSTKDVKVY-GKGNPT----KVVAVDCGIKNNVIRLLVKR----- 238
Query 96 NVVLKAVPWDYDISNDSFDGLFISNG 121
+ VPW++D + +DG+ I+ G
Sbjct 239 GAEVHLVPWNHDFTKMEYDGILIAGG 264
> CE03105
Length=2198
Score = 58.5 bits (140), Expect = 3e-09, Method: Composition-based stats.
Identities = 39/112 (34%), Positives = 56/112 (50%), Gaps = 12/112 (10%)
Query 11 QQLQQRAQRAAKLQAWAAALQAAAGRLPLDDPNSRNLVAEVSRKEPQVFVSPVNPSCHQV 70
+QL ++ + ++A A D N+ NLV VSRKEP V+ S
Sbjct 126 RQLVKKIRETGTMKAKLVIESDNAQNFDYVDVNAENLVDFVSRKEPVVYGS------GDQ 179
Query 71 KVLAIDCGMKTNIYRQFLYSNFPDCNVVLKAVPWDYDISNDS-FDGLFISNG 121
+LA+DCG+K N R F +K VPW++ I +S +DGLF+SNG
Sbjct 180 TILAVDCGLKNNQIRCLAKRGFR-----VKVVPWNHPIDTESDYDGLFLSNG 226
> 7293258
Length=2189
Score = 54.3 bits (129), Expect = 5e-08, Method: Composition-based stats.
Identities = 32/81 (39%), Positives = 44/81 (54%), Gaps = 8/81 (9%)
Query 41 DPNSRNLVAEVSRKEPQVFVSPVNPSCHQVKVLAIDCGMKTNIYRQFLYSNFPDCNVVLK 100
DPN RNL E S KE QV+ NP+ ++ +DCG+K N R L +V L
Sbjct 168 DPNVRNLAKECSVKERQVY---GNPNGKGPRIAILDCGLKLNQLRCLLQRG---ASVTL- 220
Query 101 AVPWDYDISNDSFDGLFISNG 121
+PW + ++ FD LF+SNG
Sbjct 221 -LPWSARLEDEQFDALFLSNG 240
> YOR303w
Length=411
Score = 49.3 bits (116), Expect = 2e-06, Method: Composition-based stats.
Identities = 33/87 (37%), Positives = 46/87 (52%), Gaps = 9/87 (10%)
Query 37 LPLDDPNSRNLVAEVSRKEPQVFVSPVNPSCHQVKVLAIDCGMKTNIYRQFLYSNFPDCN 96
+P +P NLVA+V+ K+P F P + V IDCG+K NI R + N
Sbjct 153 VPYVNPMKTNLVAQVTTKKP--FHVSALPGKAKANVALIDCGVKENIIRCLVKRG---AN 207
Query 97 VVLKAVPWDYDISN--DSFDGLFISNG 121
V + P+DY I + FDG+F+SNG
Sbjct 208 VTV--FPYDYRIQDVASEFDGIFLSNG 232
> SPBC56F2.09c
Length=415
Score = 47.4 bits (111), Expect = 8e-06, Method: Composition-based stats.
Identities = 29/83 (34%), Positives = 45/83 (54%), Gaps = 11/83 (13%)
Query 41 DPNSRNLVAEVSRKEPQVFVSPVNPSCHQVKVLAIDCGMKTNIYRQFLYSNFPDCNVVLK 100
+P NLV++VS +EP FVS + + + IDCG+K NI R + +
Sbjct 199 NPEEVNLVSQVSTREP-FFVSGGD---GMLNIAVIDCGVKENILRSLVSR-----GASVT 249
Query 101 AVPWDYDISN--DSFDGLFISNG 121
P+DY I N ++DG+F++NG
Sbjct 250 VFPFDYPIQNVASNYDGIFLTNG 272
> Hs5174695
Length=326
Score = 30.4 bits (67), Expect = 0.86, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 33/62 (53%), Gaps = 4/62 (6%)
Query 30 LQAAAGRLPLDDPNSRNLVAEVSRKEPQVFVSPVNPSCHQVKVLAIDCGMKTNIYRQFLY 89
L AA+ R+PL D NS ++ + EP+ +P VKV AID G + +I ++Y
Sbjct 3 LSAASHRIPLSDGNSIPIIGLGTYSEPK--STPKGACATSVKV-AIDTGYR-HIDGAYIY 58
Query 90 SN 91
N
Sbjct 59 QN 60
> At2g30360
Length=435
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 13/46 (28%), Positives = 23/46 (50%), Gaps = 3/46 (6%)
Query 13 LQQRAQRAAKLQAWAAALQA---AAGRLPLDDPNSRNLVAEVSRKE 55
L ++ AK+ W+ + AG LP +DPN N+ ++ + E
Sbjct 194 LSKKGYEGAKVDVWSCGIVLFVLVAGYLPFNDPNVMNMYKKIYKGE 239
> Hs18079323
Length=586
Score = 28.9 bits (63), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 21/74 (28%), Positives = 33/74 (44%), Gaps = 7/74 (9%)
Query 31 QAAAGRLPLDDPNSRNLVAEVSRKEPQVFVSP---VNPSCHQVKV----LAIDCGMKTNI 83
AA+ LP DDPN+ + E +R E ++ P V +C ++ L C +N
Sbjct 150 HAASSSLPRDDPNTNAVATEETRSESELLFLPDYLVLSNCETGRLHHTSLPTRCDSWSNS 209
Query 84 YRQFLYSNFPDCNV 97
R ++F D V
Sbjct 210 DRSLEQASFDDVFV 223
> At5g08100
Length=315
Score = 28.9 bits (63), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 10/25 (40%), Positives = 15/25 (60%), Gaps = 0/25 (0%)
Query 23 LQAWAAALQAAAGRLPLDDPNSRNL 47
+ WA AL AG +P+D P+ R +
Sbjct 1 MVGWAIALHGGAGDIPIDLPDERRI 25
> Hs4507295
Length=261
Score = 28.5 bits (62), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 17/47 (36%), Positives = 24/47 (51%), Gaps = 0/47 (0%)
Query 14 QQRAQRAAKLQAWAAALQAAAGRLPLDDPNSRNLVAEVSRKEPQVFV 60
+Q A+R + A A +G P D RNLV+ S+ +PQV V
Sbjct 109 RQAAEREKEFVARVRASSRVSGSFPEDSSKERNLVSWESQTQPQVQV 155
> At3g12290
Length=299
Score = 28.1 bits (61), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 17/70 (24%), Positives = 31/70 (44%), Gaps = 6/70 (8%)
Query 52 SRKEPQVFVSPVNPSCHQVKVLAIDCGMKTNIYRQFLYSNF------PDCNVVLKAVPWD 105
SRK+ Q +V+ +C +V + + D G+ + L S PD + +L +P
Sbjct 49 SRKDSQTYVNTKRKACAEVGIKSFDVGLPEEVSEADLISKVHELNSNPDVHGILVQLPLP 108
Query 106 YDISNDSFDG 115
I+ + G
Sbjct 109 KHINEEHILG 118
> At5g01420
Length=401
Score = 27.3 bits (59), Expect = 6.8, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 25/59 (42%), Gaps = 2/59 (3%)
Query 40 DDPNSRNLVAEVSRKEPQVFVSPVNPSCHQVKVLAIDCGMKTNIYRQFLYSNF--PDCN 96
++ S N V + P V P P + LA+ CG T I R+ S F PD N
Sbjct 78 EEEASGNFVGDEENVRPPVNQIPRVPCDRKESKLAVSCGSDTGIGRKSFGSGFRRPDLN 136
> At3g27740
Length=428
Score = 26.9 bits (58), Expect = 8.6, Method: Composition-based stats.
Identities = 26/85 (30%), Positives = 37/85 (43%), Gaps = 11/85 (12%)
Query 46 NLVAEVSRKEPQVFVSPVNPSC---------HQVKVLAIDCGMKTNIYRQFLYSNFPDCN 96
+L+++VS K P +V N KV+A D G+K NI R+ L S
Sbjct 207 DLISDVSCKSPYEWVDKTNAEWDFNTNSRDGKSYKVIAYDFGIKQNILRR-LSSYGCQIT 265
Query 97 VVLKAVPWDYDISNDSFDGLFISNG 121
VV P + + DG+ SNG
Sbjct 266 VVPSTFPAAEALKMNP-DGILFSNG 289
> Hs20538878
Length=1068
Score = 26.9 bits (58), Expect = 8.9, Method: Composition-based stats.
Identities = 25/90 (27%), Positives = 33/90 (36%), Gaps = 11/90 (12%)
Query 8 QQQQQLQQR---------AQRAAKLQAWAA--ALQAAAGRLPLDDPNSRNLVAEVSRKEP 56
QQQQQ R A + + Q W A L + P+D P R+ +R
Sbjct 339 QQQQQPGFREWGEMRAPGAPQVEEAQVWLAYPCLLLGSWDYPVDPPEKRSKSPTPTRVHK 398
Query 57 QVFVSPVNPSCHQVKVLAIDCGMKTNIYRQ 86
V +CHQ V D K + RQ
Sbjct 399 GPLVHQEGETCHQATVAYTDTRAKAHRSRQ 428
> SPCC1393.02c
Length=406
Score = 26.9 bits (58), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 26/39 (66%), Gaps = 0/39 (0%)
Query 1 VKREHELQQQQQLQQRAQRAAKLQAWAAALQAAAGRLPL 39
++RE EL+Q+ + +QR Q+A + + AA+ + + R PL
Sbjct 50 LQRERELRQKYEEEQRRQQAMEAKRIAASTRQTSERPPL 88
Lambda K H
0.319 0.132 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1198419392
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40