bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_3394_orf2
Length=156
Score E
Sequences producing significant alignments: (Bits) Value
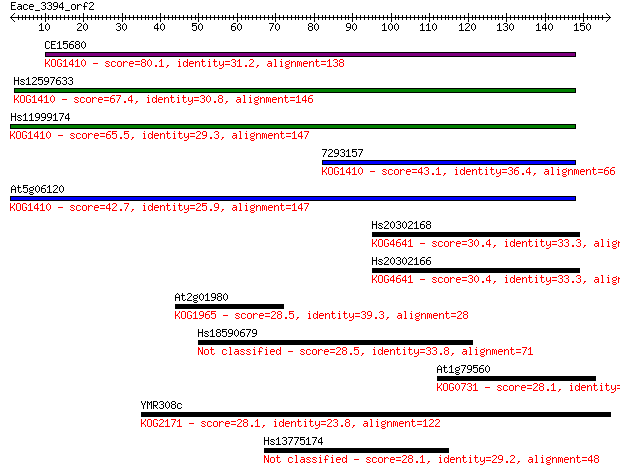
CE15680 80.1 2e-15
Hs12597633 67.4 1e-11
Hs11999174 65.5 5e-11
7293157 43.1 2e-04
At5g06120 42.7 3e-04
Hs20302168 30.4 1.6
Hs20302166 30.4 1.6
At2g01980 28.5 5.3
Hs18590679 28.5 6.2
At1g79560 28.1 6.7
YMR308c 28.1 7.0
Hs13775174 28.1 7.9
> CE15680
Length=1078
Score = 80.1 bits (196), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 43/148 (29%), Positives = 83/148 (56%), Gaps = 10/148 (6%)
Query 10 LLRCVEDGLCAQETVLMQS-CATLGHFVEFIFKYRNSE---------EKEGDAVRAFLEA 59
LLR + GL + + +++ S C++L + ++++ + EGD + ++
Sbjct 904 LLRSIHSGLSSVDAIVITSACSSLDTILNYLYRRLTRSTPPTNKVGMDPEGDNILIAIKQ 963
Query 60 QPGSLPRMLQLIFQLLMTGQLPNLFAISGVLLGLIVLQQNEYLKLKQQIIEQQIESKRPQ 119
P L +MLQ + L+M G++ +++S LLGLI++Q++ Y +K+++ QQ ++
Sbjct 964 HPDILAKMLQAVITLMMFGEVKCQWSLSRPLLGLILIQEDVYSDMKRELTSQQTYDRQAD 1023
Query 120 VEGFFIELMVNIEDDLTPANKDRFMRNL 147
+ F +LM N+E +LT NKD F +NL
Sbjct 1024 FDMLFTQLMSNVEMNLTVKNKDTFTQNL 1051
> Hs12597633
Length=1088
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 45/157 (28%), Positives = 80/157 (50%), Gaps = 11/157 (7%)
Query 2 LPPAHLAQLLRCVEDGLCAQETVLMQSCAT-LGHFVEFIFKYRNSEEKE----------G 50
L P L +L + +GL +TV+ SC T L + V ++FK+ E K+ G
Sbjct 906 LEPPVLMYVLTSISEGLTTLDTVVSSSCCTSLDYIVTYLFKHIAKEGKKPLRCREATQAG 965
Query 51 DAVRAFLEAQPGSLPRMLQLIFQLLMTGQLPNLFAISGVLLGLIVLQQNEYLKLKQQIIE 110
+ F++ P L +M+ ++ ++ N +++S LLGLI+L + + +L+ +I
Sbjct 966 QRLLHFMQQNPDVLQQMMSVLMNTIVFEDCRNQWSVSRPLLGLILLNEKYFSELRASLIN 1025
Query 111 QQIESKRPQVEGFFIELMVNIEDDLTPANKDRFMRNL 147
Q K+ + F LM +E +L+ N+DRF +NL
Sbjct 1026 SQPLPKQEVLAQCFRNLMEGVEQNLSVKNRDRFTQNL 1062
> Hs11999174
Length=1087
Score = 65.5 bits (158), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 43/156 (27%), Positives = 79/156 (50%), Gaps = 9/156 (5%)
Query 1 ELPPAHLAQLLRCVEDGLCAQETVLMQSCAT-LGHFVEFIFK--YRNSEEK------EGD 51
L P + +L + +GL A +T++ C + L H V ++FK R+++++ E D
Sbjct 907 SLEPHVIMYILSSISEGLTALDTMVCTGCCSCLDHIVTYLFKQLSRSTKKRTTPLNQESD 966
Query 52 AVRAFLEAQPGSLPRMLQLIFQLLMTGQLPNLFAISGVLLGLIVLQQNEYLKLKQQIIEQ 111
++ P + +ML + +++ N +++S LLGLI+L + + L+ I+
Sbjct 967 RFLHIMQQHPEMIQQMLSTVLNIIIFEDCRNQWSMSRPLLGLILLNEKYFSDLRNSIVNS 1026
Query 112 QIESKRPQVEGFFIELMVNIEDDLTPANKDRFMRNL 147
Q K+ + F LM IE +L N+DRF +NL
Sbjct 1027 QPPEKQQAMHLCFENLMEGIERNLLTKNRDRFTQNL 1062
> 7293157
Length=109
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 24/66 (36%), Positives = 42/66 (63%), Gaps = 0/66 (0%)
Query 82 NLFAISGVLLGLIVLQQNEYLKLKQQIIEQQIESKRPQVEGFFIELMVNIEDDLTPANKD 141
N +++S LL LI+L ++ Y LK +II Q K+ + +F +LMV IE +++ NK+
Sbjct 24 NQWSMSRPLLVLILLYEDYYRSLKDRIICAQPIEKQQTMAQWFDDLMVGIERNVSSKNKE 83
Query 142 RFMRNL 147
+F +N+
Sbjct 84 KFTQNM 89
> At5g06120
Length=1059
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 38/161 (23%), Positives = 71/161 (44%), Gaps = 16/161 (9%)
Query 1 ELPPAHLAQLLRCVEDGLCAQETVLMQSCA-TLGHFVEFIFKYRNSEEKEGDAVRAFLE- 58
+L A L+ +E GL +T + CA + + + F N E A +
Sbjct 891 KLDTATFMHLVGSLESGLKGLDTSISSQCAIAVDNLASYYFN--NITMGEAPTTPAAIRF 948
Query 59 AQ-----PGSLPRMLQLIFQLLMTGQLPNLFAISGVLLGLIVLQQNEYLKLKQQIIEQQI 113
AQ P P +L+ +F++++ N +++S +L LI++ + + LK +I+ Q
Sbjct 949 AQHIADCPSLFPEILKTLFEIVLFEDCGNQWSLSRPMLSLILISEQIFSDLKAKILSSQS 1008
Query 114 -------ESKRPQVEGFFIELMVNIEDDLTPANKDRFMRNL 147
+ ++ F LM +I L N+D+F +NL
Sbjct 1009 IILMVQPADQHQRLSACFDSLMTDISRGLDSKNRDKFTQNL 1049
> Hs20302168
Length=1041
Score = 30.4 bits (67), Expect = 1.6, Method: Composition-based stats.
Identities = 18/56 (32%), Positives = 31/56 (55%), Gaps = 6/56 (10%)
Query 95 VLQQNEYLKLKQQIIEQQIES--KRPQVEGFFIELMVNIEDDLTPANKDRFMRNLY 148
VLQ ++YL+L+Q+I + I P+ EG F + + N+ + N R+ N+Y
Sbjct 983 VLQHSQYLRLRQRICKSSILQWPDNPKAEGLFWQTLRNV---VLTENDSRY-NNMY 1034
> Hs20302166
Length=1059
Score = 30.4 bits (67), Expect = 1.6, Method: Composition-based stats.
Identities = 18/56 (32%), Positives = 31/56 (55%), Gaps = 6/56 (10%)
Query 95 VLQQNEYLKLKQQIIEQQIES--KRPQVEGFFIELMVNIEDDLTPANKDRFMRNLY 148
VLQ ++YL+L+Q+I + I P+ EG F + + N+ + N R+ N+Y
Sbjct 1001 VLQHSQYLRLRQRICKSSILQWPDNPKAEGLFWQTLRNV---VLTENDSRY-NNMY 1052
> At2g01980
Length=1162
Score = 28.5 bits (62), Expect = 5.3, Method: Composition-based stats.
Identities = 11/28 (39%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 44 NSEEKEGDAVRAFLEAQPGSLPRMLQLI 71
N EKEG+ + FLE S P++L+++
Sbjct 673 NESEKEGEEAKKFLEKVRSSFPQVLRVV 700
> Hs18590679
Length=524
Score = 28.5 bits (62), Expect = 6.2, Method: Composition-based stats.
Identities = 24/71 (33%), Positives = 34/71 (47%), Gaps = 7/71 (9%)
Query 50 GDAVRAFLEAQPGSLPRMLQLIFQLLMTGQLPNLFAISGVLLGLIVLQQNEYLKLKQQII 109
G RA AQP +L+ + TG P++F +SG L+GL L+ E L QQ
Sbjct 66 GLTTRALDHAQP-----LLEHLQPQRATGVCPSVFGVSGFLVGLDKLE--EKLPFLQQPS 118
Query 110 EQQIESKRPQV 120
E + S + V
Sbjct 119 ETVVTSAKDVV 129
> At1g79560
Length=1000
Score = 28.1 bits (61), Expect = 6.7, Method: Composition-based stats.
Identities = 12/41 (29%), Positives = 27/41 (65%), Gaps = 3/41 (7%)
Query 112 QIESKRPQVEGFFIELMVNIEDDLTPANKDRFMRNLYQKAT 152
++E +R ++ F+E ++ + +P N ++F RN+++KAT
Sbjct 178 RVERERRELTESFMEALI---PEPSPGNIEKFKRNMWRKAT 215
> YMR308c
Length=1089
Score = 28.1 bits (61), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 29/122 (23%), Positives = 52/122 (42%), Gaps = 10/122 (8%)
Query 35 FVEFIFKYRNSEEKEGDAVRAFLEAQPGSLPRMLQLIFQLLMTGQLPNLFAISGVLLGLI 94
+ + +NS+ E DA+R++LE + R+L F L+ +P L + +
Sbjct 580 LISILVALQNSDIDEDDALRSYLEQSWSRICRILGDDFVPLLPIVIPPLLITAKATQDVG 639
Query 95 VLQQNEYLKLKQQIIEQQIESKRPQVEGFFIELMVNIEDDLTPANKDRFMRNLYQKATTS 154
++++ E +Q + QV+G I + ++ DD A M L AT
Sbjct 640 LIEEEEAANFQQYPDWDVV-----QVQGKHIAIHTSVLDDKVSA-----MELLQSYATLL 689
Query 155 RG 156
RG
Sbjct 690 RG 691
> Hs13775174
Length=2789
Score = 28.1 bits (61), Expect = 7.9, Method: Composition-based stats.
Identities = 14/48 (29%), Positives = 26/48 (54%), Gaps = 0/48 (0%)
Query 67 MLQLIFQLLMTGQLPNLFAISGVLLGLIVLQQNEYLKLKQQIIEQQIE 114
+L+ FQ+L + N+F +L +++ +E + LK + IE IE
Sbjct 2017 ILKKYFQMLQDNNMDNIFITEENVLDVVINHSHEAIILKPEAIEMYIE 2064
Lambda K H
0.323 0.139 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2070320142
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40