bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_3401_orf1
Length=153
Score E
Sequences producing significant alignments: (Bits) Value
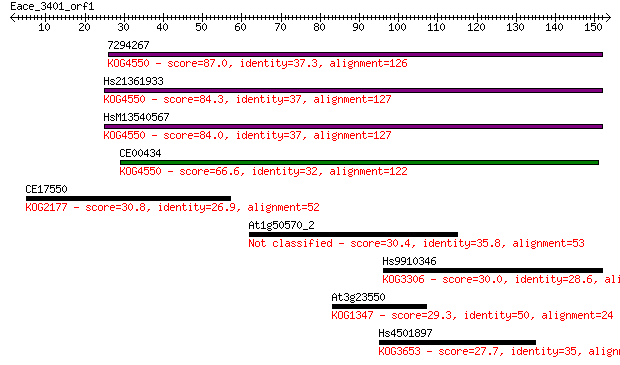
7294267 87.0 1e-17
Hs21361933 84.3 9e-17
HsM13540567 84.0 9e-17
CE00434 66.6 2e-11
CE17550 30.8 1.1
At1g50570_2 30.4 1.5
Hs9910346 30.0 1.7
At3g23550 29.3 3.2
Hs4501897 27.7 8.6
> 7294267
Length=596
Score = 87.0 bits (214), Expect = 1e-17, Method: Composition-based stats.
Identities = 47/130 (36%), Positives = 75/130 (57%), Gaps = 12/130 (9%)
Query 26 YGAAAVGATFKLTVTDLNGTKTPRTGVQLPQSAHAPLSLPFELFGLGQTNNYVEEFFLGL 85
YG G + T +G + + VQLPQ+++ L LP+ FGLG+T N+V++ +GL
Sbjct 451 YGTNLPGPRITYSTTTQDGDQQCGSSVQLPQASYFALQLPYTCFGLGRTPNFVDQLTVGL 510
Query 86 PLAQLTSHNMWVSLIPNSQVIVMPYPLEKPSSWQLELSVHPSRSFANILITTVICL---- 141
+ W LIPNSQ+IV+P PL +PS W+ +L V PS+ +++ +V+ L
Sbjct 511 G----SKLRNWTQLIPNSQIIVVPKPLNEPSHWKAQLFVTPSK----LILMSVVALGGTC 562
Query 142 LLVALIIFIL 151
L++ II +L
Sbjct 563 LVIVFIILVL 572
> Hs21361933
Length=612
Score = 84.3 bits (207), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 47/130 (36%), Positives = 71/130 (54%), Gaps = 9/130 (6%)
Query 25 PYGAAAVGATFKLTVTDLNGTKTPRTGVQLPQSAHAPLSLPFELFGLGQTNNYVEEFFLG 84
P+G G T D NG + QL QSAH L LP+ + GLG++ N+++ ++G
Sbjct 459 PFGVNQPGPYIMYTTVDANGYLKNGSAGQLSQSAHLALQLPYNVLGLGRSANFLDHLYVG 518
Query 85 LPL---AQLTSHNMWVSLIPNSQVIVMPYPLEKPSSWQLELSVHPSRSFANILITTVICL 141
+P + W ++IPNSQ+IV+PYP P SW +L + PS NI++ T I
Sbjct 519 IPRPSGEKSIRKQEWTAIIPNSQLIVIPYPHNVPRSWSAKLYLTPS----NIVLLTAIA- 573
Query 142 LLVALIIFIL 151
L+ + +FIL
Sbjct 574 -LIGVCVFIL 582
> HsM13540567
Length=609
Score = 84.0 bits (206), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 47/130 (36%), Positives = 71/130 (54%), Gaps = 9/130 (6%)
Query 25 PYGAAAVGATFKLTVTDLNGTKTPRTGVQLPQSAHAPLSLPFELFGLGQTNNYVEEFFLG 84
P+G G T D NG + QL QSAH L LP+ + GLG++ N+++ ++G
Sbjct 456 PFGVNQPGPYIMYTTVDANGYLKNGSAGQLSQSAHLALQLPYNVLGLGRSANFLDHLYVG 515
Query 85 LPL---AQLTSHNMWVSLIPNSQVIVMPYPLEKPSSWQLELSVHPSRSFANILITTVICL 141
+P + W ++IPNSQ+IV+PYP P SW +L + PS NI++ T I
Sbjct 516 IPRPSGEKSIRKQEWTAIIPNSQLIVIPYPHNVPRSWSAKLYLTPS----NIVLLTAIA- 570
Query 142 LLVALIIFIL 151
L+ + +FIL
Sbjct 571 -LIGVCVFIL 579
> CE00434
Length=599
Score = 66.6 bits (161), Expect = 2e-11, Method: Composition-based stats.
Identities = 39/126 (30%), Positives = 68/126 (53%), Gaps = 5/126 (3%)
Query 29 AAVGATFKLTVTDLNGTKTPRTGVQLPQSAHAPLSLPFELFGLGQTNNYVEEFFLGLPLA 88
+ GA ++TD G T Q+P S++ L LPF L+GLG++ N+V+E + +P
Sbjct 451 SMTGACASFSMTDGWGGSTQSVACQVPASSNRALYLPFLLYGLGRSPNFVDELNIAIPKY 510
Query 89 QLTSHNMWVSL---IPNSQVIVMPYPLEKPSSWQLELSVHPSRSFANIL-ITTVICLLLV 144
+ SL +PNS++IV+P P ++ W L V PS L + ++C +L+
Sbjct 511 ADRKEDWKHSLKQIVPNSRIIVLP-PSDQYPHWTSRLYVTPSALIVQSLAVIALVCCMLL 569
Query 145 ALIIFI 150
+++F+
Sbjct 570 MVVVFL 575
> CE17550
Length=603
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 14/53 (26%), Positives = 29/53 (54%), Gaps = 2/53 (3%)
Query 5 QQQQQNVSA-SAPQVVPDLPPPYGAAAVGATFKLTVTDLNGTKTPRTGVQLPQ 56
++++QNV+ P++V +LPPP + AT K+ + + P ++P+
Sbjct 99 RRKEQNVTVIEVPELVHNLPPPMAEVSSSAT-KIVTSQQQQQQQPAVATKMPR 150
> At1g50570_2
Length=357
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 26/53 (49%), Gaps = 12/53 (22%)
Query 62 LSLPFELFGLGQTNNYVEEFFLGLPLAQLTSHNMWVSLIPNSQVIVMPYPLEK 114
SL E+F + + NY+E+ LG SL+P S+VIV LEK
Sbjct 72 CSLKCEIFMMSRVKNYLEDQLLGF------------SLVPLSEVIVRNGKLEK 112
> Hs9910346
Length=242
Score = 30.0 bits (66), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 16/57 (28%), Positives = 34/57 (59%), Gaps = 1/57 (1%)
Query 96 WVSLIPNSQVIVMPYPLEKPSSWQLELSV-HPSRSFANILITTVICLLLVALIIFIL 151
+V+ I S+V+ +PYPL+ SS + S + + L+ ++ ++++ L+IF+L
Sbjct 122 YVNYIKTSEVVRLPYPLQMKSSGPPSYFIKRESWGWTDFLMNPMVMMMVLPLLIFVL 178
> At3g23550
Length=469
Score = 29.3 bits (64), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 12/24 (50%), Positives = 15/24 (62%), Gaps = 0/24 (0%)
Query 83 LGLPLAQLTSHNMWVSLIPNSQVI 106
LG+ +A L H+ WV L NS VI
Sbjct 347 LGVVIAILVGHDAWVGLFSNSHVI 370
> Hs4501897
Length=513
Score = 27.7 bits (60), Expect = 8.6, Method: Composition-based stats.
Identities = 14/42 (33%), Positives = 21/42 (50%), Gaps = 2/42 (4%)
Query 95 MWVSLIPNSQVIVMPYPLEKPSSWQLELSVH--PSRSFANIL 134
+W + + N V V +P++ SWQ E V+ P NIL
Sbjct 206 VWKAQLLNEYVAVKIFPIQDKQSWQNEYEVYSLPGMKHENIL 247
Lambda K H
0.319 0.135 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1961355924
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40