bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_3406_orf1
Length=138
Score E
Sequences producing significant alignments: (Bits) Value
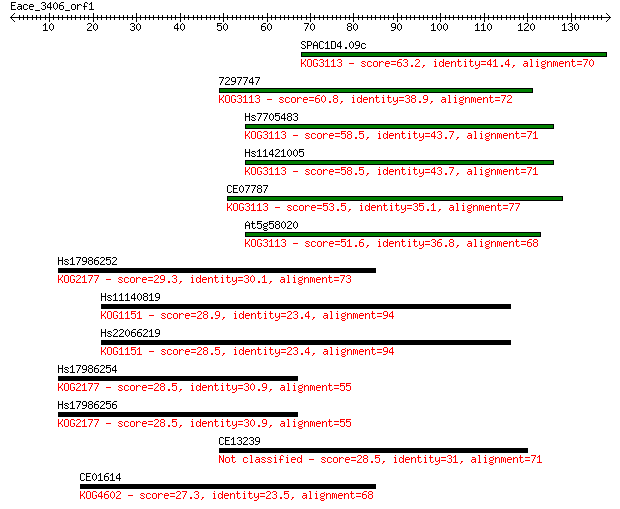
SPAC1D4.09c 63.2 1e-10
7297747 60.8 8e-10
Hs7705483 58.5 3e-09
Hs11421005 58.5 3e-09
CE07787 53.5 1e-07
At5g58020 51.6 4e-07
Hs17986252 29.3 2.1
Hs11140819 28.9 3.2
Hs22066219 28.5 3.8
Hs17986254 28.5 4.0
Hs17986256 28.5 4.1
CE13239 28.5 4.4
CE01614 27.3 8.3
> SPAC1D4.09c
Length=240
Score = 63.2 bits (152), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 29/73 (39%), Positives = 54/73 (73%), Gaps = 4/73 (5%)
Query 68 SACAFSQEKLQPPLLACRIGRLYNKEAVIKALLEK-ALPAHMKHVKSLKDMKEVQVDINA 126
S CA + E L PP+++C +G+LYNK ++++ LL++ ++P H+KSLKD+ ++QV+++
Sbjct 38 SQCAITDEPLYPPIVSCGLGKLYNKASILQMLLDRSSVPKSPSHIKSLKDVVQLQVELD- 96
Query 127 ETG--FPVCPITK 137
++G +CPIT+
Sbjct 97 DSGKVLWLCPITR 109
> 7297747
Length=299
Score = 60.8 bits (146), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 28/73 (38%), Positives = 48/73 (65%), Gaps = 1/73 (1%)
Query 49 QIRAGDERFGKNESRDLRTSACAFSQEKLQPPLLACRIGRLYNKEAVIKALLEKA-LPAH 107
+++ E+ K+ R+ R C +Q+ LQ P+ C +GRLY+K++VI+ LLEK +P
Sbjct 16 RVKQKPEQKDKDAEREFRWRHCTLTQQSLQEPIAMCGMGRLYSKQSVIERLLEKEPMPET 75
Query 108 MKHVKSLKDMKEV 120
HVKS+KD++++
Sbjct 76 AAHVKSMKDIRQL 88
> Hs7705483
Length=306
Score = 58.5 bits (140), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 31/75 (41%), Positives = 48/75 (64%), Gaps = 4/75 (5%)
Query 55 ERFGKNESRDLRTSACAFSQEKLQPPLLACRIGRLYNKEAVIKALL----EKALPAHMKH 110
E+ K+ + + C SQE L+ P++AC +GRLYNK+AVI+ LL EKAL H
Sbjct 22 EKVDKDAELVAQWNYCTLSQEILRRPIVACELGRLYNKDAVIEFLLDKSAEKALGKAASH 81
Query 111 VKSLKDMKEVQVDIN 125
+KS+K++ E+++ N
Sbjct 82 IKSIKNVTELKLSDN 96
> Hs11421005
Length=306
Score = 58.5 bits (140), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 31/75 (41%), Positives = 48/75 (64%), Gaps = 4/75 (5%)
Query 55 ERFGKNESRDLRTSACAFSQEKLQPPLLACRIGRLYNKEAVIKALL----EKALPAHMKH 110
E+ K+ + + C SQE L+ P++AC +GRLYNK+AVI+ LL EKAL H
Sbjct 22 EKVDKDAELVAQWNYCTLSQEILRRPIVACELGRLYNKDAVIEFLLDKSAEKALGKAASH 81
Query 111 VKSLKDMKEVQVDIN 125
+KS+K++ E+++ N
Sbjct 82 IKSIKNVTELKLSDN 96
> CE07787
Length=300
Score = 53.5 bits (127), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 27/79 (34%), Positives = 44/79 (55%), Gaps = 2/79 (2%)
Query 51 RAGDERFGKNESRDLRTSACAFSQEKLQPPLLACRIGRLYNKEAVIKALLEKAL--PAHM 108
R E+ K+ + C +Q L+ P++ACR G+LYNKE VI ++L K + A
Sbjct 18 RKKKEKLDKHVKNATKWRNCQLTQLPLKRPVIACRFGKLYNKEDVINSILSKTISKSASA 77
Query 109 KHVKSLKDMKEVQVDINAE 127
H+K KD E+++ +N +
Sbjct 78 SHIKGPKDFVELKLTLNKD 96
> At5g58020
Length=354
Score = 51.6 bits (122), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 25/68 (36%), Positives = 38/68 (55%), Gaps = 0/68 (0%)
Query 55 ERFGKNESRDLRTSACAFSQEKLQPPLLACRIGRLYNKEAVIKALLEKALPAHMKHVKSL 114
++ NE R + CA S E L P + +G L+NKE ++ ALL K LP ++K L
Sbjct 109 DKVDPNEQRLSKWLNCALSNEPLAEPCVIDLLGNLFNKEVLVHALLSKRLPKQFSYIKGL 168
Query 115 KDMKEVQV 122
KDM +++
Sbjct 169 KDMVNIKL 176
> Hs17986252
Length=549
Score = 29.3 bits (64), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 22/73 (30%), Positives = 29/73 (39%), Gaps = 1/73 (1%)
Query 12 PKTCSSLFSFSHRTEMVRTKGFKFLRNLGGMGYTPNTQIRAGDERFGKNESRDLRTSACA 71
P TC ++ +M K +RN G TPN G R KN R++R
Sbjct 465 PVTCGEAKGILYKKKMKHGSSVKCIRNEDGTWLTPNEFEVEGKGRNAKNWKRNIRCEGMT 524
Query 72 FSQEKLQPPLLAC 84
E L+ LL C
Sbjct 525 LG-ELLKSGLLLC 536
> Hs11140819
Length=749
Score = 28.9 bits (63), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 22/94 (23%), Positives = 39/94 (41%), Gaps = 3/94 (3%)
Query 22 SHRTEMVRTKGFKFLRNLGGMGYTPNTQIRAGDERFGKNESRDLRTSACAFSQEKLQPPL 81
S R E+ R + R MG P +++ K+++ ++ Q +
Sbjct 329 SQREEIERQRKMLAKRKPPAMGQAPPA---TNEQKQRKSKTNGAENETLTLAEYHEQEEI 385
Query 82 LACRIGRLYNKEAVIKALLEKALPAHMKHVKSLK 115
R+G L +EA I+A LE+ H++ LK
Sbjct 386 FKLRLGHLKKEEAEIQAELERLERVRNLHIRELK 419
> Hs22066219
Length=601
Score = 28.5 bits (62), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 22/94 (23%), Positives = 39/94 (41%), Gaps = 3/94 (3%)
Query 22 SHRTEMVRTKGFKFLRNLGGMGYTPNTQIRAGDERFGKNESRDLRTSACAFSQEKLQPPL 81
S R E+ R + R MG P +++ K+++ ++ Q +
Sbjct 181 SQREEIERQRKMLAKRKPPAMGQAPPA---TNEQKQRKSKTNGAENETLTLAEYHEQEEI 237
Query 82 LACRIGRLYNKEAVIKALLEKALPAHMKHVKSLK 115
R+G L +EA I+A LE+ H++ LK
Sbjct 238 FKLRLGHLKKEEAEIQAELERLERVRNLHIRELK 271
> Hs17986254
Length=713
Score = 28.5 bits (62), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 23/55 (41%), Gaps = 0/55 (0%)
Query 12 PKTCSSLFSFSHRTEMVRTKGFKFLRNLGGMGYTPNTQIRAGDERFGKNESRDLR 66
P TC ++ +M K +RN G TPN G R KN R++R
Sbjct 465 PVTCGEAKGILYKKKMKHGSSVKCIRNEDGTWLTPNEFEVEGKGRNAKNWKRNIR 519
> Hs17986256
Length=689
Score = 28.5 bits (62), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 23/55 (41%), Gaps = 0/55 (0%)
Query 12 PKTCSSLFSFSHRTEMVRTKGFKFLRNLGGMGYTPNTQIRAGDERFGKNESRDLR 66
P TC ++ +M K +RN G TPN G R KN R++R
Sbjct 465 PVTCGEAKGILYKKKMKHGSSVKCIRNEDGTWLTPNEFEVEGKGRNAKNWKRNIR 519
> CE13239
Length=381
Score = 28.5 bits (62), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 35/72 (48%), Gaps = 4/72 (5%)
Query 49 QIRAGDERFGKNESR-DLRTSACAFSQEKLQPPLLACRIGRLYNKEAVIKALLEKALPAH 107
Q++ DERF ++R +L S S K + + RLY +I L +K +
Sbjct 86 QMKLDDERFDHQKTRRELANSRLEISDLKGEAKCHDSELNRLY---TIINNLEKKVEDLN 142
Query 108 MKHVKSLKDMKE 119
+H KSL+ +KE
Sbjct 143 GEHQKSLEKLKE 154
> CE01614
Length=311
Score = 27.3 bits (59), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 16/68 (23%), Positives = 27/68 (39%), Gaps = 0/68 (0%)
Query 17 SLFSFSHRTEMVRTKGFKFLRNLGGMGYTPNTQIRAGDERFGKNESRDLRTSACAFSQEK 76
SL + + ++ F N G GY P + R G E+R+ R ++ + +
Sbjct 174 SLQEYLEQNNLLPPSTMPFGENSGFHGYNPQMNVHNNSTRNGNVENREQRNNSTSPPRGN 233
Query 77 LQPPLLAC 84
PL C
Sbjct 234 PPHPLCCC 241
Lambda K H
0.319 0.134 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1498437086
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40