bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0016_orf1
Length=158
Score E
Sequences producing significant alignments: (Bits) Value
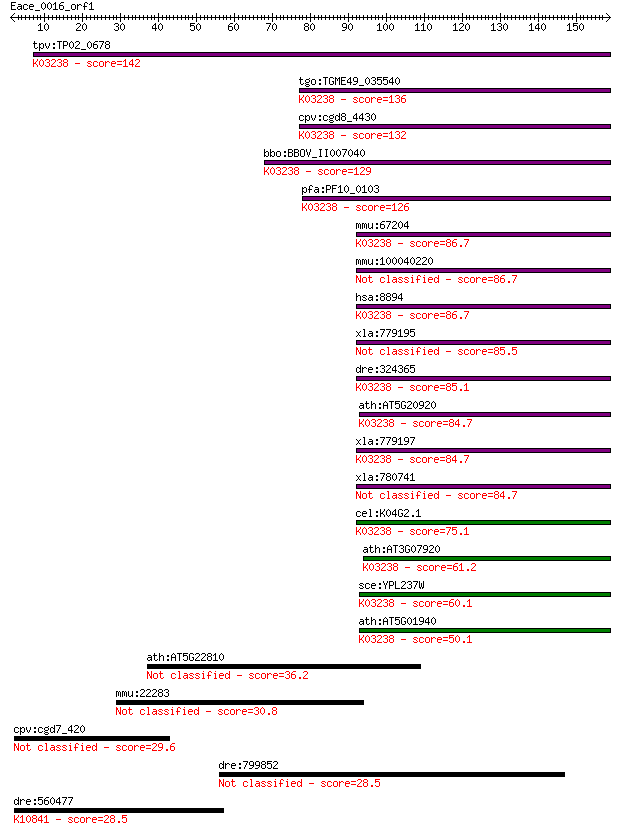
tpv:TP02_0678 eukaryotic translation initiation factor 2 subun... 142 6e-34
tgo:TGME49_035540 translation initiation factor 2 beta, putati... 136 3e-32
cpv:cgd8_4430 translation initiation factor if-2 betam beta su... 132 5e-31
bbo:BBOV_II007040 18.m06583; eukaryotic translation initiation... 129 4e-30
pfa:PF10_0103 eukaryotic translation initiation factor 2 beta,... 126 3e-29
mmu:67204 Eif2s2, 2810026E11Rik, 38kDa, AA408636, AA571381, AA... 86.7 3e-17
mmu:100040220 Gm9892; predicted gene 9892 86.7
hsa:8894 EIF2S2, DKFZp686L18198, EIF2, EIF2B, EIF2beta, MGC850... 86.7 3e-17
xla:779195 hypothetical protein MGC130708 85.5 7e-17
dre:324365 eif2s2, wu:fc26g03, wu:fu10f07, zgc:77084; eukaryot... 85.1 8e-17
ath:AT5G20920 EIF2 BETA; translation initiation factor; K03238... 84.7 1e-16
xla:779197 eif2s2, MGC130959; eukaryotic translation initiatio... 84.7 1e-16
xla:780741 eukaryote initiation factor 2 beta 84.7 1e-16
cel:K04G2.1 iftb-1; eIFTwoBeta (eIF2beta translation initiatio... 75.1 1e-13
ath:AT3G07920 translation initiation factor; K03238 translatio... 61.2 1e-09
sce:YPL237W SUI3; Beta subunit of the translation initiation f... 60.1 3e-09
ath:AT5G01940 eukaryotic translation initiation factor 2B fami... 50.1 3e-06
ath:AT5G22810 GDSL-motif lipase, putative 36.2 0.042
mmu:22283 Ush2a, A930011D15Rik, A930037M10Rik, Gm676, Gm983, M... 30.8 1.9
cpv:cgd7_420 protein with DEXDc plus ring plus HELICc; possibl... 29.6 4.0
dre:799852 si:ch211-219a4.6 28.5 9.2
dre:560477 ercc6; excision repair cross-complementing rodent r... 28.5 9.5
> tpv:TP02_0678 eukaryotic translation initiation factor 2 subunit
beta; K03238 translation initiation factor 2 subunit 2
Length=241
Score = 142 bits (357), Expect = 6e-34, Method: Compositional matrix adjust.
Identities = 71/152 (46%), Positives = 98/152 (64%), Gaps = 5/152 (3%)
Query 7 SSSSSRRGSRNSSDSNTITIISNSAGRITTATSAASSNISSKQQHQQQQQQQHRGLCVSL 66
S +S + ++S + ++N + T + S + ++++ R ++
Sbjct 5 SDVTSEKSPKDSPEEEVTDKLNNMGLKPETQDPKPADAKDSPPKFNFGEKKKKR----TV 60
Query 67 FAADAAAAEGFIDGSGQLFIRGHVCSYEEMLSRIQDLIVKNNPDLAGSKRYTIKPPQVVR 126
A D+ E IDG+GQ+F++GHV YEE+L+RIQ +I NNPDL+GSKRYTIKPPQVVR
Sbjct 61 DAGDSPKTE-IIDGTGQVFVKGHVYPYEELLARIQSMINANNPDLSGSKRYTIKPPQVVR 119
Query 127 VGSKKVAWINFKDICGIMHRPSEHVLQFVLAE 158
VGSKKVAWINF+DIC IM R +HV QFVLAE
Sbjct 120 VGSKKVAWINFRDICSIMGRSMDHVHQFVLAE 151
> tgo:TGME49_035540 translation initiation factor 2 beta, putative
; K03238 translation initiation factor 2 subunit 2
Length=231
Score = 136 bits (343), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 62/82 (75%), Positives = 70/82 (85%), Gaps = 0/82 (0%)
Query 77 FIDGSGQLFIRGHVCSYEEMLSRIQDLIVKNNPDLAGSKRYTIKPPQVVRVGSKKVAWIN 136
IDG+G+LF RG SY+EML RIQ LI K+NPDL+G+KRYTIKPPQVVRVGSKKVAWIN
Sbjct 59 IIDGTGELFKRGKEYSYQEMLQRIQTLIEKHNPDLSGAKRYTIKPPQVVRVGSKKVAWIN 118
Query 137 FKDICGIMHRPSEHVLQFVLAE 158
FKDIC IM+R +HV QFVLAE
Sbjct 119 FKDICNIMNRQPDHVHQFVLAE 140
> cpv:cgd8_4430 translation initiation factor if-2 betam beta
subunit ZnR ; K03238 translation initiation factor 2 subunit
2
Length=214
Score = 132 bits (332), Expect = 5e-31, Method: Compositional matrix adjust.
Identities = 54/82 (65%), Positives = 72/82 (87%), Gaps = 0/82 (0%)
Query 77 FIDGSGQLFIRGHVCSYEEMLSRIQDLIVKNNPDLAGSKRYTIKPPQVVRVGSKKVAWIN 136
++DGSGQLF++G + YEE+L R++ LI+++NPDL G+KRYT+KPPQVVRVGSKKVAWIN
Sbjct 42 YVDGSGQLFVKGAIYPYEELLERVRRLILEHNPDLWGAKRYTLKPPQVVRVGSKKVAWIN 101
Query 137 FKDICGIMHRPSEHVLQFVLAE 158
F++IC IM R ++HV QFVL+E
Sbjct 102 FQEICNIMQRNADHVFQFVLSE 123
> bbo:BBOV_II007040 18.m06583; eukaryotic translation initiation
factor 2, beta; K03238 translation initiation factor 2 subunit
2
Length=238
Score = 129 bits (324), Expect = 4e-30, Method: Compositional matrix adjust.
Identities = 60/91 (65%), Positives = 76/91 (83%), Gaps = 1/91 (1%)
Query 68 AADAAAAEGFIDGSGQLFIRGHVCSYEEMLSRIQDLIVKNNPDLAGSKRYTIKPPQVVRV 127
A+D+ +E IDGSGQ+F+RGHV YEE+L+RIQ LI ++ DL+G+K+YTI+PPQVVRV
Sbjct 59 ASDSVKSE-VIDGSGQVFVRGHVYQYEELLNRIQTLINAHSHDLSGNKKYTIRPPQVVRV 117
Query 128 GSKKVAWINFKDICGIMHRPSEHVLQFVLAE 158
GSKKVAWINFK++C IM R +HV QFVLAE
Sbjct 118 GSKKVAWINFKELCNIMGRTMDHVHQFVLAE 148
> pfa:PF10_0103 eukaryotic translation initiation factor 2 beta,
putative; K03238 translation initiation factor 2 subunit
2
Length=222
Score = 126 bits (317), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 59/81 (72%), Positives = 67/81 (82%), Gaps = 0/81 (0%)
Query 78 IDGSGQLFIRGHVCSYEEMLSRIQDLIVKNNPDLAGSKRYTIKPPQVVRVGSKKVAWINF 137
IDG+G++F RG V Y+E+L RIQDLI K+N DL SK+YTIKPPQVVRVGSKKVAWINF
Sbjct 50 IDGTGKVFERGAVYPYDELLHRIQDLINKHNIDLCISKKYTIKPPQVVRVGSKKVAWINF 109
Query 138 KDICGIMHRPSEHVLQFVLAE 158
KDIC IM+R EHV FVLAE
Sbjct 110 KDICTIMNRNEEHVFHFVLAE 130
> mmu:67204 Eif2s2, 2810026E11Rik, 38kDa, AA408636, AA571381,
AA986487, AW822225, D2Ertd303e, EIF2, EIF2B, MGC130606; eukaryotic
translation initiation factor 2, subunit 2 (beta); K03238
translation initiation factor 2 subunit 2
Length=331
Score = 86.7 bits (213), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 36/69 (52%), Positives = 56/69 (81%), Gaps = 2/69 (2%)
Query 92 SYEEMLSRIQDLIVKNNPDL-AGSKR-YTIKPPQVVRVGSKKVAWINFKDICGIMHRPSE 149
+YEE+L+R+ +++ + NPD+ AG KR + +KPPQVVRVG+KK +++NF DIC ++HR +
Sbjct 173 TYEELLNRVFNIMREKNPDMVAGEKRKFVMKPPQVVRVGTKKTSFVNFTDICKLLHRQPK 232
Query 150 HVLQFVLAE 158
H+L F+LAE
Sbjct 233 HLLAFLLAE 241
> mmu:100040220 Gm9892; predicted gene 9892
Length=331
Score = 86.7 bits (213), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 35/69 (50%), Positives = 56/69 (81%), Gaps = 2/69 (2%)
Query 92 SYEEMLSRIQDLIVKNNPDL-AGSKR-YTIKPPQVVRVGSKKVAWINFKDICGIMHRPSE 149
+YEE+L+R+ +++ + NPD+ AG KR + +KPPQV+RVG+KK +++NF DIC ++HR +
Sbjct 173 TYEELLNRVLNIMREKNPDMVAGEKRKFVMKPPQVIRVGTKKTSFVNFTDICKLLHRQPK 232
Query 150 HVLQFVLAE 158
H+L F+LAE
Sbjct 233 HLLAFLLAE 241
> hsa:8894 EIF2S2, DKFZp686L18198, EIF2, EIF2B, EIF2beta, MGC8508;
eukaryotic translation initiation factor 2, subunit 2 beta,
38kDa; K03238 translation initiation factor 2 subunit 2
Length=333
Score = 86.7 bits (213), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 36/69 (52%), Positives = 56/69 (81%), Gaps = 2/69 (2%)
Query 92 SYEEMLSRIQDLIVKNNPDL-AGSKR-YTIKPPQVVRVGSKKVAWINFKDICGIMHRPSE 149
+YEE+L+R+ +++ + NPD+ AG KR + +KPPQVVRVG+KK +++NF DIC ++HR +
Sbjct 175 TYEELLNRVFNIMREKNPDMVAGEKRKFVMKPPQVVRVGTKKTSFVNFTDICKLLHRQPK 234
Query 150 HVLQFVLAE 158
H+L F+LAE
Sbjct 235 HLLAFLLAE 243
> xla:779195 hypothetical protein MGC130708
Length=332
Score = 85.5 bits (210), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 35/69 (50%), Positives = 56/69 (81%), Gaps = 2/69 (2%)
Query 92 SYEEMLSRIQDLIVKNNPDL-AGSKR-YTIKPPQVVRVGSKKVAWINFKDICGIMHRPSE 149
+Y+E+L+R+ +++ + NPD+ AG KR + +KPPQVVRVG+KK +++NF DIC ++HR +
Sbjct 174 TYDELLNRVFNIMREKNPDMVAGEKRKFVMKPPQVVRVGTKKTSFVNFTDICKLLHRQPK 233
Query 150 HVLQFVLAE 158
H+L F+LAE
Sbjct 234 HLLAFLLAE 242
> dre:324365 eif2s2, wu:fc26g03, wu:fu10f07, zgc:77084; eukaryotic
translation initiation factor 2, subunit 2 beta; K03238
translation initiation factor 2 subunit 2
Length=327
Score = 85.1 bits (209), Expect = 8e-17, Method: Compositional matrix adjust.
Identities = 35/69 (50%), Positives = 56/69 (81%), Gaps = 2/69 (2%)
Query 92 SYEEMLSRIQDLIVKNNPDL-AGSKR-YTIKPPQVVRVGSKKVAWINFKDICGIMHRPSE 149
+Y+E+L+R+ +++ + NPD+ AG KR + +KPPQVVRVG+KK +++NF DIC ++HR +
Sbjct 169 TYDELLNRVFNIMREKNPDMVAGEKRKFVMKPPQVVRVGTKKTSFVNFTDICKLLHRQPK 228
Query 150 HVLQFVLAE 158
H+L F+LAE
Sbjct 229 HLLVFLLAE 237
> ath:AT5G20920 EIF2 BETA; translation initiation factor; K03238
translation initiation factor 2 subunit 2
Length=268
Score = 84.7 bits (208), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 33/67 (49%), Positives = 53/67 (79%), Gaps = 1/67 (1%)
Query 93 YEEMLSRIQDLIVKNNPDLAGSKRYTI-KPPQVVRVGSKKVAWINFKDICGIMHRPSEHV 151
Y+E+L R+ +++ +NNP+LAG +R T+ +PPQV+R G+KK ++NF D+C MHR +HV
Sbjct 118 YDELLGRVFNILRENNPELAGDRRRTVMRPPQVLREGTKKTVFVNFMDLCKTMHRQPDHV 177
Query 152 LQFVLAE 158
+Q++LAE
Sbjct 178 MQYLLAE 184
> xla:779197 eif2s2, MGC130959; eukaryotic translation initiation
factor 2, subunit 2 beta, 38kDa; K03238 translation initiation
factor 2 subunit 2
Length=335
Score = 84.7 bits (208), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 35/69 (50%), Positives = 56/69 (81%), Gaps = 2/69 (2%)
Query 92 SYEEMLSRIQDLIVKNNPDL-AGSKR-YTIKPPQVVRVGSKKVAWINFKDICGIMHRPSE 149
+Y+E+L+R+ +++ + NPD+ AG KR + +KPPQVVRVG+KK +++NF DIC ++HR +
Sbjct 177 TYDELLNRVFNIMREKNPDMVAGEKRKFVMKPPQVVRVGTKKTSFVNFTDICKLLHRQPK 236
Query 150 HVLQFVLAE 158
H+L F+LAE
Sbjct 237 HLLAFLLAE 245
> xla:780741 eukaryote initiation factor 2 beta
Length=335
Score = 84.7 bits (208), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 35/69 (50%), Positives = 56/69 (81%), Gaps = 2/69 (2%)
Query 92 SYEEMLSRIQDLIVKNNPDL-AGSKR-YTIKPPQVVRVGSKKVAWINFKDICGIMHRPSE 149
+Y+E+L+R+ +++ + NPD+ AG KR + +KPPQVVRVG+KK +++NF DIC ++HR +
Sbjct 177 TYDELLNRVFNIMREKNPDMVAGEKRKFVMKPPQVVRVGTKKTSFVNFTDICKLLHRQPK 236
Query 150 HVLQFVLAE 158
H+L F+LAE
Sbjct 237 HLLAFLLAE 245
> cel:K04G2.1 iftb-1; eIFTwoBeta (eIF2beta translation initiation
factor) family member (iftb-1); K03238 translation initiation
factor 2 subunit 2
Length=250
Score = 75.1 bits (183), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 34/68 (50%), Positives = 47/68 (69%), Gaps = 1/68 (1%)
Query 92 SYEEMLSRIQDLIVKNNPDLAGSKR-YTIKPPQVVRVGSKKVAWINFKDICGIMHRPSEH 150
+YEE L+ + ++ NPD AG K+ + IK P+V R GSKK A+ NF +IC +M R +H
Sbjct 88 TYEEALTLVYQVMKDKNPDFAGDKKKFAIKLPEVARAGSKKTAFSNFLEICRLMKRQDKH 147
Query 151 VLQFVLAE 158
VLQF+LAE
Sbjct 148 VLQFLLAE 155
> ath:AT3G07920 translation initiation factor; K03238 translation
initiation factor 2 subunit 2
Length=164
Score = 61.2 bits (147), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 30/70 (42%), Positives = 46/70 (65%), Gaps = 5/70 (7%)
Query 94 EEMLSRIQDLIVKNNPDLAGSKRYTI-KPPQVVR----VGSKKVAWINFKDICGIMHRPS 148
+E+L R+ +++ +N+P+L G TI PPQV+R G+KK ++NF D C MHR
Sbjct 17 QEILRRVFNILRENSPELVGIWLLTIIWPPQVLREETAKGTKKTVFVNFMDYCKTMHRNP 76
Query 149 EHVLQFVLAE 158
+HV+ F+LAE
Sbjct 77 DHVMAFLLAE 86
> sce:YPL237W SUI3; Beta subunit of the translation initiation
factor eIF2, involved in the identification of the start codon;
proposed to be involved in mRNA binding; K03238 translation
initiation factor 2 subunit 2
Length=285
Score = 60.1 bits (144), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 28/69 (40%), Positives = 44/69 (63%), Gaps = 4/69 (5%)
Query 93 YEEMLSRIQDLIVKNNPDLAGSK---RYTIKPPQVVRVGSKKVAWINFKDICGIMHRPSE 149
Y E+LSR +++ NNP+LAG + ++ I PP +R G KK + N +DI +HR E
Sbjct 131 YSELLSRFFNILRTNNPELAGDRSGPKFRIPPPVCLRDG-KKTIFSNIQDIAEKLHRSPE 189
Query 150 HVLQFVLAE 158
H++Q++ AE
Sbjct 190 HLIQYLFAE 198
> ath:AT5G01940 eukaryotic translation initiation factor 2B family
protein / eIF-2B family protein; K03238 translation initiation
factor 2 subunit 2
Length=231
Score = 50.1 bits (118), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 44/67 (65%), Gaps = 2/67 (2%)
Query 93 YEEMLSRIQDLIVKNNPDLAGSK-RYTIKPPQVVRVGSKKVAWINFKDICGIMHRPSEHV 151
Y+E+LS + D + + + +++ + R + PPQ++ G+ V +NF D+C MHR +HV
Sbjct 75 YKELLSMVFDRLREEDVEVSTERPRTVMMPPQLLAEGTITVC-LNFADLCRTMHRKPDHV 133
Query 152 LQFVLAE 158
++F+LA+
Sbjct 134 MKFLLAQ 140
> ath:AT5G22810 GDSL-motif lipase, putative
Length=337
Score = 36.2 bits (82), Expect = 0.042, Method: Compositional matrix adjust.
Identities = 21/76 (27%), Positives = 40/76 (52%), Gaps = 4/76 (5%)
Query 37 ATSAASSNISSKQQHQQQQQQQHRGLCVSLFAADAAAAEGFIDGSGQLF----IRGHVCS 92
AT + N+ K Q ++ +G + + A A+AA G+ DG+ +L+ + +
Sbjct 59 ATDFTAENLGFKSYPQAYLSKKAKGKNLLIGANFASAASGYYDGTAKLYSAISLPQQLEH 118
Query 93 YEEMLSRIQDLIVKNN 108
Y++ +SRIQ++ NN
Sbjct 119 YKDYISRIQEIATSNN 134
> mmu:22283 Ush2a, A930011D15Rik, A930037M10Rik, Gm676, Gm983,
Mush2a, Usherin; Usher syndrome 2A (autosomal recessive, mild)
homolog (human)
Length=5193
Score = 30.8 bits (68), Expect = 1.9, Method: Composition-based stats.
Identities = 19/65 (29%), Positives = 34/65 (52%), Gaps = 13/65 (20%)
Query 29 NSAGRITTATSAASSNISSKQQHQQQQQQQHRGLCVSLFAADAAAAEGFIDGSGQLFIRG 88
N AG +T++ +N S+K++ Q Q +LF+ D+ A+ ++D SG + G
Sbjct 4904 NEAG----STASGWTNFSTKKEMPQYQ---------ALFSVDSNASMVWVDWSGTFLLNG 4950
Query 89 HVCSY 93
H+ Y
Sbjct 4951 HLKEY 4955
> cpv:cgd7_420 protein with DEXDc plus ring plus HELICc; possible
SNF2 domain
Length=2042
Score = 29.6 bits (65), Expect = 4.0, Method: Composition-based stats.
Identities = 16/41 (39%), Positives = 26/41 (63%), Gaps = 0/41 (0%)
Query 2 SSNENSSSSSRRGSRNSSDSNTITIISNSAGRITTATSAAS 42
+SN +S++S + NS+ +N+ T SN+A ITT +S S
Sbjct 323 NSNTANSNTSNSNTANSNTANSNTSNSNTANSITTTSSNCS 363
> dre:799852 si:ch211-219a4.6
Length=840
Score = 28.5 bits (62), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 23/91 (25%), Positives = 37/91 (40%), Gaps = 1/91 (1%)
Query 56 QQQHRGLCVSLFAADAAAAEGFIDGSGQLFIRGHVCSYEEMLSRIQDLIVKNNPDLAGSK 115
Q++H L D E F G+G + I G ++ ++ K D A ++
Sbjct 526 QKEHEFLLTLQAVFDVRIEEFFRSGNGGITITGSPSGVSCAAIEVESMLCKAQNDFARAE 585
Query 116 RYTIKPPQVVRVGSKKVAWINFKDICGIMHR 146
I VVR K+ WI +I GI+ +
Sbjct 586 ERDIL-YSVVRWSCKEEPWIQTPEISGILEK 615
> dre:560477 ercc6; excision repair cross-complementing rodent
repair deficiency, complementation group 6; K10841 DNA excision
repair protein ERCC-6
Length=1390
Score = 28.5 bits (62), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 39/57 (68%), Gaps = 3/57 (5%)
Query 2 SSNENSSSSSRRGSRNSSDS--NTITIISNSAGRITTATSAASSNISSKQQHQQQQQ 56
S N N++S + + N+S+S N + SN+ G +T++TS A S+++S Q+H+++++
Sbjct 1033 SGNRNTTSVNGLPNNNTSESPNNKLESRSNANG-LTSSTSKALSSMNSPQKHREKKK 1088
Lambda K H
0.314 0.126 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3582056500
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40