bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0031_orf1
Length=192
Score E
Sequences producing significant alignments: (Bits) Value
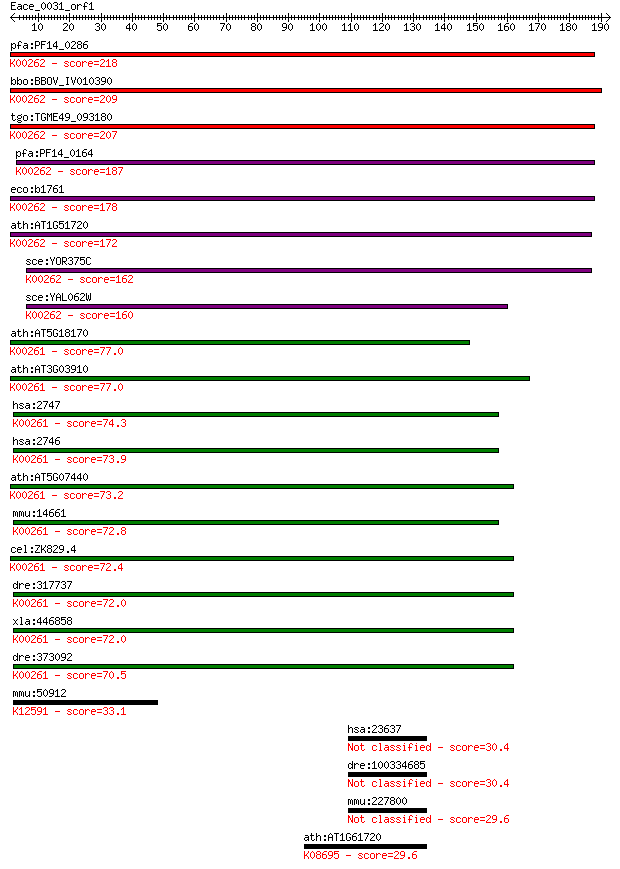
pfa:PF14_0286 glutamate dehydrogenase, putative (EC:1.4.1.3); ... 218 1e-56
bbo:BBOV_IV010390 23.m06170; glutamate dehydrogenase (EC:1.4.1... 209 5e-54
tgo:TGME49_093180 NADP-specific glutamate dehydrogenase, putat... 207 1e-53
pfa:PF14_0164 NADP-specific glutamate dehydrogenase; K00262 gl... 187 2e-47
eco:b1761 gdhA, ECK1759, JW1750; glutamate dehydrogenase, NADP... 178 1e-44
ath:AT1G51720 glutamate dehydrogenase, putative; K00262 glutam... 172 7e-43
sce:YOR375C GDH1, DHE4, URE1; Gdh1p (EC:1.4.1.4); K00262 gluta... 162 8e-40
sce:YAL062W GDH3, FUN51; Gdh3p (EC:1.4.1.4); K00262 glutamate ... 160 3e-39
ath:AT5G18170 GDH1; GDH1 (GLUTAMATE DEHYDROGENASE 1); ATP bind... 77.0 4e-14
ath:AT3G03910 GDH3; GDH3 (GLUTAMATE DEHYDROGENASE 3); binding ... 77.0 4e-14
hsa:2747 GLUD2, GDH2, GLUDP1; glutamate dehydrogenase 2 (EC:1.... 74.3 2e-13
hsa:2746 GLUD1, GDH, GDH1, GLUD, MGC132003; glutamate dehydrog... 73.9 3e-13
ath:AT5G07440 GDH2; GDH2 (GLUTAMATE DEHYDROGENASE 2); ATP bind... 73.2 5e-13
mmu:14661 Glud1, AI118167, Gdh-X, Glud, Gludl; glutamate dehyd... 72.8 7e-13
cel:ZK829.4 hypothetical protein; K00261 glutamate dehydrogena... 72.4 8e-13
dre:317737 glud1a, cb622, glud1, wu:fc33g09, wu:fc66a10, zgc:7... 72.0 1e-12
xla:446858 glud1, MGC80801; glutamate dehydrogenase 1 (EC:1.4.... 72.0 1e-12
dre:373092 glud1b, MGC192851, cb719, glud1, wu:fb16e02, wu:fb5... 70.5 3e-12
mmu:50912 Exosc10, PM-Scl, PM/Scl-100, Pmscl2, RRP6, p2, p3, p... 33.1 0.66
hsa:23637 RABGAP1, DKFZp586D2123, GAPCENA, RP11-123N4.2, TBC1D... 30.4 3.8
dre:100334685 hypothetical protein LOC100334685 30.4 4.1
mmu:227800 Rabgap1, Gapcena, KIAA4104, MGC30493, mKIAA4104; RA... 29.6 5.9
ath:AT1G61720 BAN; BAN (BANYULS); oxidoreductase (EC:1.3.1.77)... 29.6 6.9
> pfa:PF14_0286 glutamate dehydrogenase, putative (EC:1.4.1.3);
K00262 glutamate dehydrogenase (NADP+) [EC:1.4.1.4]
Length=510
Score = 218 bits (554), Expect = 1e-56, Method: Compositional matrix adjust.
Identities = 108/187 (57%), Positives = 132/187 (70%), Gaps = 1/187 (0%)
Query 1 GGSDFDPKGKSVAEIRRFCQSSMTELAKHIGPSRDVPAGDIGVGTREIGFLFGQYKRLCS 60
GGSDFDPKGKS EI +FCQS MT L ++IGP+ DVPAGDIGVG REIG+LFGQYK+L +
Sbjct 190 GGSDFDPKGKSENEILKFCQSFMTNLFRYIGPNTDVPAGDIGVGGREIGYLFGQYKKLKN 249
Query 61 NFEGALTGKDIGWGGSEIRPEATGYGCAYFAENVLDQLLYDTLKGKKCILSGCGNVAVYC 120
+FEG LTGK+I WGGS IR EATGYG YFAENVL L D L+ KKC++SG GNVA Y
Sbjct 250 SFEGVLTGKNIKWGGSNIRAEATGYGVVYFAENVLKD-LNDNLENKKCLVSGSGNVAQYL 308
Query 121 AEKLLEMGAEVLTLSDSSGTIYCSAGFSKEEIRTIKEAKATDRAVRVEAFANGESVVFER 180
EKL+E GA VLT+SDS+G I GF+KE++ I + K R E ++ +
Sbjct 309 VEKLIEKGAIVLTMSDSNGYILEPNGFTKEQLNYIMDIKNNQRLRLKEYLKYSKTAKYFE 368
Query 181 RGTPWSV 187
PW++
Sbjct 369 NQKPWNI 375
> bbo:BBOV_IV010390 23.m06170; glutamate dehydrogenase (EC:1.4.1.4);
K00262 glutamate dehydrogenase (NADP+) [EC:1.4.1.4]
Length=455
Score = 209 bits (532), Expect = 5e-54, Method: Compositional matrix adjust.
Identities = 106/190 (55%), Positives = 130/190 (68%), Gaps = 2/190 (1%)
Query 1 GGSDFDPKGKSVAEIRRFCQSSMTELAKHIGPSRDVPAGDIGVGTREIGFLFGQYKRLCS 60
GGSDFDP GKS EIR FCQS MTEL +HIGP D+PAGDIGVG REIG+LFGQY+RL +
Sbjct 134 GGSDFDPSGKSDNEIRNFCQSFMTELHRHIGPDTDIPAGDIGVGAREIGYLFGQYRRLHN 193
Query 61 NFEGALTGKDIGWGGSEIRPEATGYGCAYFAENVLDQLLYDTLKGKKCILSGCGNVAVYC 120
F+GALTGK + WGGS IRPEATGYG YF VL+ + DT+ GK+CI+SG GNVA Y
Sbjct 194 GFDGALTGKGLQWGGSNIRPEATGYGAVYFGCAVLEDIFKDTIAGKRCIVSGSGNVAQYT 253
Query 121 AEKLLEMGAEVLTLSDSSGTIYCSAGFSKEEIRTIKEAKATDRAVRVEAFAN-GESVVFE 179
EKL+E+GA +T+SDSSG I G + E +R I K + R+ + N ++ F
Sbjct 254 VEKLIELGAVPITMSDSSGYIIEPEGITLEGLRYIMAFK-NPHSRRISEYLNYSKTATFH 312
Query 180 RRGTPWSVAA 189
PW +A
Sbjct 313 PGDKPWGESA 322
> tgo:TGME49_093180 NADP-specific glutamate dehydrogenase, putative
(EC:1.4.1.4); K00262 glutamate dehydrogenase (NADP+) [EC:1.4.1.4]
Length=489
Score = 207 bits (528), Expect = 1e-53, Method: Compositional matrix adjust.
Identities = 108/189 (57%), Positives = 130/189 (68%), Gaps = 2/189 (1%)
Query 1 GGSDFDPKGKSVAEIRRFCQSSMTELAKHIGPSRDVPAGDIGVGTREIGFLFGQYKRL-C 59
GGSDFDPKG+S E+ RFCQ+ MTEL+++IGP+RDVPAGDIGVG REIG+LFGQYK L
Sbjct 165 GGSDFDPKGRSDNEVMRFCQAFMTELSRYIGPNRDVPAGDIGVGAREIGYLFGQYKLLKA 224
Query 60 SNFEGALTGKDIGWGGSEIRPEATGYGCAYFAENVLDQLLYDTLKGKKCILSGCGNVAVY 119
FEGALTGKD WGGS +RPEATGYGC YF +L + +KG +C +SG GNVA+Y
Sbjct 225 GQFEGALTGKDKNWGGSAMRPEATGYGCVYFVLELLKAQNKEGIKGMRCAISGSGNVALY 284
Query 120 CAEKLLEMGAEVLTLSDSSGTIYCSAGFSKEEIRTIKEAKATDRAVRVEAFANGESVV-F 178
EKL +GA+V+T SDSSG I GF +I+ +KE K T + RV FA S V F
Sbjct 285 AGEKLATLGAKVMTFSDSSGYIVNEKGFPLGQIQRLKEMKETRSSTRVSEFAAKYSTVKF 344
Query 179 ERRGTPWSV 187
W V
Sbjct 345 VPDKRAWEV 353
> pfa:PF14_0164 NADP-specific glutamate dehydrogenase; K00262
glutamate dehydrogenase (NADP+) [EC:1.4.1.4]
Length=470
Score = 187 bits (475), Expect = 2e-47, Method: Compositional matrix adjust.
Identities = 93/186 (50%), Positives = 123/186 (66%), Gaps = 3/186 (1%)
Query 3 SDFDPKGKSVAEIRRFCQSSMTELAKHIGPSRDVPAGDIGVGTREIGFLFGQYKRLCSNF 62
SDFDPKGKS EI +FCQ+ M EL +HIGP DVPAGDIGVG REIG+L+GQYK++ ++F
Sbjct 151 SDFDPKGKSDNEILKFCQAFMNELYRHIGPCTDVPAGDIGVGGREIGYLYGQYKKIVNSF 210
Query 63 EGALTGKDIGWGGSEIRPEATGYGCAYFAENVLDQLLYDTLKGKKCILSGCGNVAVYCAE 122
G LTGK++ WGGS +R EATGYG YF VL L K + ++SG GNVA+YC +
Sbjct 211 NGTLTGKNVKWGGSNLRVEATGYGLVYFVLEVLKSLNIPVEK-QTAVVSGSGNVALYCVQ 269
Query 123 KLLEMGAEVLTLSDSSGTIYCSAGFSKEEIRTIKEAKATDRAVRVEAFANGESVV-FERR 181
KLL + +VLTLSDS+G +Y GF+ E + + + K + R++ + N S +
Sbjct 270 KLLHLNVKVLTLSDSNGYVYEPNGFTHENLEFLIDLKEEKKG-RIKEYLNHSSTAKYFPN 328
Query 182 GTPWSV 187
PW V
Sbjct 329 EKPWGV 334
> eco:b1761 gdhA, ECK1759, JW1750; glutamate dehydrogenase, NADP-specific
(EC:1.4.1.4); K00262 glutamate dehydrogenase (NADP+)
[EC:1.4.1.4]
Length=447
Score = 178 bits (451), Expect = 1e-44, Method: Compositional matrix adjust.
Identities = 95/187 (50%), Positives = 123/187 (65%), Gaps = 2/187 (1%)
Query 1 GGSDFDPKGKSVAEIRRFCQSSMTELAKHIGPSRDVPAGDIGVGTREIGFLFGQYKRLCS 60
GGSDFDPKGKS E+ RFCQ+ MTEL +H+G DVPAGDIGVG RE+GF+ G K+L +
Sbjct 129 GGSDFDPKGKSEGEVMRFCQALMTELYRHLGADTDVPAGDIGVGGREVGFMAGMMKKLSN 188
Query 61 NFEGALTGKDIGWGGSEIRPEATGYGCAYFAENVLDQLLYDTLKGKKCILSGCGNVAVYC 120
N TGK + +GGS IRPEATGYG YF E +L + +G + +SG GNVA Y
Sbjct 189 NTACVFTGKGLSFGGSLIRPEATGYGLVYFTEAMLKRHGMG-FEGMRVSVSGSGNVAQYA 247
Query 121 AEKLLEMGAEVLTLSDSSGTIYCSAGFSKEEIRTIKEAKATDRAVRVEAFANGESVVFER 180
EK +E GA V+T SDSSGT+ +GF+KE++ + E KA+ R RV +A +V+
Sbjct 248 IEKAMEFGARVITASDSSGTVVDESGFTKEKLARLIEIKAS-RDGRVADYAKEFGLVYLE 306
Query 181 RGTPWSV 187
PWS+
Sbjct 307 GQQPWSL 313
> ath:AT1G51720 glutamate dehydrogenase, putative; K00262 glutamate
dehydrogenase (NADP+) [EC:1.4.1.4]
Length=637
Score = 172 bits (436), Expect = 7e-43, Method: Compositional matrix adjust.
Identities = 81/186 (43%), Positives = 122/186 (65%), Gaps = 2/186 (1%)
Query 1 GGSDFDPKGKSVAEIRRFCQSSMTELAKHIGPSRDVPAGDIGVGTREIGFLFGQYKRLCS 60
GGSDFDPKGKS EI RFCQS M E+ +++GP +D+P+ ++GVGTRE+G+LFGQY+RL
Sbjct 314 GGSDFDPKGKSDNEIMRFCQSFMNEMYRYMGPDKDLPSEEVGVGTREMGYLFGQYRRLAG 373
Query 61 NFEGALTGKDIGWGGSEIRPEATGYGCAYFAENVLDQLLYDTLKGKKCILSGCGNVAVYC 120
F+G+ TG I W S +R EA+GYG YFA +L + +KG +C++SGCG +A++
Sbjct 374 QFQGSFTGPRIYWAASSLRTEASGYGVVYFARLILAD-MNKEIKGLRCVVSGCGKIAMHV 432
Query 121 AEKLLEMGAEVLTLSDSSGTIYCSAGFSKEEIRTIKEAKATDRAVRVEAFANGESVVFER 180
EKL+ GA +T+SDS G + GF ++ +++ K+ R++R + + F+
Sbjct 433 VEKLIACGAHPVTVSDSKGYLVDDDGFDYMKLAFLRDIKSQQRSLRDYSKTYARAKYFDE 492
Query 181 RGTPWS 186
PW+
Sbjct 493 L-KPWN 497
> sce:YOR375C GDH1, DHE4, URE1; Gdh1p (EC:1.4.1.4); K00262 glutamate
dehydrogenase (NADP+) [EC:1.4.1.4]
Length=454
Score = 162 bits (409), Expect = 8e-40, Method: Compositional matrix adjust.
Identities = 85/189 (44%), Positives = 117/189 (61%), Gaps = 10/189 (5%)
Query 6 DPKGKSVAEIRRFCQSSMTELAKHIGPSRDVPAGDIGVGTREIGFLFGQYKRLCSNFEGA 65
D KG+S EIRR C + M EL++HIG DVPAGDIGVG REIG+LFG Y+ +++EG
Sbjct 116 DLKGRSNNEIRRICYAFMRELSRHIGQDTDVPAGDIGVGGREIGYLFGAYRSYKNSWEGV 175
Query 66 LTGKDIGWGGSEIRPEATGYGCAYFAENVLDQLLY--DTLKGKKCILSGCGNVAVYCAEK 123
LTGK + WGGS IRPEATGYG Y+ + ++D ++ +GK+ +SG GNVA Y A K
Sbjct 176 LTGKGLNWGGSLIRPEATGYGLVYYTQAMIDYATNGKESFEGKRVTISGSGNVAQYAALK 235
Query 124 LLEMGAEVLTLSDSSGTIYCSAGFSKEEIRTIKEAKATDRAVRVEAFANGESVVFERR-- 181
++E+G V++LSDS G I G + E++ I AK ++ +E N S E +
Sbjct 236 VIELGGTVVSLSDSKGCIISETGITSEQVADISSAKVNFKS--LEQIVNEYSTFSENKVQ 293
Query 182 ----GTPWS 186
PW+
Sbjct 294 YIAGARPWT 302
> sce:YAL062W GDH3, FUN51; Gdh3p (EC:1.4.1.4); K00262 glutamate
dehydrogenase (NADP+) [EC:1.4.1.4]
Length=457
Score = 160 bits (404), Expect = 3e-39, Method: Compositional matrix adjust.
Identities = 80/156 (51%), Positives = 106/156 (67%), Gaps = 2/156 (1%)
Query 6 DPKGKSVAEIRRFCQSSMTELAKHIGPSRDVPAGDIGVGTREIGFLFGQYKRLCSNFEGA 65
D KGKS EIRR C + M EL++HIG DVPAGDIGVG REIG+LFG Y+ +++EG
Sbjct 117 DLKGKSDNEIRRICYAFMRELSRHIGKDTDVPAGDIGVGGREIGYLFGAYRSYKNSWEGV 176
Query 66 LTGKDIGWGGSEIRPEATGYGCAYFAENVLDQLLY--DTLKGKKCILSGCGNVAVYCAEK 123
LTGK + WGGS IRPEATG+G Y+ + ++D ++ +GK+ +SG GNVA Y A K
Sbjct 177 LTGKGLNWGGSLIRPEATGFGLVYYTQAMIDYATNGKESFEGKRVTISGSGNVAQYAALK 236
Query 124 LLEMGAEVLTLSDSSGTIYCSAGFSKEEIRTIKEAK 159
++E+G V++LSDS G I G + E+I I AK
Sbjct 237 VIELGGIVVSLSDSKGCIISETGITSEQIHDIASAK 272
> ath:AT5G18170 GDH1; GDH1 (GLUTAMATE DEHYDROGENASE 1); ATP binding
/ glutamate dehydrogenase [NAD(P)+]/ oxidoreductase (EC:1.4.1.3);
K00261 glutamate dehydrogenase (NAD(P)+) [EC:1.4.1.3]
Length=411
Score = 77.0 bits (188), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 45/147 (30%), Positives = 76/147 (51%), Gaps = 1/147 (0%)
Query 1 GGSDFDPKGKSVAEIRRFCQSSMTELAKHIGPSRDVPAGDIGVGTREIGFLFGQYKRLCS 60
GG DP S++E+ R + ++ IG DVPA D+G G + + ++ +Y +
Sbjct 103 GGIGCDPSKLSISELERLTRVFTQKIHDLIGIHTDVPAPDMGTGPQTMAWILDEYSKFHG 162
Query 61 NFEGALTGKDIGWGGSEIRPEATGYGCAYFAENVLDQLLYDTLKGKKCILSGCGNVAVYC 120
+TGK I GGS R ATG G + E +L++ T+ G++ ++ G GNV +
Sbjct 163 YSPAVVTGKPIDLGGSLGRDAATGRGVMFGTEALLNE-HGKTISGQRFVIQGFGNVGSWA 221
Query 121 AEKLLEMGAEVLTLSDSSGTIYCSAGF 147
A+ + E G +++ +SD +G I G
Sbjct 222 AKLISEKGGKIVAVSDITGAIKNKDGI 248
> ath:AT3G03910 GDH3; GDH3 (GLUTAMATE DEHYDROGENASE 3); binding
/ catalytic/ oxidoreductase/ oxidoreductase, acting on the
CH-NH2 group of donors, NAD or NADP as acceptor (EC:1.4.1.3);
K00261 glutamate dehydrogenase (NAD(P)+) [EC:1.4.1.3]
Length=411
Score = 77.0 bits (188), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 47/166 (28%), Positives = 86/166 (51%), Gaps = 4/166 (2%)
Query 1 GGSDFDPKGKSVAEIRRFCQSSMTELAKHIGPSRDVPAGDIGVGTREIGFLFGQYKRLCS 60
GG DP S++E+ R + ++ IG DVPA D+G G + + ++ +Y +
Sbjct 103 GGIGCDPSELSLSELERLTRVFTQKIHDLIGIHTDVPAPDMGTGPQTMAWILDEYSKFHG 162
Query 61 NFEGALTGKDIGWGGSEIRPEATGYGCAYFAENVLDQLLYDTLKGKKCILSGCGNVAVYC 120
+ +TGK I GGS R ATG G + E +L++ T+ G++ + G GNV +
Sbjct 163 HSPAVVTGKPIDLGGSLGRDAATGRGVLFATEALLNE-HGKTISGQRFAIQGFGNVGSWA 221
Query 121 AEKLLEMGAEVLTLSDSSGTIYCSAGFSKEEIRTIKEAKATDRAVR 166
A+ + + G +++ +SD +G I + G +I ++ E +R ++
Sbjct 222 AKLISDKGGKIVAVSDVTGAIKNNNGI---DILSLLEHAEENRGIK 264
> hsa:2747 GLUD2, GDH2, GLUDP1; glutamate dehydrogenase 2 (EC:1.4.1.3);
K00261 glutamate dehydrogenase (NAD(P)+) [EC:1.4.1.3]
Length=558
Score = 74.3 bits (181), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 49/168 (29%), Positives = 80/168 (47%), Gaps = 13/168 (7%)
Query 2 GSDFDPKGKSVAEIRRFCQSSMTELAKH--IGPSRDVPAGDIGVGTREIGFLFGQYKRLC 59
G +PK + E+ + + ELAK IGP DVPA D+ G RE+ ++ Y
Sbjct 185 GVKINPKNYTENELEKITRRFTMELAKKGFIGPGVDVPAPDMNTGEREMSWIADTYASTI 244
Query 60 SNFE----GALTGKDIGWGGSEIRPEATGYGCAYFAENVLDQLLYDTLKG-------KKC 108
+++ +TGK I GG R ATG G + EN +++ Y ++ G K
Sbjct 245 GHYDINAHACVTGKPISQGGIHGRISATGRGVFHGIENFINEASYMSILGMTPGFRDKTF 304
Query 109 ILSGCGNVAVYCAEKLLEMGAEVLTLSDSSGTIYCSAGFSKEEIRTIK 156
++ G GNV ++ L GA+ + + +S G+I+ G +E+ K
Sbjct 305 VVQGFGNVGLHSMRYLHRFGAKCIAVGESDGSIWNPDGIDPKELEDFK 352
> hsa:2746 GLUD1, GDH, GDH1, GLUD, MGC132003; glutamate dehydrogenase
1 (EC:1.4.1.3); K00261 glutamate dehydrogenase (NAD(P)+)
[EC:1.4.1.3]
Length=558
Score = 73.9 bits (180), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 49/168 (29%), Positives = 80/168 (47%), Gaps = 13/168 (7%)
Query 2 GSDFDPKGKSVAEIRRFCQSSMTELAKH--IGPSRDVPAGDIGVGTREIGFLFGQYKRLC 59
G +PK + E+ + + ELAK IGP DVPA D+ G RE+ ++ Y
Sbjct 185 GVKINPKNYTDNELEKITRRFTMELAKKGFIGPGIDVPAPDMSTGEREMSWIADTYASTI 244
Query 60 SNFE----GALTGKDIGWGGSEIRPEATGYGCAYFAENVLDQLLYDTLKG-------KKC 108
+++ +TGK I GG R ATG G + EN +++ Y ++ G K
Sbjct 245 GHYDINAHACVTGKPISQGGIHGRISATGRGVFHGIENFINEASYMSILGMTPGFGDKTF 304
Query 109 ILSGCGNVAVYCAEKLLEMGAEVLTLSDSSGTIYCSAGFSKEEIRTIK 156
++ G GNV ++ L GA+ + + +S G+I+ G +E+ K
Sbjct 305 VVQGFGNVGLHSMRYLHRFGAKCIAVGESDGSIWNPDGIDPKELEDFK 352
> ath:AT5G07440 GDH2; GDH2 (GLUTAMATE DEHYDROGENASE 2); ATP binding
/ glutamate dehydrogenase [NAD(P)+]/ glutamate dehydrogenase/
oxidoreductase (EC:1.4.1.3); K00261 glutamate dehydrogenase
(NAD(P)+) [EC:1.4.1.3]
Length=411
Score = 73.2 bits (178), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 47/161 (29%), Positives = 79/161 (49%), Gaps = 3/161 (1%)
Query 1 GGSDFDPKGKSVAEIRRFCQSSMTELAKHIGPSRDVPAGDIGVGTREIGFLFGQYKRLCS 60
GG P+ S++E+ R + ++ IG DVPA D+G + + ++ +Y +
Sbjct 103 GGIGCSPRDLSLSELERLTRVFTQKIHDLIGIHTDVPAPDMGTNAQTMAWILDEYSKFHG 162
Query 61 NFEGALTGKDIGWGGSEIRPEATGYGCAYFAENVLDQLLYDTLKGKKCILSGCGNVAVYC 120
+ +TGK I GGS R ATG G + E +L + +++G ++ G GNV +
Sbjct 163 HSPAVVTGKPIDLGGSLGREAATGRGVVFATEALLAE-YGKSIQGLTFVIQGFGNVGTWA 221
Query 121 AEKLLEMGAEVLTLSDSSGTIYCSAGFSKEEIRTIKEAKAT 161
A+ + E G +V+ +SD +G I G + IK AT
Sbjct 222 AKLIHEKGGKVVAVSDITGAIRNPEGIDINAL--IKHKDAT 260
> mmu:14661 Glud1, AI118167, Gdh-X, Glud, Gludl; glutamate dehydrogenase
1 (EC:1.4.1.3); K00261 glutamate dehydrogenase (NAD(P)+)
[EC:1.4.1.3]
Length=558
Score = 72.8 bits (177), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 49/168 (29%), Positives = 80/168 (47%), Gaps = 13/168 (7%)
Query 2 GSDFDPKGKSVAEIRRFCQSSMTELAKH--IGPSRDVPAGDIGVGTREIGFLFGQYKRLC 59
G +PK + E+ + + ELAK IGP DVPA D+ G RE+ ++ Y
Sbjct 185 GVKINPKNYTDNELEKITRRFTMELAKKGFIGPGIDVPAPDMSTGEREMSWIADTYASTI 244
Query 60 SNFE----GALTGKDIGWGGSEIRPEATGYGCAYFAENVLDQLLYDTLKG-------KKC 108
+++ +TGK I GG R ATG G + EN +++ Y ++ G K
Sbjct 245 GHYDINAHACVTGKPISQGGIHGRISATGRGVFHGIENFINEASYMSILGMTPGFGDKTF 304
Query 109 ILSGCGNVAVYCAEKLLEMGAEVLTLSDSSGTIYCSAGFSKEEIRTIK 156
++ G GNV ++ L GA+ + + +S G+I+ G +E+ K
Sbjct 305 VVQGFGNVGLHSMRYLHRFGAKCVGVGESDGSIWNPDGIDPKELEDFK 352
> cel:ZK829.4 hypothetical protein; K00261 glutamate dehydrogenase
(NAD(P)+) [EC:1.4.1.3]
Length=536
Score = 72.4 bits (176), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 55/174 (31%), Positives = 80/174 (45%), Gaps = 13/174 (7%)
Query 1 GGSDFDPKGKSVAEIRRFCQSSMTELAKH--IGPSRDVPAGDIGVGTREIGFLFGQYKRL 58
GG DPK + EI + + E AK +GP DVPA D+G G RE+G++ Y +
Sbjct 162 GGVKIDPKQYTDYEIEKITRRIAIEFAKKGFLGPGVDVPAPDMGTGEREMGWIADTYAQT 221
Query 59 CSNFE----GALTGKDIGWGGSEIRPEATGYGCAYFAENVLDQLLY------DT-LKGKK 107
+ + +TGK I GG R ATG G E + Y DT L GK
Sbjct 222 IGHLDRDASACITGKPIVSGGIHGRVSATGRGVWKGLEVFTNDADYMKMVGLDTGLAGKT 281
Query 108 CILSGCGNVAVYCAEKLLEMGAEVLTLSDSSGTIYCSAGFSKEEIRTIKEAKAT 161
I+ G GNV ++ L G++V+ + + +Y G +E+ K+A T
Sbjct 282 AIIQGFGNVGLHTHRYLHRAGSKVIGIQEYDCAVYNPDGIHPKELEDWKDANGT 335
> dre:317737 glud1a, cb622, glud1, wu:fc33g09, wu:fc66a10, zgc:77186;
glutamate dehydrogenase 1a; K00261 glutamate dehydrogenase
(NAD(P)+) [EC:1.4.1.3]
Length=544
Score = 72.0 bits (175), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 52/173 (30%), Positives = 79/173 (45%), Gaps = 13/173 (7%)
Query 2 GSDFDPKGKSVAEIRRFCQSSMTELAKH--IGPSRDVPAGDIGVGTREIGFLFGQYKRLC 59
G +PK S E+ + + ELAK IGP DVPA D+ G RE+ ++ Y
Sbjct 171 GVKINPKNYSDTELEKITRRFTIELAKKGFIGPGIDVPAPDMSTGEREMSWIADTYANTM 230
Query 60 S----NFEGALTGKDIGWGGSEIRPEATGYGCAYFAENVLDQLLYDTLKG-------KKC 108
N +TGK I GG R ATG G + EN +++ Y + G K
Sbjct 231 GHHDINAHACVTGKPISQGGIHGRISATGRGVFHGIENFVNEAAYMSQLGLTPGFGDKTF 290
Query 109 ILSGCGNVAVYCAEKLLEMGAEVLTLSDSSGTIYCSAGFSKEEIRTIKEAKAT 161
++ G GNV ++ L GA+ + + + G+I+ +G +E+ K A T
Sbjct 291 VIQGFGNVGLHSMRYLHRYGAKCVGIGELDGSIWNPSGIDPKELEDYKLANGT 343
> xla:446858 glud1, MGC80801; glutamate dehydrogenase 1 (EC:1.4.1.3);
K00261 glutamate dehydrogenase (NAD(P)+) [EC:1.4.1.3]
Length=540
Score = 72.0 bits (175), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 52/173 (30%), Positives = 78/173 (45%), Gaps = 13/173 (7%)
Query 2 GSDFDPKGKSVAEIRRFCQSSMTELAKH--IGPSRDVPAGDIGVGTREIGFLFGQYKRLC 59
G +P+ S AE+ + + ELAK IGP DVPA D+ G RE+ ++ Y
Sbjct 167 GVKINPRNFSDAELEKITRRFTIELAKKGFIGPGIDVPAPDMSTGEREMSWIADTYANTI 226
Query 60 S----NFEGALTGKDIGWGGSEIRPEATGYGCAYFAENVLDQLLYDTLKG-------KKC 108
N +TGK I GG R ATG G + EN +++ Y + G K
Sbjct 227 GHTDINAHACVTGKPISQGGIHGRISATGRGVFHGIENFINEASYMSQLGMTPGFGDKTF 286
Query 109 ILSGCGNVAVYCAEKLLEMGAEVLTLSDSSGTIYCSAGFSKEEIRTIKEAKAT 161
++ G GNV ++ L GA+ + + + GTI+ G +E+ K T
Sbjct 287 VIQGFGNVGLHSMRYLHRFGAKCVGIGEIDGTIWNPNGIDPKELEDYKLQHGT 339
> dre:373092 glud1b, MGC192851, cb719, glud1, wu:fb16e02, wu:fb58f12,
wu:fe37f03, wu:fj43f02, zgc:192851, zgc:55630; glutamate
dehydrogenase 1b (EC:1.4.1.3); K00261 glutamate dehydrogenase
(NAD(P)+) [EC:1.4.1.3]
Length=542
Score = 70.5 bits (171), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 51/173 (29%), Positives = 79/173 (45%), Gaps = 13/173 (7%)
Query 2 GSDFDPKGKSVAEIRRFCQSSMTELAKH--IGPSRDVPAGDIGVGTREIGFLFGQYKRLC 59
G +P+ S E+ + + ELAK IGP DVPA D+ G RE+ ++ Y
Sbjct 169 GVKINPRNYSDNELEKITRRFTIELAKKGFIGPGIDVPAPDMSTGEREMSWIADTYANTI 228
Query 60 S----NFEGALTGKDIGWGGSEIRPEATGYGCAYFAENVLDQLLYDTLKG-------KKC 108
+ N +TGK I GG R ATG G + EN +++ Y + G K
Sbjct 229 AHTDINAHACVTGKPISQGGIHGRISATGRGVFHGIENFINEASYMSKLGLTPGFADKTF 288
Query 109 ILSGCGNVAVYCAEKLLEMGAEVLTLSDSSGTIYCSAGFSKEEIRTIKEAKAT 161
I+ G GNV ++ L GA+ + +++ G+I+ G +E+ K T
Sbjct 289 IIQGFGNVGLHSMRYLHRYGAKCVGIAEIDGSIWNPNGMDPKELEDYKLQHGT 341
> mmu:50912 Exosc10, PM-Scl, PM/Scl-100, Pmscl2, RRP6, p2, p3,
p4; exosome component 10; K12591 exosome complex exonuclease
RRP6 [EC:3.1.13.-]
Length=887
Score = 33.1 bits (74), Expect = 0.66, Method: Composition-based stats.
Identities = 14/46 (30%), Positives = 24/46 (52%), Gaps = 0/46 (0%)
Query 2 GSDFDPKGKSVAEIRRFCQSSMTELAKHIGPSRDVPAGDIGVGTRE 47
G DF P S ++ R F S ++ + P++ P+G GVG ++
Sbjct 814 GKDFSPYDYSQSDFRAFAGDSKSKPSSQFDPNKLAPSGKKGVGAKK 859
> hsa:23637 RABGAP1, DKFZp586D2123, GAPCENA, RP11-123N4.2, TBC1D11;
RAB GTPase activating protein 1
Length=1069
Score = 30.4 bits (67), Expect = 3.8, Method: Composition-based stats.
Identities = 13/25 (52%), Positives = 19/25 (76%), Gaps = 0/25 (0%)
Query 109 ILSGCGNVAVYCAEKLLEMGAEVLT 133
+LSG G+V+ CAEK+LE E+L+
Sbjct 522 LLSGSGDVSKECAEKILETWGELLS 546
> dre:100334685 hypothetical protein LOC100334685
Length=612
Score = 30.4 bits (67), Expect = 4.1, Method: Composition-based stats.
Identities = 13/25 (52%), Positives = 19/25 (76%), Gaps = 0/25 (0%)
Query 109 ILSGCGNVAVYCAEKLLEMGAEVLT 133
+LSG G+V+ CAEK+LE E+L+
Sbjct 482 LLSGSGDVSKECAEKILETWGELLS 506
> mmu:227800 Rabgap1, Gapcena, KIAA4104, MGC30493, mKIAA4104;
RAB GTPase activating protein 1
Length=809
Score = 29.6 bits (65), Expect = 5.9, Method: Composition-based stats.
Identities = 13/25 (52%), Positives = 19/25 (76%), Gaps = 0/25 (0%)
Query 109 ILSGCGNVAVYCAEKLLEMGAEVLT 133
+LSG G+V+ CAEK+LE E+L+
Sbjct 517 LLSGFGDVSKECAEKILETWGELLS 541
> ath:AT1G61720 BAN; BAN (BANYULS); oxidoreductase (EC:1.3.1.77);
K08695 anthocyanidin reductase [EC:1.3.1.77]
Length=340
Score = 29.6 bits (65), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 95 LDQLLYDTLKGKKCILSGCGNVAVYCAEKLLEMGAEVLT 133
+DQ L T K C++ G GN+A + LL+ G +V T
Sbjct 1 MDQTLTHTGSKKACVIGGTGNLASILIKHLLQSGYKVNT 39
Lambda K H
0.318 0.136 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5558593832
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40