bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0041_orf1
Length=483
Score E
Sequences producing significant alignments: (Bits) Value
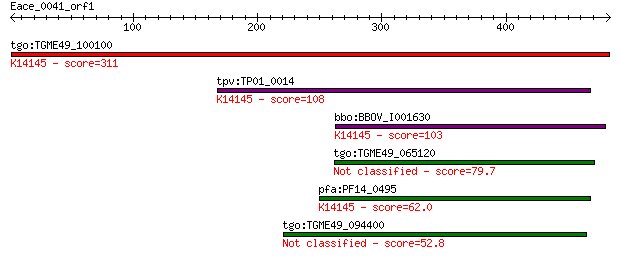
tgo:TGME49_100100 rhoptry neck protein 2 (EC:3.2.1.39); K14145... 311 3e-84
tpv:TP01_0014 hypothetical protein; K14145 rhoptry neck protein 2 108 5e-23
bbo:BBOV_I001630 19.m02019; hypothetical protein; K14145 rhopt... 103 1e-21
tgo:TGME49_065120 hypothetical protein 79.7 2e-14
pfa:PF14_0495 PfRON2; rhoptry neck protein 2; K14145 rhoptry n... 62.0 5e-09
tgo:TGME49_094400 hypothetical protein 52.8 3e-06
> tgo:TGME49_100100 rhoptry neck protein 2 (EC:3.2.1.39); K14145
rhoptry neck protein 2
Length=1404
Score = 311 bits (798), Expect = 3e-84, Method: Compositional matrix adjust.
Identities = 182/483 (37%), Positives = 271/483 (56%), Gaps = 72/483 (14%)
Query 2 AQIGNMLEDGGSRYYDTRAEQWMEIISKPAANNDILEKAFYYDNREMISKSKSAIQKAFD 61
QI + + S + TR W ++++ P A +I E+ ++D+RE+++KS A+ + +D
Sbjct 584 GQITGSIAEPFSHSWRTR---WGKVVADPTAYGEIFERTLWFDDRELMAKSSGALFRQYD 640
Query 62 AFARRITALAGKASVSFSEATTIFEAIQKDVAAQRPRKWYKREPPINKKNLFRVAVILFL 121
A K S+SF +F ++ +
Sbjct 641 RIA--------KDSMSFG----VFMNVENGL----------------------------- 659
Query 122 KSRLACDMRSSFAEKFEAFTQFLFKSREAGGRRQPALQRSLLAFFRTNKAQSFYAMCSFD 181
L DMRS F+ K Q+S A R ++ D
Sbjct 660 ---LKKDMRSKLEAYISQRKSFVEKR-----------QQSRFAKLRKKIPEN-------D 698
Query 182 PLSLSYLFLYRFVLMGSDAGRTHDKQHAAMSRTRLSTRLLGSKWTPSILKSVMMGFKRKS 241
P +L F+ + A HD+QH +SR + + R+L SKWTP++LK +M K
Sbjct 699 PYALRAAI---FLALNYPAA-LHDRQHTRVSRNKKTMRILNSKWTPAVLKKLMRKVNHKH 754
Query 242 LVQTAKQLLLESLDPSILSSLLTSFDWIVHTQATLQVNQSASMYHDLEAPSQQYKTTGEK 301
+ + AK LLL SLDP++LSS++T+FD+I HTQA L+VNQ+A MYH++ A ++ EK
Sbjct 755 MAREAKALLLRSLDPTVLSSIVTAFDFITHTQANLEVNQNAFMYHEVRAREVSRQSAAEK 814
Query 302 KEGYRLSSGGLVKTTDKQLEDWAEYGIPASLSRKLRRGEKLPKSVAFEKIATPDLTKWEQ 361
+RL GLV+ TD ++ WAE+GIP + R+L RGEKLP+ ++F I P+LT W+
Sbjct 815 GS-HRLHERGLVRETDDMIKRWAEHGIPGDIKRRLARGEKLPEGMSFGGIPIPNLTNWDA 873
Query 362 QLNNKWLDALSAYLEHPYGRAALAARDPVALLVKESKDRLEAEVDSTIFLGRVVKVPRMS 421
QLN+KWL+A +AYL HPYGRAAL ARDPVALLVK+S+DRL+AE + TIFLGR+ K S
Sbjct 874 QLNSKWLEAYNAYLRHPYGRAALNARDPVALLVKDSRDRLQAEAEGTIFLGRIAKRVHQS 933
Query 422 K--FKKAMKAISNFFRAILRAPEQSEYAVWMGVKVNVDKVLAVMKAVNNAAEVAKSSGVY 479
K ++A +A+ FF ++LR E+SEYAVW GVKV++ +V+ + +N+ AEV K+ +Y
Sbjct 934 KNLLRRAGRALKTFFLSLLRENERSEYAVWFGVKVDMRQVIQTCRQINSVAEVVKNDRLY 993
Query 480 EHI 482
+ I
Sbjct 994 DFI 996
> tpv:TP01_0014 hypothetical protein; K14145 rhoptry neck protein
2
Length=788
Score = 108 bits (270), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 82/313 (26%), Positives = 153/313 (48%), Gaps = 28/313 (8%)
Query 168 TNKAQSFYAMCSFDPLSLSYLFLYRFVLMGSDAGRTHDKQHAAMSRTRLSTRLL--GSKW 225
+N+ + FY S + L++LFLY +L + + K+ + + +R + S +
Sbjct 112 SNRFREFYDSSS---VGLAHLFLYN-ILSKKYPAQDYLKELEKLGKADPFSRFVDASSIF 167
Query 226 TPSILKSVMMGFKRKSLV-----QTAKQLLLESLDPSILSSLLTSFDWIVHTQATLQVNQ 280
P L+ V+ F + S V + K L + + +L L S ++ H+ A LQV Q
Sbjct 168 APEKLRKVVKWFLKGSFVKQFMREKTKLTLKQLVRSEVLRDALESITFVTHSMANLQVIQ 227
Query 281 SASMYHDLEAPSQQYKTTGEKKEGYRLSSGGLVKTTDKQLEDWAEYGIPASLSRKLRRGE 340
+A +Y + GG++ D +++W++ G S+S KL+
Sbjct 228 NA-----------EYWGRAYPNSSFYFRRGGVISVIDNTIKNWSDDGYTRSISEKLKNHV 276
Query 341 KLP----KSVAFEKIATPDLTKWEQQLNNKWLDALSAYLEHPYGRAALAARDPVALLVKE 396
L K F+ I + KW++ LN + L + + +LE +A + +VK+
Sbjct 277 DLNEDDLKIADFQHIHNMESVKWDKSLNTEILKSFNEFLELGSVKALEGRNHILYEVVKD 336
Query 397 SKDRLEAEVDSTIFLGRVVKVPRM-SKFKKAMKAISNFFRAIL-RAPEQSEYAVWMGVKV 454
S+D LE +++TIF GR++ P++ S +K ++ NF IL RA + ++A+W G+K+
Sbjct 337 SRDNLEQNIENTIFFGRILNSPKIESNLRKYLRRAVNFGNFILDRAEKNVDHAIWFGIKL 396
Query 455 NVDKVLAVMKAVN 467
N+DKV+ V+ ++
Sbjct 397 NLDKVMRVVDELH 409
> bbo:BBOV_I001630 19.m02019; hypothetical protein; K14145 rhoptry
neck protein 2
Length=1365
Score = 103 bits (258), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 62/224 (27%), Positives = 116/224 (51%), Gaps = 15/224 (6%)
Query 263 LTSFDWIVHTQATLQVNQSASMY-HDLEAPSQQYKTTGEKKEGYRLSSGGLVKTTDKQLE 321
++ ++ H+ A +Q+NQ+A ++ L A + K +GG + D ++
Sbjct 775 ISCITFVTHSLADIQINQNAEVFGRGLLADKDRIKK--------HFVAGGYINHVDSIIK 826
Query 322 DWAEYGIPASLSRKLRRGEKLPK----SVAFEKIATPDLTKWEQQLNNKWLDALSAYLEH 377
+W++ G ++++K+++GE L K I + +WE+ LN+ LD +A+L+
Sbjct 827 EWSDEGYTDAIAKKVKQGEDLNKEDMEDANIHNIVHTESLRWEKHLNSLILDGYNAFLDL 886
Query 378 PYGRAALAARDPVALLVKESKDRLEAEVDSTIFLGRVVKVPRM-SKFKKAMKAISNFFRA 436
P + + +VK+S+D LE +D TIF GRVV P +K+K+A + ++
Sbjct 887 PSIKVLDGRHSLIYEIVKDSRDNLEQHLDETIFFGRVVNPPEYNNKWKRAFDRAAKVTKS 946
Query 437 IL-RAPEQSEYAVWMGVKVNVDKVLAVMKAVNNAAEVAKSSGVY 479
IL R + E+AVW GVK+N ++ V++ +N + V + Y
Sbjct 947 ILNRTQHKVEHAVWFGVKLNYQHIIEVVEELNRISNVISPTESY 990
> tgo:TGME49_065120 hypothetical protein
Length=620
Score = 79.7 bits (195), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 52/216 (24%), Positives = 104/216 (48%), Gaps = 11/216 (5%)
Query 262 LLTSFDWIVHTQATLQVNQSASMYHDLEAPSQQYKTTGEKKEGYRLSSGGLVKTTDKQLE 321
++ +F + H+ A +QV S++ D Q + + + ++K D+Q+
Sbjct 18 VMDAFAFTAHSAALMQVVNYHSLFRDRTLNDLQEASYLATPPAFSIEDQSVMKRCDEQIY 77
Query 322 DWAEYG-----IPASLSRKLRRGEKLPKSVAFEKIATPDLTKWEQQLNNKWLDALSAYLE 376
+W+ +G + A L + +G+ +++ E + TPD +W ++L+ LD+L Y
Sbjct 78 EWSRFGFSPDVLEADLHNETFKGKW--ENLRLETMPTPDTHQWWRRLHRAILDSLEIYQA 135
Query 377 HPYGRAALAARDPVALLVKESKDRLE--AEVDSTIFLGRVVKVPRMSKFKKAMKAISNFF 434
H R D + LV+++ L+ + D TIF GR+ + R S +K + + FF
Sbjct 136 HE--RHLKGMSDNLYTLVEDTIKNLQDGSSRDDTIFFGRIARGARESVGQKMWRGVKTFF 193
Query 435 RAILRAPEQSEYAVWMGVKVNVDKVLAVMKAVNNAA 470
++R S++AVW GV N ++ +++ + A
Sbjct 194 LDLVRDVPGSDHAVWFGVGFNFPRIFQILRKMKEIA 229
> pfa:PF14_0495 PfRON2; rhoptry neck protein 2; K14145 rhoptry
neck protein 2
Length=2189
Score = 62.0 bits (149), Expect = 5e-09, Method: Composition-based stats.
Identities = 57/256 (22%), Positives = 108/256 (42%), Gaps = 45/256 (17%)
Query 250 LLESLDPSILSSLLTSFDWIVHTQATLQVNQSASMY--HDLEAPSQQYKTTGEKKEGYRL 307
LL+ + +L ++ +S + HT AT+Q+ Q+A H+ QY + G
Sbjct 1548 LLQHIPAHMLENITSSIRFTTHTIATMQIIQNAKYMSKHNF----SQYDSKGMLARQI-F 1602
Query 308 SSGGLVKTTDKQLEDWAEYG-------------IPASLSRKLRRGEKLPKSVAFEKIATP 354
+ GG + D + W G + S+ +L+ E+ ++ + E+ A
Sbjct 1603 TKGGFAEYADNLMAKWFSKGFEEYKREQIENFKMENSIDSELKDSEREDENDSSEESAKK 1662
Query 355 DLT----------------------KWEQQLNNKWLDALSAYLEHPYGRAALAARDPVAL 392
L KW+Q +N + + AL +LE + A V
Sbjct 1663 KLQDLQLEEREKMKKENSLLFNQSDKWDQFINKELVRALGLWLE--FNDNPTNASSFVYK 1720
Query 393 LVKESKDRLEAEVDSTIFLGRVVKVPRMSKFKKAMKAISNFFRAILRAPE-QSEYAVWMG 451
+V++SK LE +D+ I R VK + + F++ I + +LR P + E+A+W G
Sbjct 1721 VVEDSKHLLENNIDNNIIFSRTVKPTKQTAFRRFFNKILSLGNMLLRKPSFRVEHALWFG 1780
Query 452 VKVNVDKVLAVMKAVN 467
+++ K +++ V+
Sbjct 1781 ATIDIKKAFILLEKVS 1796
> tgo:TGME49_094400 hypothetical protein
Length=1490
Score = 52.8 bits (125), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 63/262 (24%), Positives = 105/262 (40%), Gaps = 32/262 (12%)
Query 221 LGSKWTPSILKSVMMGFKRKSLVQTAKQLLLESLDPSILSSLLTSFDWIVHTQATLQVNQ 280
L S+ T S+L+ + K+ QLL + L P + + IVH A +NQ
Sbjct 808 LFSRLTGSLLRRLGTRLAEKAYQPYMMQLLHQKLRPETRKKMFQAIQVIVHPLALKHLNQ 867
Query 281 SASMYHDLEAPSQQYKTTGEKKEGYRLSSGGLVKTTDKQLEDWAEYGIPASLSRKLRRGE 340
A +Y D +G GG + D L +W+EYG+ L +L +G+
Sbjct 868 MAPLYSD--------------SKGNLNKEGGFAEEIDNTLREWSEYGMNEKLIDRLEKGQ 913
Query 341 KLPKS----VAFEKIATPDLTKWEQQLNNKWLDALSAYLEHP----YGRAA---LAARDP 389
L S V + P + + + + AL E+P RAA L+
Sbjct 914 NLSASDFAGVNILTMPIPSRERMDSLVAERLNGALKRLQENPENVETSRAACDFLSHASN 973
Query 390 VALLVKESKDRLEA-EVDSTIFLGRVVKVPRMSK-----FKKAMKAISNFFRAILRAPE- 442
+ +S+ L ++ +FLG++VK + F ++ + + ILR P
Sbjct 974 SIYVTNDSRYLLNKFGPENLLFLGKIVKRSETLEKPQGFFARSWAGVEKTLKGILRLPPY 1033
Query 443 QSEYAVWMGVKVNVDKVLAVMK 464
+ V+ VKV D V V++
Sbjct 1034 MVRHFVFAAVKVREDLVSEVVR 1055
Lambda K H
0.319 0.130 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 22994080296
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40