bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0045_orf2
Length=154
Score E
Sequences producing significant alignments: (Bits) Value
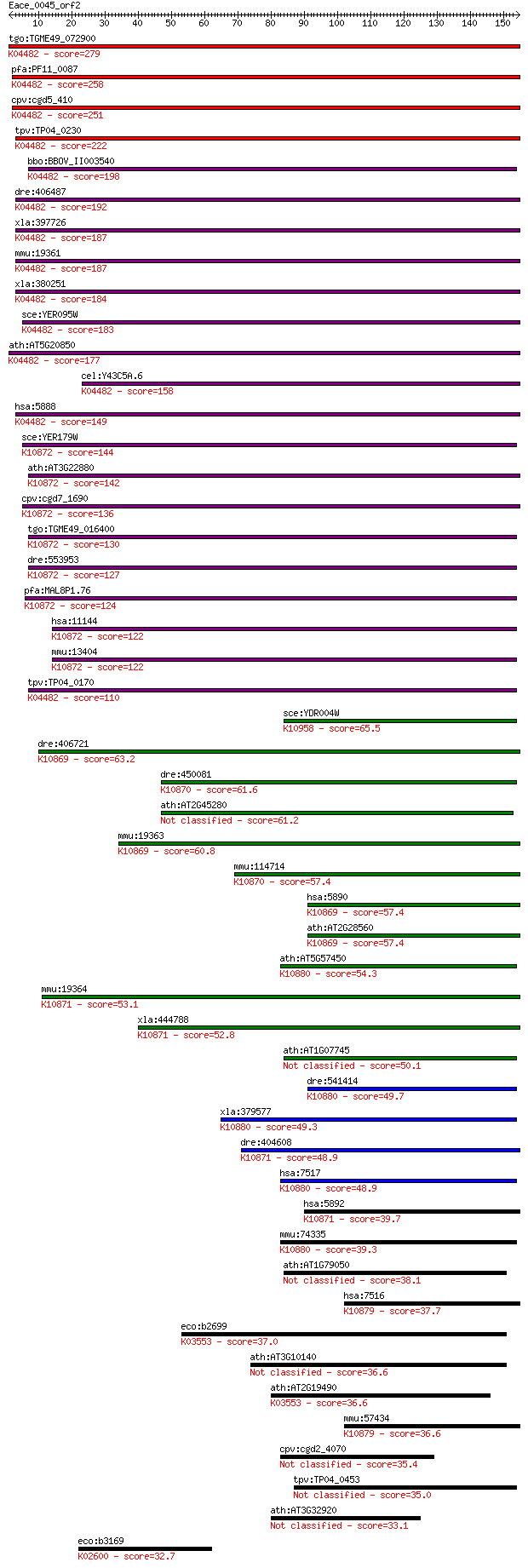
tgo:TGME49_072900 DNA repair protein, putative (EC:3.6.3.8); K... 279 3e-75
pfa:PF11_0087 rad51; Rad51 homolog; K04482 DNA repair protein ... 258 4e-69
cpv:cgd5_410 Rad51 ; K04482 DNA repair protein RAD51 251 6e-67
tpv:TP04_0230 DNA repair protein Rad51; K04482 DNA repair prot... 222 4e-58
bbo:BBOV_II003540 18.m06297; Rad51 protein; K04482 DNA repair ... 198 7e-51
dre:406487 rad51, wu:fb38e12, zgc:77754; RAD51 homolog (RecA h... 192 2e-49
xla:397726 rad51-a, MGC84850, brcc5, rad51, rad51a, reca, xrad... 187 1e-47
mmu:19361 Rad51, AV304093, Rad51a, Reca; RAD51 homolog (S. cer... 187 2e-47
xla:380251 rad51-b, MGC130792, MGC52570, brcc5, rad51, rad51a,... 184 7e-47
sce:YER095W RAD51, MUT5; Rad51p; K04482 DNA repair protein RAD51 183 2e-46
ath:AT5G20850 ATRAD51; ATP binding / DNA binding / DNA-depende... 177 2e-44
cel:Y43C5A.6 rad-51; RADiation sensitivity abnormal/yeast RAD-... 158 6e-39
hsa:5888 RAD51, BRCC5, HRAD51, HsRad51, HsT16930, RAD51A, RECA... 149 5e-36
sce:YER179W DMC1, ISC2; Dmc1p; K10872 meiotic recombination pr... 144 1e-34
ath:AT3G22880 DMC1; DMC1 (DISRUPTION OF MEIOTIC CONTROL 1); AT... 142 3e-34
cpv:cgd7_1690 meiotic recombination protein DMC1-like protein ... 136 3e-32
tgo:TGME49_016400 meiotic recombination protein DMC1-like prot... 130 1e-30
dre:553953 dmc1, MGC136628, zgc:136628; DMC1 dosage suppressor... 127 2e-29
pfa:MAL8P1.76 meiotic recombination protein dmc1-like protein;... 124 9e-29
hsa:11144 DMC1, DMC1H, HsLim15, LIM15, MGC150472, MGC150473, d... 122 4e-28
mmu:13404 Dmc1, Dmc1h, MGC151144, Mei11, sgdp; DMC1 dosage sup... 122 5e-28
tpv:TP04_0170 meiotic recombination protein DMC1; K04482 DNA r... 110 3e-24
sce:YDR004W RAD57; Protein that stimulates strand exchange by ... 65.5 7e-11
dre:406721 rad51l1, wu:fd07f04, zgc:56581; RAD51-like 1 (S. ce... 63.2 4e-10
dre:450081 rad51c, zgc:101596; rad51 homolog C (S. cerevisiae)... 61.6 1e-09
ath:AT2G45280 ATRAD51C; ATP binding / damaged DNA binding / pr... 61.2 1e-09
mmu:19363 Rad51l1, AI553500, R51H2, Rad51b, mREC2; RAD51-like ... 60.8 2e-09
mmu:114714 Rad51c, R51H3, Rad51l2; RAD51 homolog c (S. cerevis... 57.4 2e-08
hsa:5890 RAD51L1, MGC34245, R51H2, RAD51B, REC2, hREC2; RAD51-... 57.4 2e-08
ath:AT2G28560 RAD51B; RAD51B; recombinase; K10869 RAD51-like p... 57.4 2e-08
ath:AT5G57450 XRCC3; XRCC3; ATP binding / damaged DNA binding ... 54.3 2e-07
mmu:19364 Rad51l3, DKFZp586D0122, R51H3, Rad51d, Trad-d2, Trad... 53.1 4e-07
xla:444788 rad51l3, MGC82048; RAD51-like 3; K10871 RAD51-like ... 52.8 5e-07
ath:AT1G07745 RAD51D; RAD51D (ARABIDOPSIS HOMOLOG OF RAD51 D);... 50.1 3e-06
dre:541414 xrcc3, im:7142103, si:dkey-11b8.1, zgc:101608; X-ra... 49.7 4e-06
xla:379577 xrcc3, MGC69118; X-ray repair complementing defecti... 49.3 4e-06
dre:404608 MGC77165; zgc:77165; K10871 RAD51-like protein 3 48.9
hsa:7517 XRCC3, CMM6; X-ray repair complementing defective rep... 48.9 7e-06
hsa:5892 RAD51L3, R51H3, RAD51D, TRAD; RAD51-like 3 (S. cerevi... 39.7 0.004
mmu:74335 Xrcc3, 4432412E01Rik, AI182522, AW537713; X-ray repa... 39.3 0.005
ath:AT1G79050 DNA repair protein recA 38.1 0.012
hsa:7516 XRCC2, DKFZp781P0919; X-ray repair complementing defe... 37.7 0.016
eco:b2699 recA, ECK2694, JW2669, lexB, recH, rnmB, srf, tif, u... 37.0 0.023
ath:AT3G10140 RECA3; RECA3 (recA homolog 3); ATP binding / DNA... 36.6 0.030
ath:AT2G19490 recA family protein; K03553 recombination protei... 36.6 0.034
mmu:57434 Xrcc2, 4921524O04Rik, 8030409M04Rik, RAD51, RecA; X-... 36.6 0.036
cpv:cgd2_4070 hypothetical protein 35.4 0.066
tpv:TP04_0453 hypothetical protein 35.0 0.088
ath:AT3G32920 ATP binding / DNA binding / DNA-dependent ATPase... 33.1 0.33
eco:b3169 nusA, ECK3158, JW3138; transcription termination/ant... 32.7 0.46
> tgo:TGME49_072900 DNA repair protein, putative (EC:3.6.3.8);
K04482 DNA repair protein RAD51
Length=354
Score = 279 bits (713), Expect = 3e-75, Method: Compositional matrix adjust.
Identities = 130/154 (84%), Positives = 145/154 (94%), Gaps = 0/154 (0%)
Query 1 QTGPLKLEHLLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVS 60
Q+GPLKLEHLLAKG TK+DL+LLK+ G TVEC+AFAP+K L+A+KG+SEQK KLK+ S
Sbjct 32 QSGPLKLEHLLAKGFTKRDLELLKDAGYQTVECIAFAPVKNLVAVKGLSEQKVEKLKKAS 91
Query 61 KELCSLGFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCH 120
KELC+LGFCSAQEYLEAR NLI+FTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCH
Sbjct 92 KELCNLGFCSAQEYLEARENLIRFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCH 151
Query 121 TLAVSCQLPVEQSGGEGKCLWIDTEGTFRPERIV 154
TLAV+CQLP+EQ+GGEGKCLWIDTEGTFRPERIV
Sbjct 152 TLAVTCQLPIEQAGGEGKCLWIDTEGTFRPERIV 185
> pfa:PF11_0087 rad51; Rad51 homolog; K04482 DNA repair protein
RAD51
Length=350
Score = 258 bits (660), Expect = 4e-69, Method: Compositional matrix adjust.
Identities = 122/153 (79%), Positives = 139/153 (90%), Gaps = 0/153 (0%)
Query 2 TGPLKLEHLLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSK 61
TGPLK+E LLAKG K+DL+LLKEGGL TVECVA+AP++TL AIKGISEQKA KLK+ K
Sbjct 30 TGPLKIEQLLAKGFVKRDLELLKEGGLQTVECVAYAPMRTLCAIKGISEQKAEKLKKACK 89
Query 62 ELCSLGFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHT 121
ELC+ GFC+A +Y +AR NLIKFTTGS QLD+LLKGGIETG +TELFGEFRTGK+QLCHT
Sbjct 90 ELCNSGFCNAIDYHDARQNLIKFTTGSKQLDALLKGGIETGGITELFGEFRTGKSQLCHT 149
Query 122 LAVSCQLPVEQSGGEGKCLWIDTEGTFRPERIV 154
LA++CQLP+EQSGGEGKCLWIDTEGTFRPERIV
Sbjct 150 LAITCQLPIEQSGGEGKCLWIDTEGTFRPERIV 182
> cpv:cgd5_410 Rad51 ; K04482 DNA repair protein RAD51
Length=347
Score = 251 bits (641), Expect = 6e-67, Method: Compositional matrix adjust.
Identities = 116/153 (75%), Positives = 133/153 (86%), Gaps = 0/153 (0%)
Query 2 TGPLKLEHLLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSK 61
GPLKLEHLL GLTK+DL++L+E G HT+EC+A+AP K LL++KGISEQK K+K K
Sbjct 26 NGPLKLEHLLPSGLTKRDLEILRENGYHTIECLAYAPKKALLSVKGISEQKCDKIKSACK 85
Query 62 ELCSLGFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHT 121
EL ++GFCS EYLEAR NLIKFTTGS QLD LL+GGIETG++TE+FGEFRTGKTQLCHT
Sbjct 86 ELVAMGFCSGTEYLEARTNLIKFTTGSSQLDRLLQGGIETGSITEIFGEFRTGKTQLCHT 145
Query 122 LAVSCQLPVEQSGGEGKCLWIDTEGTFRPERIV 154
LAV+CQLPVE GGEGKCLWIDTEGTFRPERIV
Sbjct 146 LAVTCQLPVEHKGGEGKCLWIDTEGTFRPERIV 178
> tpv:TP04_0230 DNA repair protein Rad51; K04482 DNA repair protein
RAD51
Length=343
Score = 222 bits (565), Expect = 4e-58, Method: Compositional matrix adjust.
Identities = 110/152 (72%), Positives = 131/152 (86%), Gaps = 0/152 (0%)
Query 3 GPLKLEHLLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSKE 62
P +LE LL+KGL ++DLDLL+E G T+ECVA+AP K LL IKG+SEQK K+K +E
Sbjct 24 NPQRLECLLSKGLLQRDLDLLREAGYSTLECVAYAPQKNLLVIKGLSEQKVLKIKAACRE 83
Query 63 LCSLGFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTL 122
LC LGFCS Q+YLEAR NLIKFTTGS QLD LL+GG+ETG++TE+ GEF+TGK+QLCHTL
Sbjct 84 LCHLGFCSGQDYLEARGNLIKFTTGSAQLDKLLQGGVETGSITEIIGEFKTGKSQLCHTL 143
Query 123 AVSCQLPVEQSGGEGKCLWIDTEGTFRPERIV 154
AV+CQLPVEQSGGEGKCLW+D+EGTFRPERIV
Sbjct 144 AVTCQLPVEQSGGEGKCLWVDSEGTFRPERIV 175
> bbo:BBOV_II003540 18.m06297; Rad51 protein; K04482 DNA repair
protein RAD51
Length=346
Score = 198 bits (503), Expect = 7e-51, Method: Compositional matrix adjust.
Identities = 90/147 (61%), Positives = 119/147 (80%), Gaps = 0/147 (0%)
Query 7 LEHLLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSKELCSL 66
+E LL+KG ++D+D+LK G T++ +A KTLL +KG+SEQK AK+K++ KELC
Sbjct 28 VECLLSKGFLQRDIDVLKAAGYVTLDSIAQVASKTLLEVKGLSEQKVAKIKEIVKELCPP 87
Query 67 GFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSC 126
C+A EYLE R NLIKFTTGS LD+LL+GGIE+G++TE+ G+F TGKTQLCHTLA++
Sbjct 88 DICTAAEYLECRLNLIKFTTGSTALDALLQGGIESGSITEIIGDFSTGKTQLCHTLAITS 147
Query 127 QLPVEQSGGEGKCLWIDTEGTFRPERI 153
QLP+EQ+GGEGKCLWIDT+ +FRPER+
Sbjct 148 QLPIEQNGGEGKCLWIDTQNSFRPERL 174
> dre:406487 rad51, wu:fb38e12, zgc:77754; RAD51 homolog (RecA
homolog, E. coli) (S. cerevisiae); K04482 DNA repair protein
RAD51
Length=338
Score = 192 bits (489), Expect = 2e-49, Method: Compositional matrix adjust.
Identities = 88/152 (57%), Positives = 118/152 (77%), Gaps = 0/152 (0%)
Query 3 GPLKLEHLLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSKE 62
GP + L G++ D+ L++GG HTVE VA+AP K LL IKGISE KA K+ + +
Sbjct 20 GPQPVSRLEQSGISSSDIKKLEDGGFHTVEAVAYAPKKELLNIKGISEAKADKILTEAAK 79
Query 63 LCSLGFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTL 122
+ +GF +A E+ + RA +I+ +TGS +LD LL+GGIETG++TE+FGEFRTGKTQLCHTL
Sbjct 80 MVPMGFTTATEFHQRRAEIIQISTGSKELDKLLQGGIETGSITEMFGEFRTGKTQLCHTL 139
Query 123 AVSCQLPVEQSGGEGKCLWIDTEGTFRPERIV 154
AV+CQLP++Q GGEGK ++IDTEGTFRPER++
Sbjct 140 AVTCQLPIDQGGGEGKAMYIDTEGTFRPERLL 171
> xla:397726 rad51-a, MGC84850, brcc5, rad51, rad51a, reca, xrad51;
RAD51 homolog (RecA homolog); K04482 DNA repair protein
RAD51
Length=336
Score = 187 bits (474), Expect = 1e-47, Method: Compositional matrix adjust.
Identities = 86/152 (56%), Positives = 116/152 (76%), Gaps = 0/152 (0%)
Query 3 GPLKLEHLLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSKE 62
GP + L G+ D+ L+E G HTVE VA+AP K LL IKGISE KA K+ + +
Sbjct 18 GPQAISRLEQCGINANDVKKLEEAGFHTVEAVAYAPKKELLNIKGISEAKAEKILAEAAK 77
Query 63 LCSLGFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTL 122
L +GF +A E+ + R+ +I+ +TGS +LD LL+GG+ETG++TE+FGEFRTGKTQLCHTL
Sbjct 78 LVPMGFTTATEFHQRRSEIIQISTGSKELDKLLQGGVETGSITEMFGEFRTGKTQLCHTL 137
Query 123 AVSCQLPVEQSGGEGKCLWIDTEGTFRPERIV 154
AV+CQLP+++ GGEGK ++IDTEGTFRPER++
Sbjct 138 AVTCQLPIDRGGGEGKAMYIDTEGTFRPERLL 169
> mmu:19361 Rad51, AV304093, Rad51a, Reca; RAD51 homolog (S. cerevisiae);
K04482 DNA repair protein RAD51
Length=339
Score = 187 bits (474), Expect = 2e-47, Method: Compositional matrix adjust.
Identities = 86/152 (56%), Positives = 116/152 (76%), Gaps = 0/152 (0%)
Query 3 GPLKLEHLLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSKE 62
GP + L G+ D+ L+E G HTVE VA+AP K L+ IKGISE KA K+ + +
Sbjct 21 GPQPISRLEQCGINANDVKKLEEAGYHTVEAVAYAPKKELINIKGISEAKADKILTEAAK 80
Query 63 LCSLGFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTL 122
L +GF +A E+ + R+ +I+ TTGS +LD LL+GGIETG++TE+FGEFRTGKTQ+CHTL
Sbjct 81 LVPMGFTTATEFHQRRSEIIQITTGSKELDKLLQGGIETGSITEMFGEFRTGKTQICHTL 140
Query 123 AVSCQLPVEQSGGEGKCLWIDTEGTFRPERIV 154
AV+CQLP+++ GGEGK ++IDTEGTFRPER++
Sbjct 141 AVTCQLPIDRGGGEGKAMYIDTEGTFRPERLL 172
> xla:380251 rad51-b, MGC130792, MGC52570, brcc5, rad51, rad51a,
reca, xrad51; RAD51 homolog (RecA homolog); K04482 DNA repair
protein RAD51
Length=336
Score = 184 bits (468), Expect = 7e-47, Method: Compositional matrix adjust.
Identities = 86/152 (56%), Positives = 115/152 (75%), Gaps = 0/152 (0%)
Query 3 GPLKLEHLLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSKE 62
GP + L G+ D+ L++ G HTVE VA+AP K LL IKGISE KA K+ + +
Sbjct 18 GPQAITRLEQCGINANDVKKLEDAGFHTVEAVAYAPKKELLNIKGISEAKAEKILAEAAK 77
Query 63 LCSLGFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTL 122
L +GF +A E+ + R+ +I+ TGS +LD LL+GGIETG++TE+FGEFRTGKTQLCHTL
Sbjct 78 LVPMGFTTATEFHQRRSEIIQIGTGSKELDKLLQGGIETGSITEMFGEFRTGKTQLCHTL 137
Query 123 AVSCQLPVEQSGGEGKCLWIDTEGTFRPERIV 154
AV+CQLP+++ GGEGK ++IDTEGTFRPER++
Sbjct 138 AVTCQLPIDRGGGEGKAMYIDTEGTFRPERLL 169
> sce:YER095W RAD51, MUT5; Rad51p; K04482 DNA repair protein RAD51
Length=400
Score = 183 bits (465), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 89/150 (59%), Positives = 112/150 (74%), Gaps = 0/150 (0%)
Query 5 LKLEHLLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSKELC 64
+ +E L G+T D+ L+E GLHT E VA+AP K LL IKGISE KA KL + L
Sbjct 81 VPIEKLQVNGITMADVKKLRESGLHTAEAVAYAPRKDLLEIKGISEAKADKLLNEAARLV 140
Query 65 SLGFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAV 124
+GF +A ++ R+ LI TTGS LD+LL GG+ETG++TELFGEFRTGK+QLCHTLAV
Sbjct 141 PMGFVTAADFHMRRSELICLTTGSKNLDTLLGGGVETGSITELFGEFRTGKSQLCHTLAV 200
Query 125 SCQLPVEQSGGEGKCLWIDTEGTFRPERIV 154
+CQ+P++ GGEGKCL+IDTEGTFRP R+V
Sbjct 201 TCQIPLDIGGGEGKCLYIDTEGTFRPVRLV 230
> ath:AT5G20850 ATRAD51; ATP binding / DNA binding / DNA-dependent
ATPase/ damaged DNA binding / nucleoside-triphosphatase/
nucleotide binding / protein binding / sequence-specific DNA
binding; K04482 DNA repair protein RAD51
Length=342
Score = 177 bits (448), Expect = 2e-44, Method: Compositional matrix adjust.
Identities = 83/154 (53%), Positives = 113/154 (73%), Gaps = 0/154 (0%)
Query 1 QTGPLKLEHLLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVS 60
Q GP +E L A G+ D+ L++ GL TVE VA+ P K LL IKGIS+ K K+ + +
Sbjct 22 QHGPFPVEQLQAAGIASVDVKKLRDAGLCTVEGVAYTPRKDLLQIKGISDAKVDKIVEAA 81
Query 61 KELCSLGFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCH 120
+L LGF SA + R +I+ T+GS +LD +L+GGIETG++TEL+GEFR+GKTQLCH
Sbjct 82 SKLVPLGFTSASQLHAQRQEIIQITSGSRELDKVLEGGIETGSITELYGEFRSGKTQLCH 141
Query 121 TLAVSCQLPVEQSGGEGKCLWIDTEGTFRPERIV 154
TL V+CQLP++Q GGEGK ++ID EGTFRP+R++
Sbjct 142 TLCVTCQLPMDQGGGEGKAMYIDAEGTFRPQRLL 175
> cel:Y43C5A.6 rad-51; RADiation sensitivity abnormal/yeast RAD-related
family member (rad-51); K04482 DNA repair protein
RAD51
Length=395
Score = 158 bits (400), Expect = 6e-39, Method: Compositional matrix adjust.
Identities = 71/132 (53%), Positives = 96/132 (72%), Gaps = 0/132 (0%)
Query 23 LKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSKELCSLGFCSAQEYLEARANLI 82
LKE G +T E +AF + L +KGIS+QKA K+ + + + +GF + E R+ L+
Sbjct 95 LKEAGYYTYESLAFTTRRELRNVKGISDQKAEKIMKEAMKFVQMGFTTGAEVHVKRSQLV 154
Query 83 KFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGKCLWI 142
+ TGS LD LL GGIETG++TE++GE+RTGKTQLCH+LAV CQLP++ GGEGKC++I
Sbjct 155 QIRTGSASLDRLLGGGIETGSITEVYGEYRTGKTQLCHSLAVLCQLPIDMGGGEGKCMYI 214
Query 143 DTEGTFRPERIV 154
DT TFRPERI+
Sbjct 215 DTNATFRPERII 226
> hsa:5888 RAD51, BRCC5, HRAD51, HsRad51, HsT16930, RAD51A, RECA;
RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae); K04482
DNA repair protein RAD51
Length=340
Score = 149 bits (375), Expect = 5e-36, Method: Compositional matrix adjust.
Identities = 80/163 (49%), Positives = 106/163 (65%), Gaps = 21/163 (12%)
Query 3 GPLKLEHLLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKL----KQ 58
GP + L G+ D+ L+E G HTVE VA+AP K L+ IKGISE KA K+ +
Sbjct 21 GPQPISRLEQCGINANDVKKLEEAGFHTVEAVAYAPKKELINIKGISEAKADKILTESRS 80
Query 59 VSKELCS---LGFC----SAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEF 111
V++ C+ L +C S A A+ + TTG GIETG++TE+FGEF
Sbjct 81 VARLECNSVILVYCTLRLSGSSDSPASASRVVGTTG----------GIETGSITEMFGEF 130
Query 112 RTGKTQLCHTLAVSCQLPVEQSGGEGKCLWIDTEGTFRPERIV 154
RTGKTQ+CHTLAV+CQLP+++ GGEGK ++IDTEGTFRPER++
Sbjct 131 RTGKTQICHTLAVTCQLPIDRGGGEGKAMYIDTEGTFRPERLL 173
> sce:YER179W DMC1, ISC2; Dmc1p; K10872 meiotic recombination
protein DMC1
Length=334
Score = 144 bits (363), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 74/149 (49%), Positives = 97/149 (65%), Gaps = 0/149 (0%)
Query 5 LKLEHLLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSKELC 64
L ++ L G+ DL LK GG++TV V + L IKG+SE K K+K+ + ++
Sbjct 17 LSVDELQNYGINASDLQKLKSGGIYTVNTVLSTTRRHLCKIKGLSEVKVEKIKEAAGKII 76
Query 65 SLGFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAV 124
+GF A L+ R + +TGS QLDS+L GGI T ++TE+FGEFR GKTQ+ HTL V
Sbjct 77 QVGFIPATVQLDIRQRVYSLSTGSKQLDSILGGGIMTMSITEVFGEFRCGKTQMSHTLCV 136
Query 125 SCQLPVEQSGGEGKCLWIDTEGTFRPERI 153
+ QLP E GGEGK +IDTEGTFRPERI
Sbjct 137 TTQLPREMGGGEGKVAYIDTEGTFRPERI 165
> ath:AT3G22880 DMC1; DMC1 (DISRUPTION OF MEIOTIC CONTROL 1);
ATP binding / DNA binding / DNA-dependent ATPase/ damaged DNA
binding / nucleoside-triphosphatase/ nucleotide binding /
protein binding; K10872 meiotic recombination protein DMC1
Length=344
Score = 142 bits (359), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 69/148 (46%), Positives = 97/148 (65%), Gaps = 0/148 (0%)
Query 7 LEHLLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSKELCSL 66
++ L+A+G+ D+ L+E G+HT + K L IKG+SE K K+ + ++++ +
Sbjct 31 IDKLIAQGINAGDVKKLQEAGIHTCNGLMMHTKKNLTGIKGLSEAKVDKICEAAEKIVNF 90
Query 67 GFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSC 126
G+ + + L R +++K TTG LD LL GGIET +TE FGEFR+GKTQL HTL V+
Sbjct 91 GYMTGSDALIKRKSVVKITTGCQALDDLLGGGIETSAITEAFGEFRSGKTQLAHTLCVTT 150
Query 127 QLPVEQSGGEGKCLWIDTEGTFRPERIV 154
QLP GG GK +IDTEGTFRP+RIV
Sbjct 151 QLPTNMKGGNGKVAYIDTEGTFRPDRIV 178
> cpv:cgd7_1690 meiotic recombination protein DMC1-like protein
; K10872 meiotic recombination protein DMC1
Length=342
Score = 136 bits (342), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 72/151 (47%), Positives = 100/151 (66%), Gaps = 1/151 (0%)
Query 5 LKLEHLLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSKELC 64
++++ L + G+ D++ LK GL TV + A K L IKG+SE K K+ + +++L
Sbjct 25 VEIDKLQSAGINVADINKLKTAGLCTVLSIIQATKKELCNIKGLSEAKVEKIVEAAQKLD 84
Query 65 -SLGFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLA 123
S F S E + R N+++ TTGS Q D +L GG E+ +TE+FGE R GKTQ+CHTL
Sbjct 85 QSSSFQSGSEVMSRRQNILRITTGSEQFDKMLMGGFESMCITEIFGENRCGKTQICHTLC 144
Query 124 VSCQLPVEQSGGEGKCLWIDTEGTFRPERIV 154
V+ QLP+E +GG GK +IDTEGTFRPERIV
Sbjct 145 VAAQLPLEMNGGNGKVCFIDTEGTFRPERIV 175
> tgo:TGME49_016400 meiotic recombination protein DMC1-like protein,
putative (EC:2.7.11.1); K10872 meiotic recombination
protein DMC1
Length=349
Score = 130 bits (328), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 73/151 (48%), Positives = 97/151 (64%), Gaps = 7/151 (4%)
Query 7 LEHLLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSKELCSL 66
++ L A G+ D++ LK+ G TV + K L +KGISE AK++++ + L
Sbjct 34 IDKLQAAGINAADINKLKQAGYCTVLSIVQTTKKELCLVKGISE---AKVEKIVEAAAKL 90
Query 67 GFCSA----QEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTL 122
G C+A E ++ R +IK TTGS QLD LL GG ET ++TELFGE R GKTQLCHT+
Sbjct 91 GMCNAFITGVELVQKRGRVIKITTGSDQLDQLLGGGFETMSITELFGENRCGKTQLCHTV 150
Query 123 AVSCQLPVEQSGGEGKCLWIDTEGTFRPERI 153
V+ QLP + GG GK +IDTEGTFRPE+I
Sbjct 151 CVTAQLPRDMKGGCGKVCYIDTEGTFRPEKI 181
> dre:553953 dmc1, MGC136628, zgc:136628; DMC1 dosage suppressor
of mck1 homolog, meiosis-specific homologous recombination
(yeast); K10872 meiotic recombination protein DMC1
Length=342
Score = 127 bits (318), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 68/147 (46%), Positives = 92/147 (62%), Gaps = 0/147 (0%)
Query 7 LEHLLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSKELCSL 66
+E L G+ D+ LK G+ TV+ + + L IKG+SE K K+K+ + +L +
Sbjct 26 IELLQKHGINVADIKKLKSVGICTVKGIQMTTRRALCNIKGLSEAKVDKIKEAAGKLLTC 85
Query 67 GFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSC 126
GF +A EY R + TTGS++ D LL GG+E+ +TE FGEFRTGKTQL HTL V+
Sbjct 86 GFQTASEYCIKRKQVFHITTGSLEFDKLLGGGVESMAITEAFGEFRTGKTQLSHTLCVTA 145
Query 127 QLPVEQSGGEGKCLWIDTEGTFRPERI 153
QLP E GK ++IDTE TFRPER+
Sbjct 146 QLPGEYGYTGGKVIFIDTENTFRPERL 172
> pfa:MAL8P1.76 meiotic recombination protein dmc1-like protein;
K10872 meiotic recombination protein DMC1
Length=347
Score = 124 bits (312), Expect = 9e-29, Method: Compositional matrix adjust.
Identities = 69/150 (46%), Positives = 96/150 (64%), Gaps = 3/150 (2%)
Query 6 KLEHLLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSKEL-- 63
++E L G+ D++ LK G T+ + K L +KGISE K K+ +V+ ++
Sbjct 31 EIEKLQDLGINAADINKLKGSGYCTILSLIQTTKKELCNVKGISEAKVDKILEVASKIEN 90
Query 64 CSLGFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLA 123
CS F +A E ++ R+ ++K TTGS D L GGIE+ +TELFGE R GKTQ+CHTLA
Sbjct 91 CS-SFITANELVQKRSKVLKITTGSTVFDQTLGGGIESMCITELFGENRCGKTQVCHTLA 149
Query 124 VSCQLPVEQSGGEGKCLWIDTEGTFRPERI 153
V+ QLP +GG GK +IDTEGTFRPE++
Sbjct 150 VTAQLPKSLNGGNGKVCYIDTEGTFRPEKV 179
> hsa:11144 DMC1, DMC1H, HsLim15, LIM15, MGC150472, MGC150473,
dJ199H16.1; DMC1 dosage suppressor of mck1 homolog, meiosis-specific
homologous recombination (yeast); K10872 meiotic recombination
protein DMC1
Length=340
Score = 122 bits (307), Expect = 4e-28, Method: Compositional matrix adjust.
Identities = 64/140 (45%), Positives = 87/140 (62%), Gaps = 0/140 (0%)
Query 14 GLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSKELCSLGFCSAQE 73
G+ D+ LK G+ T++ + + L +KG+SE K K+K+ + +L GF +A E
Sbjct 31 GINVADIKKLKSVGICTIKGIQMTTRRALCNVKGLSEAKVDKIKEAANKLIEPGFLTAFE 90
Query 74 YLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQS 133
Y E R + TTGS + D LL GGIE+ +TE FGEFRTGKTQL HTL V+ QLP
Sbjct 91 YSEKRKMVFHITTGSQEFDKLLGGGIESMAITEAFGEFRTGKTQLSHTLCVTAQLPGAGG 150
Query 134 GGEGKCLWIDTEGTFRPERI 153
GK ++IDTE TFRP+R+
Sbjct 151 YPGGKIIFIDTENTFRPDRL 170
> mmu:13404 Dmc1, Dmc1h, MGC151144, Mei11, sgdp; DMC1 dosage suppressor
of mck1 homolog, meiosis-specific homologous recombination
(yeast); K10872 meiotic recombination protein DMC1
Length=340
Score = 122 bits (306), Expect = 5e-28, Method: Compositional matrix adjust.
Identities = 63/140 (45%), Positives = 87/140 (62%), Gaps = 0/140 (0%)
Query 14 GLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSKELCSLGFCSAQE 73
G+ D+ LK G+ T++ + + L +KG+SE K K+K+ + +L GF +A +
Sbjct 31 GINMADIKKLKSVGICTIKGIQMTTRRALCNVKGLSEAKVEKIKEAANKLIEPGFLTAFQ 90
Query 74 YLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQS 133
Y E R + TTGS + D LL GGIE+ +TE FGEFRTGKTQL HTL V+ QLP
Sbjct 91 YSERRKMVFHITTGSQEFDKLLGGGIESMAITEAFGEFRTGKTQLSHTLCVTAQLPGTGG 150
Query 134 GGEGKCLWIDTEGTFRPERI 153
GK ++IDTE TFRP+R+
Sbjct 151 YSGGKIIFIDTENTFRPDRL 170
> tpv:TP04_0170 meiotic recombination protein DMC1; K04482 DNA
repair protein RAD51
Length=346
Score = 110 bits (274), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 66/150 (44%), Positives = 91/150 (60%), Gaps = 7/150 (4%)
Query 7 LEHLLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSKEL-CS 65
+E L G+ D++ LK G+ TV V K L IKG++E K K+ + +L +
Sbjct 33 IERLEELGINVTDINKLKAAGICTVLGVIQTTKKDLCNIKGLTELKVDKISDCASKLEVT 92
Query 66 LGFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVS 125
F SA E + R +++K TGS L+ LL GGIET ++TELFGE RTGKTQ+CHT++V+
Sbjct 93 NSFISASELYKIRKSILKINTGSEMLNRLLNGGIETMSITELFGENRTGKTQICHTISVT 152
Query 126 CQL--PVEQSGGEGKCLWIDTEGTFRPERI 153
Q+ P E K +IDTE TFRPE+I
Sbjct 153 SQIINPTEP----FKVCYIDTENTFRPEKI 178
> sce:YDR004W RAD57; Protein that stimulates strand exchange by
stabilizing the binding of Rad51p to single-stranded DNA;
involved in the recombinational repair of double-strand breaks
in DNA during vegetative growth and meiosis; forms heterodimer
with Rad55p; K10958 DNA repair protein RAD57
Length=460
Score = 65.5 bits (158), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 35/70 (50%), Positives = 44/70 (62%), Gaps = 0/70 (0%)
Query 84 FTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGKCLWID 143
FTT V +D LL GGI T +TE+FGE TGK+QL LA+S QL G GKC++I
Sbjct 100 FTTADVAMDELLGGGIFTHGITEIFGESSTGKSQLLMQLALSVQLSEPAGGLGGKCVYIT 159
Query 144 TEGTFRPERI 153
TEG +R+
Sbjct 160 TEGDLPTQRL 169
> dre:406721 rad51l1, wu:fd07f04, zgc:56581; RAD51-like 1 (S.
cerevisiae); K10869 RAD51-like protein 1
Length=373
Score = 63.2 bits (152), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 47/145 (32%), Positives = 70/145 (48%), Gaps = 1/145 (0%)
Query 10 LLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSKELCSLGFC 69
L G++ + LK L T + V L + G+S A L+++ + C+
Sbjct 6 LRRSGVSADLCERLKRHQLETCQDVLSVTQVELSRLAGLSYPAALNLQRLVSKACAPAVI 65
Query 70 SAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLP 129
+A + + + L F+T LD LL GG+ G LTE+ G GKTQLC L+V LP
Sbjct 66 TALDLWKRKEELC-FSTSLPALDRLLHGGLPRGALTEVTGPSGCGKTQLCMMLSVLATLP 124
Query 130 VEQSGGEGKCLWIDTEGTFRPERIV 154
G + ++IDTE F ER+V
Sbjct 125 KSLGGLDSGVIYIDTESAFSAERLV 149
> dre:450081 rad51c, zgc:101596; rad51 homolog C (S. cerevisiae);
K10870 RAD51-like protein 2
Length=362
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 41/120 (34%), Positives = 64/120 (53%), Gaps = 19/120 (15%)
Query 47 GISEQKAAKLKQVSKE-------------LCSLGFCSAQEYLEARANLIKFTTGSVQLDS 93
GIS+++A +L Q+ ++ + +L ++ L +++ F +G LD
Sbjct 42 GISQEEAVELLQMLRDDAQPQQQRAAADGVTALDLLHQEQTL---GSIVTFCSG---LDD 95
Query 94 LLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGKCLWIDTEGTFRPERI 153
+ GG+ G TE+ G GKTQLC LAV Q+PV G GK L+IDTEG+F +R+
Sbjct 96 AIGGGVPVGKTTEICGAPGVGKTQLCMQLAVDVQIPVFFGGLGGKALYIDTEGSFLVQRV 155
> ath:AT2G45280 ATRAD51C; ATP binding / damaged DNA binding /
protein binding / recombinase/ single-stranded DNA binding
Length=363
Score = 61.2 bits (147), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 40/113 (35%), Positives = 64/113 (56%), Gaps = 7/113 (6%)
Query 47 GISEQKAAKLKQVSKELCSLGFCS-------AQEYLEARANLIKFTTGSVQLDSLLKGGI 99
I+E++A ++ +++ + C G S A + L +L + TT LD++L GGI
Sbjct 61 NITEEEAFEILKLANQSCCNGSRSLINGAKNAWDMLHEEESLPRITTSCSDLDNILGGGI 120
Query 100 ETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGKCLWIDTEGTFRPER 152
++TE+ G GKTQ+ L+V+ Q+P E G GK ++IDTEG+F ER
Sbjct 121 SCRDVTEIGGVPGIGKTQIGIQLSVNVQIPRECGGLGGKAIYIDTEGSFMVER 173
> mmu:19363 Rad51l1, AI553500, R51H2, Rad51b, mREC2; RAD51-like
1 (S. cerevisiae); K10869 RAD51-like protein 1
Length=350
Score = 60.8 bits (146), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 40/124 (32%), Positives = 63/124 (50%), Gaps = 4/124 (3%)
Query 34 VAFAPLKTLLAIKGISEQKAAKLKQVSKELCSLGFCSAQEYLEARANLIK---FTTGSVQ 90
++ +PL+ L+ + G+S + +L + C+ +A E R+ + +T
Sbjct 31 LSLSPLE-LMKVTGLSYRGVHELLHTVSKACAPQMQTAYELKTRRSAHLSPAFLSTTLCA 89
Query 91 LDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGKCLWIDTEGTFRP 150
LD L GG+ G+LTE+ G GKTQ C ++V LP G EG ++IDTE F
Sbjct 90 LDEALHGGVPCGSLTEITGPPGCGKTQFCIMMSVLATLPTSLGGLEGAVVYIDTESAFTA 149
Query 151 ERIV 154
ER+V
Sbjct 150 ERLV 153
> mmu:114714 Rad51c, R51H3, Rad51l2; RAD51 homolog c (S. cerevisiae);
K10870 RAD51-like protein 2
Length=366
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 36/86 (41%), Positives = 47/86 (54%), Gaps = 0/86 (0%)
Query 69 CSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQL 128
C+A E LE T LD++L GGI TE+ G GKTQLC LAV Q+
Sbjct 76 CTALELLEQEHTQGFIITFCSALDNILGGGIPLMKTTEVCGVPGVGKTQLCMQLAVDVQI 135
Query 129 PVEQSGGEGKCLWIDTEGTFRPERIV 154
P G G+ ++IDTEG+F +R+V
Sbjct 136 PECFGGVAGEAVFIDTEGSFMVDRVV 161
> hsa:5890 RAD51L1, MGC34245, R51H2, RAD51B, REC2, hREC2; RAD51-like
1 (S. cerevisiae); K10869 RAD51-like protein 1
Length=350
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 28/64 (43%), Positives = 37/64 (57%), Gaps = 0/64 (0%)
Query 91 LDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGKCLWIDTEGTFRP 150
LD L GG+ G+LTE+ G GKTQ C +++ LP G EG ++IDTE F
Sbjct 90 LDEALHGGVACGSLTEITGPPGCGKTQFCIMMSILATLPTNMGGLEGAVVYIDTESAFSA 149
Query 151 ERIV 154
ER+V
Sbjct 150 ERLV 153
> ath:AT2G28560 RAD51B; RAD51B; recombinase; K10869 RAD51-like
protein 1
Length=370
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 28/64 (43%), Positives = 36/64 (56%), Gaps = 0/64 (0%)
Query 91 LDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGKCLWIDTEGTFRP 150
LD L GGI G LTEL G GK+Q C LA+S PV G +G+ ++ID E F
Sbjct 91 LDDTLCGGIPFGVLTELVGPPGIGKSQFCMKLALSASFPVAYGGLDGRVIYIDVESKFSS 150
Query 151 ERIV 154
R++
Sbjct 151 RRVI 154
> ath:AT5G57450 XRCC3; XRCC3; ATP binding / damaged DNA binding
/ protein binding / single-stranded DNA binding; K10880 DNA-repair
protein XRCC3
Length=304
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 30/71 (42%), Positives = 40/71 (56%), Gaps = 0/71 (0%)
Query 83 KFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGKCLWI 142
K TTG LD L+GGI +LTE+ E GKTQLC L++ QLP+ G G L++
Sbjct 20 KLTTGCEILDGCLRGGISCDSLTEIVAESGCGKTQLCLQLSLCTQLPISHGGLNGSSLYL 79
Query 143 DTEGTFRPERI 153
+E F R+
Sbjct 80 HSEFPFPFRRL 90
> mmu:19364 Rad51l3, DKFZp586D0122, R51H3, Rad51d, Trad-d2, Trad-d3,
Trad-d4, Trad-d6, Trad-d7; RAD51-like 3 (S. cerevisiae);
K10871 RAD51-like protein 3
Length=329
Score = 53.1 bits (126), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 41/145 (28%), Positives = 73/145 (50%), Gaps = 6/145 (4%)
Query 11 LAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVS-KELCSLGFC 69
L GLT++ + LL+ + TV +A A L+ + G+S + L++V + +
Sbjct 8 LCPGLTEETVQLLRGRKIKTVADLAAADLEEVAQKCGLSYKALVALRRVLLAQFSAFPLN 67
Query 70 SAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLP 129
A Y E + + +TG LD LL G+ TG +TE+ G +GKTQ+C +A +
Sbjct 68 GADLYEELKTSTAILSTGIGSLDKLLDAGLYTGEVTEIVGGPGSGKTQVCLCVAANVAHS 127
Query 130 VEQSGGEGKCLWIDTEGTFRPERIV 154
++Q+ L++D+ G R++
Sbjct 128 LQQN-----VLYVDSNGGMTASRLL 147
> xla:444788 rad51l3, MGC82048; RAD51-like 3; K10871 RAD51-like
protein 3
Length=324
Score = 52.8 bits (125), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 35/115 (30%), Positives = 60/115 (52%), Gaps = 15/115 (13%)
Query 40 KTLLAIKGISEQKAAKLKQVSKELCSLGFCSAQEYLEARANLIKFTTGSVQLDSLLKGGI 99
KTL+A++ + L Q S + F A Y E +++ T + +LD LL G+
Sbjct 48 KTLMAVRRV------LLAQYS----AFPFSGADVYEELKSSTAILPTANRKLDILLDSGL 97
Query 100 ETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGKCLWIDTEGTFRPERIV 154
TG +TE+ G +GKTQ+C ++AV+ ++Q+ L++DT G R++
Sbjct 98 YTGEVTEIAGAAGSGKTQMCQSIAVNVAYSLKQT-----VLYVDTTGGLTASRLL 147
> ath:AT1G07745 RAD51D; RAD51D (ARABIDOPSIS HOMOLOG OF RAD51 D);
ATP binding / DNA binding / DNA-dependent ATPase
Length=304
Score = 50.1 bits (118), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 28/70 (40%), Positives = 36/70 (51%), Gaps = 5/70 (7%)
Query 84 FTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGKCLWID 143
+TG + DSLL+GG G LTEL G +GKTQ C A S G+ L++D
Sbjct 71 LSTGDKETDSLLQGGFREGQLTELVGPSSSGKTQFCMQAAASV-----AENHLGRVLYLD 125
Query 144 TEGTFRPERI 153
T +F RI
Sbjct 126 TGNSFSARRI 135
> dre:541414 xrcc3, im:7142103, si:dkey-11b8.1, zgc:101608; X-ray
repair complementing defective repair in Chinese hamster
cells 3; K10880 DNA-repair protein XRCC3
Length=352
Score = 49.7 bits (117), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 26/63 (41%), Positives = 36/63 (57%), Gaps = 0/63 (0%)
Query 91 LDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGKCLWIDTEGTFRP 150
LD L++GG+ +TEL GE GKTQ C L +S Q P E G ++I TE +F
Sbjct 89 LDGLMRGGLPLRGITELAGESAAGKTQFCLQLCLSVQYPQEHGGLNSGAVYICTEDSFPI 148
Query 151 ERI 153
+R+
Sbjct 149 KRL 151
> xla:379577 xrcc3, MGC69118; X-ray repair complementing defective
repair in Chinese hamster cells 3; K10880 DNA-repair protein
XRCC3
Length=350
Score = 49.3 bits (116), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 33/92 (35%), Positives = 47/92 (51%), Gaps = 3/92 (3%)
Query 65 SLGFCSAQEYLEAR---ANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHT 121
+LG + Q Y E + K + G LD+ L+GGI +TE+ GE GKTQ+
Sbjct 60 NLGVTALQMYSEKAKFPSQHQKLSLGCKVLDNFLRGGIPLVGITEIAGESSAGKTQIGLQ 119
Query 122 LAVSCQLPVEQSGGEGKCLWIDTEGTFRPERI 153
L +S Q PVE G ++I TE F +R+
Sbjct 120 LCLSVQYPVEYGGLASGAVYICTEDAFPSKRL 151
> dre:404608 MGC77165; zgc:77165; K10871 RAD51-like protein 3
Length=327
Score = 48.9 bits (115), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 30/84 (35%), Positives = 48/84 (57%), Gaps = 5/84 (5%)
Query 71 AQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPV 130
A Y E ++ +TGS LD LL G+ TG +TEL G +GKTQ+C ++AV+ +
Sbjct 69 ADLYEELLSSTAILSTGSPSLDKLLDSGLYTGEITELTGSPGSGKTQVCFSVAVNISHQL 128
Query 131 EQSGGEGKCLWIDTEGTFRPERIV 154
+Q+ ++IDT+G R++
Sbjct 129 KQT-----VVYIDTKGGMCANRLL 147
> hsa:7517 XRCC3, CMM6; X-ray repair complementing defective repair
in Chinese hamster cells 3; K10880 DNA-repair protein
XRCC3
Length=346
Score = 48.9 bits (115), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 26/71 (36%), Positives = 39/71 (54%), Gaps = 0/71 (0%)
Query 83 KFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGKCLWI 142
+ + G LD+LL+GG+ +TEL G GKTQL L ++ Q P + G E ++I
Sbjct 81 RLSLGCPVLDALLRGGLPLDGITELAGRSSAGKTQLALQLCLAVQFPRQHGGLEAGAVYI 140
Query 143 DTEGTFRPERI 153
TE F +R+
Sbjct 141 CTEDAFPHKRL 151
> hsa:5892 RAD51L3, R51H3, RAD51D, TRAD; RAD51-like 3 (S. cerevisiae);
K10871 RAD51-like protein 3
Length=348
Score = 39.7 bits (91), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 37/65 (56%), Gaps = 5/65 (7%)
Query 90 QLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGKCLWIDTEGTFR 149
+LD LL G+ TG +TE+ G +GKTQ+C +A + ++Q+ L++D+ G
Sbjct 108 RLDKLLDAGLYTGEVTEIVGGPGSGKTQVCLCMAANVAHGLQQN-----VLYVDSNGGLT 162
Query 150 PERIV 154
R++
Sbjct 163 ASRLL 167
> mmu:74335 Xrcc3, 4432412E01Rik, AI182522, AW537713; X-ray repair
complementing defective repair in Chinese hamster cells
3; K10880 DNA-repair protein XRCC3
Length=349
Score = 39.3 bits (90), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 35/71 (49%), Gaps = 0/71 (0%)
Query 83 KFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGKCLWI 142
+ + G LD L GG+ +T L G GKTQL L ++ Q P + G E ++I
Sbjct 81 RLSLGCPVLDQFLGGGLPLEGITGLAGCSSAGKTQLALQLCLAVQFPRQYGGLEAGAVYI 140
Query 143 DTEGTFRPERI 153
TE F +R+
Sbjct 141 CTEDAFPSKRL 151
> ath:AT1G79050 DNA repair protein recA
Length=343
Score = 38.1 bits (87), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 36/67 (53%), Gaps = 6/67 (8%)
Query 84 FTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGKCLWID 143
F++G + LD L GG+ G + E++G +GKT TLA+ V++ G G + +D
Sbjct 117 FSSGILTLDLALGGGLPKGRVVEIYGPESSGKT----TLALHAIAEVQKLG--GNAMLVD 170
Query 144 TEGTFRP 150
E F P
Sbjct 171 AEHAFDP 177
> hsa:7516 XRCC2, DKFZp781P0919; X-ray repair complementing defective
repair in Chinese hamster cells 2; K10879 DNA-repair
protein XRCC2
Length=280
Score = 37.7 bits (86), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 30/53 (56%), Gaps = 0/53 (0%)
Query 102 GNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGKCLWIDTEGTFRPERIV 154
G++ E G TGKT++ + L C LP + G E + L+IDT+ F R+V
Sbjct 41 GDILEFHGPEGTGKTEMLYHLTARCILPKSEGGLEVEVLFIDTDYHFDMLRLV 93
> eco:b2699 recA, ECK2694, JW2669, lexB, recH, rnmB, srf, tif,
umuB, umuR, zab; DNA strand exchange and recombination protein
with protease and nuclease activity; K03553 recombination
protein RecA
Length=353
Score = 37.0 bits (84), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 33/101 (32%), Positives = 49/101 (48%), Gaps = 13/101 (12%)
Query 53 AAKLKQVSKELCSLGFCSAQEYLEARA-NLIKFTTGSVQLD-SLLKGGIETGNLTELFGE 110
AA L Q+ K+ G S E R+ ++ +TGS+ LD +L GG+ G + E++G
Sbjct 12 AAALGQIEKQF---GKGSIMRLGEDRSMDVETISTGSLSLDIALGAGGLPMGRIVEIYGP 68
Query 111 FRTGKTQLCHTLAVSCQLPVEQSGGEGK-CLWIDTEGTFRP 150
+GKT L + + Q EGK C +ID E P
Sbjct 69 ESSGKTTLTLQVIAAAQR-------EGKTCAFIDAEHALDP 102
> ath:AT3G10140 RECA3; RECA3 (recA homolog 3); ATP binding / DNA
binding / DNA-dependent ATPase/ nucleoside-triphosphatase/
nucleotide binding
Length=389
Score = 36.6 bits (83), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 43/78 (55%), Gaps = 9/78 (11%)
Query 74 YLEARANLIKFTTGSVQLD-SLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQ 132
Y + R ++I +TGS+ LD +L GG+ G + E++G+ +GKT TLA+ ++
Sbjct 89 YRKRRVSVI--STGSLNLDLALGVGGLPKGRMVEVYGKEASGKT----TLALHIIKEAQK 142
Query 133 SGGEGKCLWIDTEGTFRP 150
G G C ++D E P
Sbjct 143 LG--GYCAYLDAENAMDP 158
> ath:AT2G19490 recA family protein; K03553 recombination protein
RecA
Length=430
Score = 36.6 bits (83), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 24/67 (35%), Positives = 37/67 (55%), Gaps = 7/67 (10%)
Query 80 NLIKFTTGSVQLDSLLK-GGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGK 138
N+ F+TGS LD L GG+ G + E++G +GKT TLA+ ++ G G
Sbjct 89 NVPVFSTGSFALDVALGVGGLPKGRVVEIYGPEASGKT----TLALHVIAEAQKQG--GT 142
Query 139 CLWIDTE 145
C+++D E
Sbjct 143 CVFVDAE 149
> mmu:57434 Xrcc2, 4921524O04Rik, 8030409M04Rik, RAD51, RecA;
X-ray repair complementing defective repair in Chinese hamster
cells 2; K10879 DNA-repair protein XRCC2
Length=278
Score = 36.6 bits (83), Expect = 0.036, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 30/53 (56%), Gaps = 0/53 (0%)
Query 102 GNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGKCLWIDTEGTFRPERIV 154
G++ E G TGKT++ + L C LP + G + + L+IDT+ F R+V
Sbjct 41 GDIFEFHGPEGTGKTEMLYHLTARCILPKSEGGLQIEVLFIDTDYHFDMLRLV 93
> cpv:cgd2_4070 hypothetical protein
Length=304
Score = 35.4 bits (80), Expect = 0.066, Method: Compositional matrix adjust.
Identities = 21/46 (45%), Positives = 25/46 (54%), Gaps = 0/46 (0%)
Query 83 KFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQL 128
+ +TGS +D GGI L E+ GE TGKTQ C TL S L
Sbjct 37 RLSTGSNVVDKAFNGGIPKRILFEITGEAGTGKTQWCLTLITSVLL 82
> tpv:TP04_0453 hypothetical protein
Length=286
Score = 35.0 bits (79), Expect = 0.088, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 33/67 (49%), Gaps = 0/67 (0%)
Query 87 GSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGKCLWIDTEG 146
G ++D L GG+ G + E++G +GKTQ +L + + L+I T G
Sbjct 31 GVKEIDQALNGGLLLGKVCEIYGPSGSGKTQFALSLTSEVLINNLIHSKDYVVLYIYTNG 90
Query 147 TFRPERI 153
TF ER+
Sbjct 91 TFPIERL 97
> ath:AT3G32920 ATP binding / DNA binding / DNA-dependent ATPase/
nucleoside-triphosphatase/ nucleotide binding
Length=226
Score = 33.1 bits (74), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 20/47 (42%), Positives = 28/47 (59%), Gaps = 2/47 (4%)
Query 80 NLIKFTTGSVQLDSLLK-GGIETGNLTELFGEFRTGKTQLC-HTLAV 124
N+ F+TGS LD L GG+ G L E++G +GKT L H L++
Sbjct 11 NVPVFSTGSFALDVALGVGGLPKGRLVEIYGPEASGKTALALHMLSM 57
> eco:b3169 nusA, ECK3158, JW3138; transcription termination/antitermination
L factor; K02600 N utilization substance protein
A
Length=495
Score = 32.7 bits (73), Expect = 0.46, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 25/40 (62%), Gaps = 0/40 (0%)
Query 22 LLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSK 61
+L E G T+E +A+ P+K LL I+G+ E L++ +K
Sbjct 372 VLVEEGFSTLEELAYVPMKELLEIEGLDEPTVEALRERAK 411
Lambda K H
0.317 0.136 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3321543300
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40