bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0065_orf2
Length=126
Score E
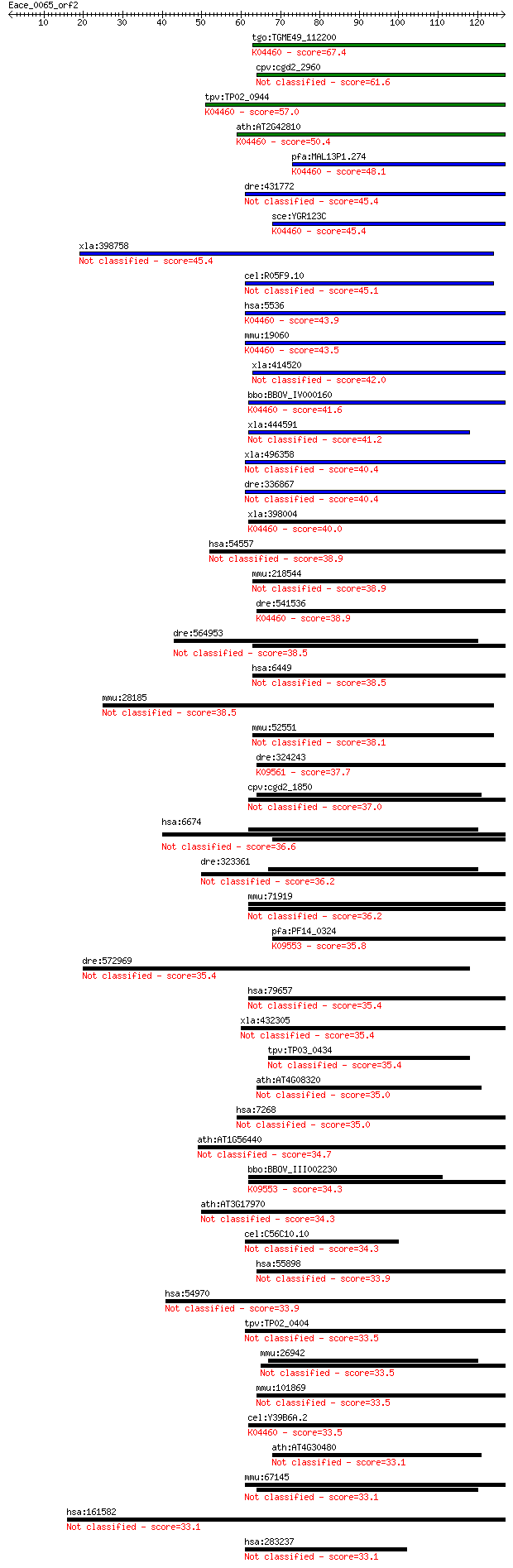
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_112200 serine/threonine protein phosphatase, putati... 67.4 1e-11
cpv:cgd2_2960 phosphoprotein phosphatase related 61.6 6e-10
tpv:TP02_0944 serine/threonine protein phosphatase; K04460 pro... 57.0 2e-08
ath:AT2G42810 PP5.2; PP5.2 (PROTEIN PHOSPHATASE 5.2); phosphop... 50.4 1e-06
pfa:MAL13P1.274 PfPP5; serine/threonine protein phosphatase (E... 48.1 6e-06
dre:431772 sgta, zgc:92462; small glutamine-rich tetratricopep... 45.4 4e-05
sce:YGR123C PPT1; Ppt1p (EC:3.1.3.16); K04460 protein phosphat... 45.4 4e-05
xla:398758 tomm70a, MGC68780; translocase of outer mitochondri... 45.4 5e-05
cel:R05F9.10 sgt-1; Small Glutamine-rich Tetratrico repeat pro... 45.1 5e-05
hsa:5536 PPP5C, FLJ36922, FLJ55954, PP5, PPP5, PPT; protein ph... 43.9 1e-04
mmu:19060 Ppp5c, AU020526, PP5; protein phosphatase 5, catalyt... 43.5 2e-04
xla:414520 hypothetical protein MGC81394 42.0 4e-04
bbo:BBOV_IV000160 21.m02802; serine/threonine protein phosphat... 41.6 7e-04
xla:444591 sgtb, MGC84046; small glutamine-rich tetratricopept... 41.2 9e-04
xla:496358 sgta; small glutamine-rich tetratricopeptide repeat... 40.4 0.001
dre:336867 fk20d10, wu:fa05b08, wu:fc52b05, wu:fk20d10, zgc:77... 40.4 0.001
xla:398004 ppp5c, pp5; protein phosphatase 5, catalytic subuni... 40.0 0.002
hsa:54557 SGTB, FLJ39002, SGT2; small glutamine-rich tetratric... 38.9 0.004
mmu:218544 Sgtb, C630001O05Rik, MGC27660; small glutamine-rich... 38.9 0.004
dre:541536 fc83f08, im:7146608, ppp5c, wu:fc83f08; zgc:110801 ... 38.9 0.004
dre:564953 spag1, MGC162178, cb1089, wu:fj78g10; sperm associa... 38.5 0.005
hsa:6449 SGTA, SGT, alphaSGT, hSGT; small glutamine-rich tetra... 38.5 0.005
mmu:28185 Tomm70a, 2610044B22Rik, D16Ium22, D16Ium22e, D16Wsu1... 38.5 0.005
mmu:52551 Sgta, 5330427H01Rik, AI194281, D10Ertd190e, MGC6336,... 38.1 0.007
dre:324243 stub1, wu:fc22f04, zgc:56076; STIP1 homology and U-... 37.7 0.009
cpv:cgd2_1850 stress-induced protein sti1-like protein 37.0 0.015
hsa:6674 SPAG1, FLJ32920, HSD-3.8, SP75, TPIS; sperm associate... 36.6 0.021
dre:323361 tomm34, wu:fb96b08, zgc:56645; translocase of outer... 36.2 0.024
mmu:71919 Rpap3, 2310042P20Rik, D15Ertd682e; RNA polymerase II... 36.2 0.026
pfa:PF14_0324 Hsp70/Hsp90 organizing protein, putative; K09553... 35.8 0.033
dre:572969 tomm70a, KIAA0719, MGC73188, wu:fj58b04, zgc:73188;... 35.4 0.040
hsa:79657 RPAP3, FLJ21908; RNA polymerase II associated protein 3 35.4
xla:432305 ttc4, MGC80137; tetratricopeptide repeat domain 4 35.4
tpv:TP03_0434 hypothetical protein 35.4 0.049
ath:AT4G08320 tetratricopeptide repeat (TPR)-containing protein 35.0 0.061
hsa:7268 TTC4, DKFZp781B0622, FLJ41930, MGC5097; tetratricopep... 35.0 0.063
ath:AT1G56440 serine/threonine protein phosphatase-related 34.7 0.069
bbo:BBOV_III002230 17.m07215; tetratricopeptide repeat domain ... 34.3 0.093
ath:AT3G17970 atToc64-III (Arabidopsis thaliana translocon at ... 34.3 0.100
cel:C56C10.10 hypothetical protein 34.3 0.10
hsa:55898 UNC45A, FLJ10043, GC-UNC45, GCUNC-45, GCUNC45, IRO03... 33.9 0.13
hsa:54970 TTC12, FLJ13859, FLJ20535, TPARM; tetratricopeptide ... 33.9 0.15
tpv:TP02_0404 hypothetical protein 33.5 0.15
mmu:26942 Spag1, tpis; sperm associated antigen 1 33.5
mmu:101869 Unc45a, AW538196; unc-45 homolog A (C. elegans) 33.5 0.16
cel:Y39B6A.2 pph-5; Protein PHosphatase family member (pph-5);... 33.5 0.18
ath:AT4G30480 tetratricopeptide repeat (TPR)-containing protein 33.1 0.20
mmu:67145 Tomm34, 2610100K07Rik, TOM34, Tomm34a, Tomm34b; tran... 33.1 0.21
hsa:161582 DYX1C1, DYX1, DYXC1, EKN1, FLJ37882, MGC70618, RD; ... 33.1 0.23
hsa:283237 TTC9C, MGC29649; tetratricopeptide repeat domain 9C 33.1 0.23
> tgo:TGME49_112200 serine/threonine protein phosphatase, putative
(EC:3.1.3.16); K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=548
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 32/65 (49%), Positives = 45/65 (69%), Gaps = 1/65 (1%)
Query 63 KAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQ-LHVYLCNRSFAHLRME 121
+AE+ KTEGNE FK + +AV KY+AAID I +++ + Q L V LCNR+F + +E
Sbjct 60 EAESLKTEGNEFFKTRLFHQAVEKYTAAIDLICSNTMTAQTKQILQVLLCNRAFCQINLE 119
Query 122 NFGSA 126
N+GSA
Sbjct 120 NYGSA 124
> cpv:cgd2_2960 phosphoprotein phosphatase related
Length=525
Score = 61.6 bits (148), Expect = 6e-10, Method: Composition-based stats.
Identities = 28/63 (44%), Positives = 42/63 (66%), Gaps = 1/63 (1%)
Query 64 AEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRMENF 123
+E +K +GNE FK +Y EA+ Y+ AI T ++S + + LH+Y NR+ H+R+ENF
Sbjct 14 SEQYKIKGNESFKSGKYNEAIEYYTLAIKT-SQASNETQNKNLHIYYSNRALCHIRLENF 72
Query 124 GSA 126
GSA
Sbjct 73 GSA 75
> tpv:TP02_0944 serine/threonine protein phosphatase; K04460 protein
phosphatase 5 [EC:3.1.3.16]
Length=548
Score = 57.0 bits (136), Expect = 2e-08, Method: Composition-based stats.
Identities = 30/90 (33%), Positives = 51/90 (56%), Gaps = 16/90 (17%)
Query 51 GKNDALGCSPLEKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSL----------- 99
GKN+ + LEKAE K EGN++F ++ + A+ YS +I +++S L
Sbjct 46 GKNNKV--QALEKAELKKLEGNKMFSENNFLSAIEHYSESIRLVEDSHLVSNFKKEGYNW 103
Query 100 ---DPKETQLHVYLCNRSFAHLRMENFGSA 126
+ ++T LH Y NR+ ++++EN+GSA
Sbjct 104 ITPELRKTNLHQYYSNRAICNIKIENYGSA 133
> ath:AT2G42810 PP5.2; PP5.2 (PROTEIN PHOSPHATASE 5.2); phosphoprotein
phosphatase/ protein binding / protein serine/threonine
phosphatase; K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=538
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 26/68 (38%), Positives = 41/68 (60%), Gaps = 9/68 (13%)
Query 59 SPLEKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHL 118
S + +AE FK++ NE FK H+Y+ A+ Y+ AI+ ++ VY NR+FAH
Sbjct 8 SDVSRAEEFKSQANEAFKGHKYSSAIDLYTKAIELNSNNA---------VYWANRAFAHT 58
Query 119 RMENFGSA 126
++E +GSA
Sbjct 59 KLEEYGSA 66
> pfa:MAL13P1.274 PfPP5; serine/threonine protein phosphatase
(EC:3.1.3.16); K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=658
Score = 48.1 bits (113), Expect = 6e-06, Method: Composition-based stats.
Identities = 24/54 (44%), Positives = 33/54 (61%), Gaps = 8/54 (14%)
Query 73 ELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRMENFGSA 126
ELFK++ A+SK S I KET LH+Y NRSF H+++EN+G+A
Sbjct 199 ELFKEYYNKSAISKKSDFISI--------KETDLHIYYTNRSFCHIKLENYGTA 244
> dre:431772 sgta, zgc:92462; small glutamine-rich tetratricopeptide
repeat (TPR)-containing, alpha
Length=306
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 26/66 (39%), Positives = 37/66 (56%), Gaps = 9/66 (13%)
Query 61 LEKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRM 120
+E+AE K EGN K+ Y+ AV Y+ AI+ LD + VY CNR+ AH ++
Sbjct 84 IERAEQLKNEGNNHMKEENYSSAVDCYTKAIE------LDQRNA---VYYCNRAAAHSKL 134
Query 121 ENFGSA 126
EN+ A
Sbjct 135 ENYTEA 140
> sce:YGR123C PPT1; Ppt1p (EC:3.1.3.16); K04460 protein phosphatase
5 [EC:3.1.3.16]
Length=513
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 23/59 (38%), Positives = 35/59 (59%), Gaps = 9/59 (15%)
Query 68 KTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRMENFGSA 126
K EGN K+ + +A+ KY+ AID LD ++ +Y NR+FAH +++NF SA
Sbjct 16 KNEGNVFVKEKHFLKAIEKYTEAID------LDSTQS---IYFSNRAFAHFKVDNFQSA 65
> xla:398758 tomm70a, MGC68780; translocase of outer mitochondrial
membrane 70 homolog A
Length=576
Score = 45.4 bits (106), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 34/107 (31%), Positives = 54/107 (50%), Gaps = 11/107 (10%)
Query 19 RKEAAKRRDGEMAVDGGSSCAAKEAGDGAERDGKNDALGCSPLEKAEAFKTEGNELFKQH 78
R +AAK +G +S E G +D + SP+EKA+A K +GN+ FK
Sbjct 40 RHKAAKSDKQRRTPEGSASPVPSEGGSNNPQDAPQE---LSPIEKAQAAKNKGNKYFKAS 96
Query 79 QYAEAVSKYSAAIDTIDESSLDP--KETQLHVYLCNRSFAHLRMENF 123
+Y +A+ Y+ AI SL P ++ L + NR+ AH + +N+
Sbjct 97 KYEQAIQCYTEAI------SLCPAHNKSDLSTFYQNRAAAHEQSQNW 137
> cel:R05F9.10 sgt-1; Small Glutamine-rich Tetratrico repeat protein
family member (sgt-1)
Length=337
Score = 45.1 bits (105), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 25/63 (39%), Positives = 35/63 (55%), Gaps = 10/63 (15%)
Query 61 LEKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRM 120
+ +A K EGN+L K Q+ AV KY+AAI + DP VY CNR+ A+ R+
Sbjct 102 ISQANKLKEEGNDLMKASQFEAAVQKYNAAIKL----NRDP------VYFCNRAAAYCRL 151
Query 121 ENF 123
E +
Sbjct 152 EQY 154
> hsa:5536 PPP5C, FLJ36922, FLJ55954, PP5, PPP5, PPT; protein
phosphatase 5, catalytic subunit (EC:3.1.3.16); K04460 protein
phosphatase 5 [EC:3.1.3.16]
Length=477
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 26/66 (39%), Positives = 36/66 (54%), Gaps = 9/66 (13%)
Query 61 LEKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRM 120
L++AE KT+ N+ FK Y A+ YS AI+ L+P +Y NRS A+LR
Sbjct 25 LKRAEELKTQANDYFKAKDYENAIKFYSQAIE------LNPSNA---IYYGNRSLAYLRT 75
Query 121 ENFGSA 126
E +G A
Sbjct 76 ECYGYA 81
> mmu:19060 Ppp5c, AU020526, PP5; protein phosphatase 5, catalytic
subunit (EC:3.1.3.16); K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=499
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 26/66 (39%), Positives = 36/66 (54%), Gaps = 9/66 (13%)
Query 61 LEKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRM 120
L++AE KT+ N+ FK Y A+ YS AI+ L+P +Y NRS A+LR
Sbjct 25 LKRAEELKTQANDYFKAKDYENAIKFYSQAIE------LNPGNA---IYYGNRSLAYLRT 75
Query 121 ENFGSA 126
E +G A
Sbjct 76 ECYGYA 81
> xla:414520 hypothetical protein MGC81394
Length=312
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 23/64 (35%), Positives = 37/64 (57%), Gaps = 9/64 (14%)
Query 63 KAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRMEN 122
+AE+ KTEGNE K + AV+ Y+ A++ L+P+ VY CNR+ A+ ++ N
Sbjct 87 EAESLKTEGNEQMKVENFESAVTYYTKALE------LNPRNA---VYYCNRAAAYSKLGN 137
Query 123 FGSA 126
+ A
Sbjct 138 YAGA 141
> bbo:BBOV_IV000160 21.m02802; serine/threonine protein phosphatase
5 (EC:3.1.3.16); K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=545
Score = 41.6 bits (96), Expect = 7e-04, Method: Composition-based stats.
Identities = 24/81 (29%), Positives = 40/81 (49%), Gaps = 16/81 (19%)
Query 62 EKAEAFKTEGNELFKQHQYAEAVSKYSAAI----DTIDESSLD------------PKETQ 105
++A+ + EGN+ F + Y AV Y+ AI I+E+++ +T
Sbjct 23 QRADEKRLEGNKFFGEGDYPVAVELYTQAIGILRKAIEEANVRQNSENSTNVDTLSSQTN 82
Query 106 LHVYLCNRSFAHLRMENFGSA 126
+H NR+ H+RMEN+G A
Sbjct 83 IHQLYTNRALCHIRMENYGLA 103
> xla:444591 sgtb, MGC84046; small glutamine-rich tetratricopeptide
repeat (TPR)-containing, beta
Length=308
Score = 41.2 bits (95), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 26/56 (46%), Positives = 29/56 (51%), Gaps = 9/56 (16%)
Query 62 EKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAH 117
EKAE K EGN L K+ Y AV YS AI+ LDP VY CNR+ A
Sbjct 87 EKAEQLKDEGNGLMKEQNYEAAVDCYSQAIE------LDPNNA---VYYCNRAAAQ 133
> xla:496358 sgta; small glutamine-rich tetratricopeptide repeat
(TPR)-containing, alpha
Length=302
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/66 (36%), Positives = 36/66 (54%), Gaps = 9/66 (13%)
Query 61 LEKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRM 120
L +AE KTEGNE K + A+S Y+ A++ L+P VY CNR+ A+ ++
Sbjct 73 LAEAERLKTEGNEQMKVENFESAISYYTKALE------LNPANA---VYYCNRAAAYSKL 123
Query 121 ENFGSA 126
N+ A
Sbjct 124 GNYAGA 129
> dre:336867 fk20d10, wu:fa05b08, wu:fc52b05, wu:fk20d10, zgc:77080;
zgc:55741
Length=320
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/66 (36%), Positives = 36/66 (54%), Gaps = 9/66 (13%)
Query 61 LEKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRM 120
L +AE KT+GN+ K ++ AV YS AI L+P+ VY CNR+ A+ ++
Sbjct 88 LAEAERLKTDGNDQMKVENFSAAVEFYSKAIQ------LNPQNA---VYFCNRAAAYSKL 138
Query 121 ENFGSA 126
N+ A
Sbjct 139 GNYAGA 144
> xla:398004 ppp5c, pp5; protein phosphatase 5, catalytic subunit
(EC:3.1.3.16); K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=493
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 23/65 (35%), Positives = 33/65 (50%), Gaps = 9/65 (13%)
Query 62 EKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRME 121
+ AE K + NE F+ Y AV Y+ AID +++ +Y NRS A+LR E
Sbjct 20 KTAEELKEQANEYFRVKDYDHAVQYYTQAIDLSPDTA---------IYYGNRSLAYLRTE 70
Query 122 NFGSA 126
+G A
Sbjct 71 CYGYA 75
> hsa:54557 SGTB, FLJ39002, SGT2; small glutamine-rich tetratricopeptide
repeat (TPR)-containing, beta
Length=304
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 30/80 (37%), Positives = 40/80 (50%), Gaps = 14/80 (17%)
Query 52 KNDALGCS---P--LEKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQL 106
KND L S P + KA+ K EGN K+ YA AV Y+ AI+ LDP
Sbjct 68 KNDVLPLSNSVPEDVGKADQLKDEGNNHMKEENYAAAVDCYTQAIE------LDPNNA-- 119
Query 107 HVYLCNRSFAHLRMENFGSA 126
VY CNR+ A ++ ++ A
Sbjct 120 -VYYCNRAAAQSKLGHYTDA 138
> mmu:218544 Sgtb, C630001O05Rik, MGC27660; small glutamine-rich
tetratricopeptide repeat (TPR)-containing, beta
Length=304
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 24/64 (37%), Positives = 33/64 (51%), Gaps = 9/64 (14%)
Query 63 KAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRMEN 122
KA+ K EGN K+ YA AV Y+ AI+ LDP VY CNR+ A ++ +
Sbjct 84 KADQLKDEGNNHMKEENYAAAVDCYTQAIE------LDPNNA---VYYCNRAAAQSKLSH 134
Query 123 FGSA 126
+ A
Sbjct 135 YTDA 138
> dre:541536 fc83f08, im:7146608, ppp5c, wu:fc83f08; zgc:110801
(EC:3.1.3.16); K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=481
Score = 38.9 bits (89), Expect = 0.004, Method: Composition-based stats.
Identities = 22/63 (34%), Positives = 32/63 (50%), Gaps = 9/63 (14%)
Query 64 AEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRMENF 123
AE K + N+ FK Y A+ Y+ A+D L+P +Y NRS ++LR E +
Sbjct 10 AEKLKEKANDYFKDKDYENAIKYYTEALD------LNPTNP---IYYSNRSLSYLRTECY 60
Query 124 GSA 126
G A
Sbjct 61 GYA 63
> dre:564953 spag1, MGC162178, cb1089, wu:fj78g10; sperm associated
antigen 1
Length=386
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 28/77 (36%), Positives = 39/77 (50%), Gaps = 5/77 (6%)
Query 43 AGDGAERDGKNDALGCSPLEKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPK 102
AG+ D AL PL + K +GN LFK Q+ +A+ KY+ AID E+ +D
Sbjct 67 AGESCNLDAPCGALP-PPLAR---LKNQGNMLFKNGQFGDALEKYTQAIDGCIEAGIDSP 122
Query 103 ETQLHVYLCNRSFAHLR 119
E L V NR+ L+
Sbjct 123 ED-LCVLYSNRAACFLK 138
Score = 35.0 bits (79), Expect = 0.052, Method: Compositional matrix adjust.
Identities = 24/68 (35%), Positives = 33/68 (48%), Gaps = 13/68 (19%)
Query 63 KAEA----FKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHL 118
KAEA K EGNEL K Q+ A KYS + ++ P E ++ NR+ L
Sbjct 256 KAEARFTILKQEGNELVKNSQFQGASEKYSECL------AIKPNECAIYT---NRALCFL 306
Query 119 RMENFGSA 126
++E F A
Sbjct 307 KLERFAEA 314
> hsa:6449 SGTA, SGT, alphaSGT, hSGT; small glutamine-rich tetratricopeptide
repeat (TPR)-containing, alpha
Length=313
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 24/64 (37%), Positives = 33/64 (51%), Gaps = 9/64 (14%)
Query 63 KAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRMEN 122
+AE KTEGNE K + AV Y AI+ L+P VY CNR+ A+ ++ N
Sbjct 90 EAERLKTEGNEQMKVENFEAAVHFYGKAIE------LNPANA---VYFCNRAAAYSKLGN 140
Query 123 FGSA 126
+ A
Sbjct 141 YAGA 144
> mmu:28185 Tomm70a, 2610044B22Rik, D16Ium22, D16Ium22e, D16Wsu109e,
Tom70, mKIAA0719; translocase of outer mitochondrial
membrane 70 homolog A (yeast)
Length=611
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 31/101 (30%), Positives = 51/101 (50%), Gaps = 11/101 (10%)
Query 25 RRDGEMAVDGGSSCAAKEAGDGAERDGKNDALGCSPLEKAEAFKTEGNELFKQHQYAEAV 84
+R+ E G + A +G DG D+L S L++A+A K +GN+ FK +Y +A+
Sbjct 81 KRNSERKTPEGRASPALGSG---HHDGSGDSLEMSSLDRAQAAKNKGNKYFKAGKYEQAI 137
Query 85 SKYSAAIDTIDESSLDP--KETQLHVYLCNRSFAHLRMENF 123
Y+ AI SL P K L + NR+ A +++ +
Sbjct 138 QCYTEAI------SLCPTEKNVDLSTFYQNRAAAFEQLQKW 172
> mmu:52551 Sgta, 5330427H01Rik, AI194281, D10Ertd190e, MGC6336,
Sgt, Stg; small glutamine-rich tetratricopeptide repeat (TPR)-containing,
alpha
Length=315
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 23/61 (37%), Positives = 32/61 (52%), Gaps = 9/61 (14%)
Query 63 KAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRMEN 122
+AE KTEGNE K + AV Y AI+ L+P VY CNR+ A+ ++ N
Sbjct 91 EAERLKTEGNEQMKLENFEAAVHLYGKAIE------LNPANA---VYFCNRAAAYSKLGN 141
Query 123 F 123
+
Sbjct 142 Y 142
> dre:324243 stub1, wu:fc22f04, zgc:56076; STIP1 homology and
U-Box containing protein 1; K09561 STIP1 homology and U-box
containing protein 1 [EC:6.3.2.19]
Length=284
Score = 37.7 bits (86), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 33/63 (52%), Gaps = 9/63 (14%)
Query 64 AEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRMENF 123
A+ K +GN LF +Y EAV+ YS AI+ + + VY NR+ +++++ +
Sbjct 11 AQELKEQGNRLFLSRKYQEAVTCYSKAIN---------RNPSVAVYYTNRALCYVKLQQY 61
Query 124 GSA 126
A
Sbjct 62 DKA 64
> cpv:cgd2_1850 stress-induced protein sti1-like protein
Length=326
Score = 37.0 bits (84), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 20/57 (35%), Positives = 33/57 (57%), Gaps = 9/57 (15%)
Query 64 AEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRM 120
AE +K +GNEL+KQ ++ EA+ +Y AI+ +DP + +L N+ +L M
Sbjct 5 AEFYKNKGNELYKQKKFDEALVQYDLAIE------IDPNDIS---FLTNKGAVYLEM 52
Score = 32.7 bits (73), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 35/65 (53%), Gaps = 9/65 (13%)
Query 62 EKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRME 121
E AE + EGN+LFKQ Y A +Y AI +P +++L+ NR+ ++++
Sbjct 138 ELAEKHRIEGNDLFKQKNYPAAKKEYDEAIKR------NPSDSRLY---SNRAACYMQLL 188
Query 122 NFGSA 126
+ SA
Sbjct 189 EYPSA 193
> hsa:6674 SPAG1, FLJ32920, HSD-3.8, SP75, TPIS; sperm associated
antigen 1
Length=926
Score = 36.6 bits (83), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 21/58 (36%), Positives = 34/58 (58%), Gaps = 1/58 (1%)
Query 62 EKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLR 119
E K++GNELF+ Q+AEA KYSAAI ++ + + + L + NR+ +L+
Sbjct 443 ENPAGLKSQGNELFRSGQFAEAAGKYSAAIALLEPAGSEIAD-DLSILYSNRAACYLK 499
Score = 28.9 bits (63), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 23/87 (26%), Positives = 42/87 (48%), Gaps = 13/87 (14%)
Query 40 AKEAGDGAERDGKNDALGCSPLEKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSL 99
+K+AGD + + G + + +A K EGN+ Y +A+SKYS + +
Sbjct 603 SKQAGDSSSHRQQ----GITDEKTFKALKEEGNQCVNDKNYKDALSKYSECL------KI 652
Query 100 DPKETQLHVYLCNRSFAHLRMENFGSA 126
+ KE ++ NR+ +L++ F A
Sbjct 653 NNKECAIYT---NRALCYLKLCQFEEA 676
Score = 28.1 bits (61), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 18/59 (30%), Positives = 28/59 (47%), Gaps = 10/59 (16%)
Query 68 KTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRMENFGSA 126
K +GNE F Y EAV Y+ +I + V NR+ A ++++N+ SA
Sbjct 213 KEKGNEAFNSGDYEEAVMYYTRSISALPTV----------VAYNNRAQAEIKLQNWNSA 261
> dre:323361 tomm34, wu:fb96b08, zgc:56645; translocase of outer
mitochondrial membrane 34
Length=305
Score = 36.2 bits (82), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 21/53 (39%), Positives = 30/53 (56%), Gaps = 1/53 (1%)
Query 67 FKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLR 119
K GNE FK QY EAV+ YS AI +++S K L + NR+ ++L+
Sbjct 13 LKQAGNECFKAGQYGEAVTLYSQAIQQLEKSG-QKKTEDLGILYSNRAASYLK 64
Score = 33.1 bits (74), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 21/77 (27%), Positives = 38/77 (49%), Gaps = 9/77 (11%)
Query 50 DGKNDALGCSPLEKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVY 109
D K A G ++K K EGN L K+ ++ +A+ KY+ ++ + DP E +
Sbjct 176 DDKKKAPGPDAVKKGRTLKEEGNALVKKGEHKKAMEKYTQSL------AQDPTEVTTYT- 228
Query 110 LCNRSFAHLRMENFGSA 126
NR+ +L ++ + A
Sbjct 229 --NRALCYLALKMYKDA 243
> mmu:71919 Rpap3, 2310042P20Rik, D15Ertd682e; RNA polymerase
II associated protein 3
Length=660
Score = 36.2 bits (82), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 33/65 (50%), Gaps = 9/65 (13%)
Query 62 EKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRME 121
+KA K +GN+ FKQ +Y EA+ Y+ +D DP V NR+ A+ R++
Sbjct 132 QKALVLKEKGNKYFKQGKYDEAIECYTKGMDA------DPYNP---VLPTNRASAYFRLK 182
Query 122 NFGSA 126
F A
Sbjct 183 KFAVA 187
Score = 28.1 bits (61), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 19/65 (29%), Positives = 33/65 (50%), Gaps = 9/65 (13%)
Query 62 EKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRME 121
+KA A K GN FK+ +Y +A+ Y+ I ++L P NR+ A+L+++
Sbjct 282 QKAIAEKDLGNGFFKEGKYEQAIECYTRGIAADRTNALLP---------ANRAMAYLKIQ 332
Query 122 NFGSA 126
+ A
Sbjct 333 RYEEA 337
> pfa:PF14_0324 Hsp70/Hsp90 organizing protein, putative; K09553
stress-induced-phosphoprotein 1
Length=564
Score = 35.8 bits (81), Expect = 0.033, Method: Compositional matrix adjust.
Identities = 18/59 (30%), Positives = 32/59 (54%), Gaps = 9/59 (15%)
Query 68 KTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRMENFGSA 126
K +GNE +KQ ++ EA+ +Y AI ++P + H N++ H+ M+N+ A
Sbjct 247 KLKGNEFYKQKKFDEALKEYEEAI------QINPNDIMYHY---NKAAVHIEMKNYDKA 296
> dre:572969 tomm70a, KIAA0719, MGC73188, wu:fj58b04, zgc:73188;
translocase of outer mitochondrial membrane 70 homolog A
(yeast)
Length=578
Score = 35.4 bits (80), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 30/100 (30%), Positives = 49/100 (49%), Gaps = 12/100 (12%)
Query 20 KEAAKRRDGEMAVDGGSSCAAKEAGDGAERDGKNDALGCSPLEKAEAFKTEGNELFKQHQ 79
KE +R+GE GS+ + + +N SPL++A++ K +GN+ FK +
Sbjct 44 KEKQGKRNGERKTPEGSASPVQGQHGATNPELEN----LSPLDRAQSAKNKGNKYFKAGK 99
Query 80 YAEAVSKYSAAIDTIDESSLDPKETQ--LHVYLCNRSFAH 117
Y A+ Y+ AI L PKE + L + NR+ A+
Sbjct 100 YDHAIKCYTEAI------GLCPKEKKGDLSTFYQNRAAAY 133
> hsa:79657 RPAP3, FLJ21908; RNA polymerase II associated protein
3
Length=631
Score = 35.4 bits (80), Expect = 0.042, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 33/65 (50%), Gaps = 9/65 (13%)
Query 62 EKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRME 121
+KA K +GN+ FKQ +Y EA+ Y+ +D DP V NR+ A+ R++
Sbjct 131 QKALVLKEKGNKYFKQGKYDEAIDCYTKGMDA------DPYNP---VLPTNRASAYFRLK 181
Query 122 NFGSA 126
F A
Sbjct 182 KFAVA 186
> xla:432305 ttc4, MGC80137; tetratricopeptide repeat domain 4
Length=384
Score = 35.4 bits (80), Expect = 0.044, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 39/68 (57%), Gaps = 7/68 (10%)
Query 60 PLEKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYL-CNRSFAHL 118
P E+A+++K EGNE FK+ Y +A++ Y+ I + K+ +L+ L NR+ A
Sbjct 71 PEEQAKSYKDEGNEYFKEKDYNKAITSYTEGIKK------NCKDQELNAILYTNRAAAQF 124
Query 119 RMENFGSA 126
+ N+ SA
Sbjct 125 YLGNYRSA 132
> tpv:TP03_0434 hypothetical protein
Length=282
Score = 35.4 bits (80), Expect = 0.049, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 28/51 (54%), Gaps = 3/51 (5%)
Query 67 FKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAH 117
+K GNE FK + Y EA+ Y+ A++ ++ S D Q+ CNR+ H
Sbjct 102 YKERGNECFKDNNYNEAIDWYTKALERLEFSEDDNLRAQI---FCNRAACH 149
> ath:AT4G08320 tetratricopeptide repeat (TPR)-containing protein
Length=427
Score = 35.0 bits (79), Expect = 0.061, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 31/57 (54%), Gaps = 9/57 (15%)
Query 64 AEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRM 120
AE K +GN+ + + Y EAV YS AI D+++ V+ CNR+ A+ ++
Sbjct 175 AETLKCQGNKAMQSNLYLEAVELYSFAIALTDKNA---------VFYCNRAAAYTQI 222
> hsa:7268 TTC4, DKFZp781B0622, FLJ41930, MGC5097; tetratricopeptide
repeat domain 4
Length=387
Score = 35.0 bits (79), Expect = 0.063, Method: Compositional matrix adjust.
Identities = 24/68 (35%), Positives = 36/68 (52%), Gaps = 5/68 (7%)
Query 59 SPLEKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHL 118
SP E+A+ +K EGN+ FK+ Y +AV Y+ + + + DP V NR+ A
Sbjct 74 SPEEQAKTYKDEGNDYFKEKDYKKAVISYT---EGLKKKCADPDLNA--VLYTNRAAAQY 128
Query 119 RMENFGSA 126
+ NF SA
Sbjct 129 YLGNFRSA 136
> ath:AT1G56440 serine/threonine protein phosphatase-related
Length=476
Score = 34.7 bits (78), Expect = 0.069, Method: Compositional matrix adjust.
Identities = 25/78 (32%), Positives = 41/78 (52%), Gaps = 11/78 (14%)
Query 49 RDGKNDALGCSPLEKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHV 108
RD + +G S L+ + K +GNE FKQ ++ EA+ YS +I +L P V
Sbjct 70 RDLSSSLIGESLLDSSSE-KEQGNEFFKQKKFNEAIDCYSRSI------ALSPN----AV 118
Query 109 YLCNRSFAHLRMENFGSA 126
NR+ A+L+++ + A
Sbjct 119 TYANRAMAYLKIKRYREA 136
> bbo:BBOV_III002230 17.m07215; tetratricopeptide repeat domain
containing protein; K09553 stress-induced-phosphoprotein 1
Length=546
Score = 34.3 bits (77), Expect = 0.093, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 31/49 (63%), Gaps = 1/49 (2%)
Query 62 EKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYL 110
+KA+ +K EGN+L+KQ ++ EA+ Y AI+ D +L + + VYL
Sbjct 223 QKAKEYKEEGNKLYKQKRFEEALEMYKKAIEH-DPDNLLLENNKAAVYL 270
Score = 30.0 bits (66), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 34/65 (52%), Gaps = 9/65 (13%)
Query 62 EKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRME 121
+KAE + +GN FK+ Q+ EA +Y AI +P + +L+ NR+ A ++
Sbjct 358 QKAEEHREKGNAFFKKFQFPEAKKEYDEAIRR------NPSDIKLYT---NRAAALTKLG 408
Query 122 NFGSA 126
+ SA
Sbjct 409 EYPSA 413
> ath:AT3G17970 atToc64-III (Arabidopsis thaliana translocon at
the outer membrane of chloroplasts 64-III); binding / carbon-nitrogen
ligase, with glutamine as amido-N-donor
Length=589
Score = 34.3 bits (77), Expect = 0.100, Method: Composition-based stats.
Identities = 23/77 (29%), Positives = 36/77 (46%), Gaps = 9/77 (11%)
Query 50 DGKNDALGCSPLEKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVY 109
D K+ + E AE K +GN+ FK+ + +A+ YS AI D ++ Y
Sbjct 460 DPKSSKKAITKEESAEIAKEKGNQAFKEKLWQKAIGLYSEAIKLSDNNA---------TY 510
Query 110 LCNRSFAHLRMENFGSA 126
NR+ A+L + F A
Sbjct 511 YSNRAAAYLELGGFLQA 527
> cel:C56C10.10 hypothetical protein
Length=342
Score = 34.3 bits (77), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 16/39 (41%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 61 LEKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSL 99
L+ EA + +GNELF Q Y EA+ Y A+ +D L
Sbjct 188 LKSVEALRQKGNELFVQKDYKEAIDAYRDALTRLDTLIL 226
> hsa:55898 UNC45A, FLJ10043, GC-UNC45, GCUNC-45, GCUNC45, IRO039700,
SMAP-1, SMAP1; unc-45 homolog A (C. elegans)
Length=929
Score = 33.9 bits (76), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 22/63 (34%), Positives = 35/63 (55%), Gaps = 6/63 (9%)
Query 64 AEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRMENF 123
E + EGNELFK Y A++ Y+ A+ +D + D + LH NR+ HL++E++
Sbjct 6 VEQLRKEGNELFKCGDYGGALAAYTQALG-LDATPQD--QAVLHR---NRAACHLKLEDY 59
Query 124 GSA 126
A
Sbjct 60 DKA 62
> hsa:54970 TTC12, FLJ13859, FLJ20535, TPARM; tetratricopeptide
repeat domain 12
Length=705
Score = 33.9 bits (76), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 23/86 (26%), Positives = 44/86 (51%), Gaps = 16/86 (18%)
Query 41 KEAGDGAERDGKNDALGCSPLEKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLD 100
K+A + A+R +N L A+A K +GNE F + Y A+ +YS ++ + +
Sbjct 90 KDAKERAKRRRENKVL-------ADALKEKGNEAFAEGNYETAILRYSEGLEKLKD---- 138
Query 101 PKETQLHVYLCNRSFAHLRMENFGSA 126
+ V NR+ A++++E++ A
Sbjct 139 -----MKVLYTNRAQAYMKLEDYEKA 159
> tpv:TP02_0404 hypothetical protein
Length=483
Score = 33.5 bits (75), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 21/68 (30%), Positives = 35/68 (51%), Gaps = 2/68 (2%)
Query 61 LEKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDE-SSLDPKETQ-LHVYLCNRSFAHL 118
L KA AFK+ GNE +K ++ EA Y++ + + + S +P + L V N +L
Sbjct 26 LSKATAFKSAGNEFYKLAKFQEASDSYNSGVSWMKKMSDFEPSHRELLSVLYSNLCATYL 85
Query 119 RMENFGSA 126
+ N+ A
Sbjct 86 ELSNYSKA 93
> mmu:26942 Spag1, tpis; sperm associated antigen 1
Length=901
Score = 33.5 bits (75), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 20/57 (35%), Positives = 32/57 (56%), Gaps = 9/57 (15%)
Query 67 FKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKET----QLHVYLCNRSFAHLR 119
K GNELF+ Q+AEA ++YS AI + L+P + +L + NR+ +L+
Sbjct 433 LKRRGNELFRGGQFAEAAAQYSVAI-----AQLEPTGSANADELSILYSNRAACYLK 484
Score = 32.3 bits (72), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 18/62 (29%), Positives = 32/62 (51%), Gaps = 9/62 (14%)
Query 65 EAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRMENFG 124
+A K EGN+L K Y +A+SKY+ + ++ K ++ NR+ +L++ F
Sbjct 607 QALKEEGNQLVKDKNYKDAISKYNECL------KINSKACAIYT---NRALCYLKLGQFE 657
Query 125 SA 126
A
Sbjct 658 EA 659
> mmu:101869 Unc45a, AW538196; unc-45 homolog A (C. elegans)
Length=944
Score = 33.5 bits (75), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 32/63 (50%), Gaps = 6/63 (9%)
Query 64 AEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRMENF 123
AE + EGNELFK Y A++ Y+ A+ SL + NR+ HL++E++
Sbjct 21 AEQLRKEGNELFKCGDYEGALTAYTQAL------SLGATPQDQAILHRNRAACHLKLEDY 74
Query 124 GSA 126
A
Sbjct 75 SKA 77
> cel:Y39B6A.2 pph-5; Protein PHosphatase family member (pph-5);
K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=496
Score = 33.5 bits (75), Expect = 0.18, Method: Composition-based stats.
Identities = 24/65 (36%), Positives = 31/65 (47%), Gaps = 10/65 (15%)
Query 62 EKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRME 121
EKA K E N+ FK Y A YS AI+ + P V NR+ A+L+ E
Sbjct 27 EKAGMIKDEANQFFKDQVYDVAADLYSVAIE------IHPTA----VLYGNRAQAYLKKE 76
Query 122 NFGSA 126
+GSA
Sbjct 77 LYGSA 81
> ath:AT4G30480 tetratricopeptide repeat (TPR)-containing protein
Length=161
Score = 33.1 bits (74), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 19/54 (35%), Positives = 30/54 (55%), Gaps = 4/54 (7%)
Query 68 KTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLH-VYLCNRSFAHLRM 120
K EGN+LF Y EA+SKY+ A++ + E P+ +L + NR L++
Sbjct 109 KAEGNKLFVNGLYEEALSKYAFALELVQEL---PESIELRSICYLNRGVCFLKL 159
> mmu:67145 Tomm34, 2610100K07Rik, TOM34, Tomm34a, Tomm34b; translocase
of outer mitochondrial membrane 34
Length=309
Score = 33.1 bits (74), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 21/66 (31%), Positives = 37/66 (56%), Gaps = 9/66 (13%)
Query 61 LEKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRM 120
+E+A+A K EGN+L K+ + +A+ KYS +++ SSL+ NR+ HL +
Sbjct 190 VERAKALKEEGNDLVKKGNHKKAIEKYS---ESLLCSSLE------SATYSNRALCHLVL 240
Query 121 ENFGSA 126
+ + A
Sbjct 241 KQYKEA 246
Score = 28.5 bits (62), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 29/57 (50%), Gaps = 3/57 (5%)
Query 64 AEAFKTEGNELFKQHQYAEAVSKYSAAIDTID-ESSLDPKETQLHVYLCNRSFAHLR 119
E + GN+ F+ QYAEA + Y A+ + S DP+E V NR+ +L+
Sbjct 9 VEELRAAGNQSFRNGQYAEASALYERALRLLQARGSADPEEES--VLYSNRAACYLK 63
> hsa:161582 DYX1C1, DYX1, DYXC1, EKN1, FLJ37882, MGC70618, RD;
dyslexia susceptibility 1 candidate 1
Length=376
Score = 33.1 bits (74), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 33/111 (29%), Positives = 47/111 (42%), Gaps = 28/111 (25%)
Query 16 ESLRKEAAKRRDGEMAVDGGSSCAAKEAGDGAERDGKNDALGCSPLEKAEAFKTEGNELF 75
E L K+A RR M D C KE + KN E K +GN+LF
Sbjct 261 EWLHKQAEARR--AMNTDIAELCDLKE-------EEKN----------PEWLKDKGNKLF 301
Query 76 KQHQYAEAVSKYSAAIDTIDESSLDPKETQLHVYLCNRSFAHLRMENFGSA 126
Y A++ Y+ AI ++ L +YL NR+ HL+++N A
Sbjct 302 ATENYLAAINAYNLAIRLNNKMPL--------LYL-NRAACHLKLKNLHKA 343
> hsa:283237 TTC9C, MGC29649; tetratricopeptide repeat domain
9C
Length=171
Score = 33.1 bits (74), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 15/41 (36%), Positives = 28/41 (68%), Gaps = 2/41 (4%)
Query 61 LEKAEAFKTEGNELFKQHQYAEAVSKYSAAIDTIDESSLDP 101
L++A+ +K EGN+ +++ +Y +AVS+Y A+ + LDP
Sbjct 5 LQEAQLYKEEGNQRYREGKYRDAVSRYHRAL--LQLRGLDP 43
Lambda K H
0.308 0.124 0.342
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2059772308
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40