bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0067_orf1
Length=172
Score E
Sequences producing significant alignments: (Bits) Value
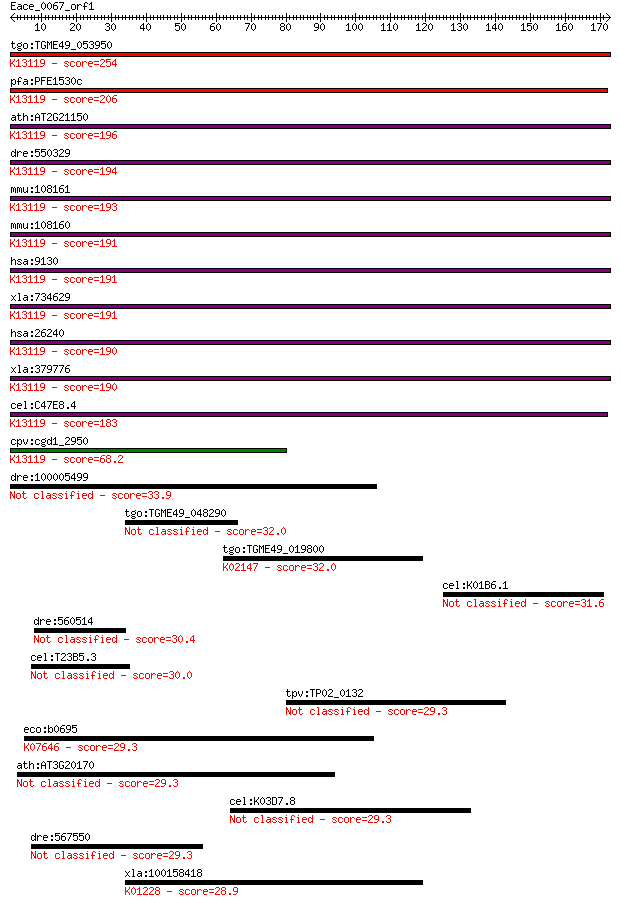
tgo:TGME49_053950 XAP5 protein, putative ; K13119 protein FAM50 254 1e-67
pfa:PFE1530c XAP-5 DNA binding protein, putative; K13119 prote... 206 3e-53
ath:AT2G21150 XCT; XCT (XAP5 CIRCADIAN TIMEKEEPER); K13119 pro... 196 3e-50
dre:550329 fam50a, mg:db03h09, wu:ft29b08, zgc:110236; family ... 194 1e-49
mmu:108161 Fam50b, D0H6S2654E, X5L, XAP-5-like; family with se... 193 2e-49
mmu:108160 Fam50a, D0HXS9928E, XAP-5; family with sequence sim... 191 1e-48
hsa:9130 FAM50A, 9F, DXS9928E, HXC-26, XAP5; family with seque... 191 1e-48
xla:734629 fam50a-b, MGC114853, fam50-b, hxc-26, xap5; family ... 191 1e-48
hsa:26240 FAM50B, D6S2654E, X5L; family with sequence similari... 190 2e-48
xla:379776 fam50a-a, MGC132143, MGC52848, fam50-a, fam50a, hxc... 190 3e-48
cel:C47E8.4 hypothetical protein; K13119 protein FAM50 183 2e-46
cpv:cgd1_2950 hypothetical protein ; K13119 protein FAM50 68.2 1e-11
dre:100005499 hypothetical LOC100005499 33.9 0.29
tgo:TGME49_048290 WD domain-containing protein 32.0 1.1
tgo:TGME49_019800 vacuolar ATP synthase subunit B, putative (E... 32.0 1.1
cel:K01B6.1 fozi-1; FOrm-homology and ZInc finger domains fami... 31.6 1.5
dre:560514 osbpl11, si:dkey-101k6.4; oxysterol binding protein... 30.4 3.0
cel:T23B5.3 hypothetical protein 30.0 4.1
tpv:TP02_0132 hypothetical protein 29.3 6.6
eco:b0695 kdpD, ECK0683, JW0683, kac; fused sensory histidine ... 29.3 6.9
ath:AT3G20170 armadillo/beta-catenin repeat family protein 29.3 6.9
cel:K03D7.8 hypothetical protein 29.3 6.9
dre:567550 novel ubiquitin-protein ligase protein-like 29.3 6.9
xla:100158418 mogs; mannosyl-oligosaccharide glucosidase (EC:3... 28.9 8.7
> tgo:TGME49_053950 XAP5 protein, putative ; K13119 protein FAM50
Length=462
Score = 254 bits (648), Expect = 1e-67, Method: Compositional matrix adjust.
Identities = 114/172 (66%), Positives = 148/172 (86%), Gaps = 0/172 (0%)
Query 1 DTSYLPDKERDAKLAAERQRLIEEYRRKEEEEKRMPLTVTFSYWDGSGHRRRIQILRGLT 60
DT +LPD+ERDA++ AER+RLIEEY R E+E K+ PL +T+S+WDG+GHRRR + +G +
Sbjct 267 DTDFLPDEERDAQILAERKRLIEEYHRLEDEAKKEPLWITYSFWDGTGHRRRTCVRKGSS 326
Query 61 VGEFLERAKRELEKEFVELRSVPTESLMYVKEDLILPHNITFFELIKSKARGKSGPLFHF 120
+ EFL++A++ELEKEF+ELR + LMY+KEDLILPH++TF++LIK+KARGKSGPLF+F
Sbjct 327 ILEFLDQARKELEKEFIELRGIGAIDLMYIKEDLILPHSLTFYDLIKTKARGKSGPLFNF 386
Query 121 DAFDDVRVFNDVRKEKEDSHAGKIVDKKWFERNKHIFPASRWEVFNPTKTYD 172
+DVR+ ND R EKEDSHAGK+V++KWFERNKHIFPASRWEVFNP KTY+
Sbjct 387 SVHEDVRMTNDSRVEKEDSHAGKVVERKWFERNKHIFPASRWEVFNPNKTYE 438
> pfa:PFE1530c XAP-5 DNA binding protein, putative; K13119 protein
FAM50
Length=447
Score = 206 bits (524), Expect = 3e-53, Method: Compositional matrix adjust.
Identities = 92/171 (53%), Positives = 134/171 (78%), Gaps = 0/171 (0%)
Query 1 DTSYLPDKERDAKLAAERQRLIEEYRRKEEEEKRMPLTVTFSYWDGSGHRRRIQILRGLT 60
+TS+L DKERD K+ +++ L E Y + E E+K + +T+SY+DGSGHRR+I + + T
Sbjct 265 NTSFLKDKERDKKIELKKKELRELYYKLENEQKEKVIDITYSYYDGSGHRRKISVKQKNT 324
Query 61 VGEFLERAKRELEKEFVELRSVPTESLMYVKEDLILPHNITFFELIKSKARGKSGPLFHF 120
+G+F+ + L+ EF++LRS E+LM+VKED+ILP+ ITF+ELIK+KA+GK+GPLF F
Sbjct 325 IGQFINKCVDNLKNEFIQLRSASCETLMFVKEDVILPNYITFYELIKNKAQGKTGPLFSF 384
Query 121 DAFDDVRVFNDVRKEKEDSHAGKIVDKKWFERNKHIFPASRWEVFNPTKTY 171
DA +++ D+RKEK D+HAGK+V+KKW+E+NKHIFPAS+WE++ P KTY
Sbjct 385 DAVENLSGITDIRKEKTDTHAGKLVEKKWYEKNKHIFPASKWEIYKPLKTY 435
> ath:AT2G21150 XCT; XCT (XAP5 CIRCADIAN TIMEKEEPER); K13119 protein
FAM50
Length=337
Score = 196 bits (499), Expect = 3e-50, Method: Compositional matrix adjust.
Identities = 93/172 (54%), Positives = 136/172 (79%), Gaps = 0/172 (0%)
Query 1 DTSYLPDKERDAKLAAERQRLIEEYRRKEEEEKRMPLTVTFSYWDGSGHRRRIQILRGLT 60
+T++LPD ER+A+ AER+RL +++ R++E+ K PL +T+SYWDG+GHRR IQ+ +G
Sbjct 159 ETNFLPDSEREAEEQAERERLKKQWLREQEQIKNEPLEITYSYWDGTGHRRVIQVRKGDP 218
Query 61 VGEFLERAKRELEKEFVELRSVPTESLMYVKEDLILPHNITFFELIKSKARGKSGPLFHF 120
+G FL +++L +F E+R+ E+L+YVKEDLI+PH +F+ELI +KARGKSGPLFHF
Sbjct 219 IGNFLRAVQQQLAPDFREIRTASVENLLYVKEDLIIPHQHSFYELIINKARGKSGPLFHF 278
Query 121 DAFDDVRVFNDVRKEKEDSHAGKIVDKKWFERNKHIFPASRWEVFNPTKTYD 172
D +DVR D EK++SHAGK+V++ W+E+NKHIFPASRWE+++PTK ++
Sbjct 279 DVHEDVRTIADATIEKDESHAGKVVERHWYEKNKHIFPASRWEIYDPTKKWE 330
> dre:550329 fam50a, mg:db03h09, wu:ft29b08, zgc:110236; family
with sequence similarity 50, member A; K13119 protein FAM50
Length=341
Score = 194 bits (494), Expect = 1e-49, Method: Compositional matrix adjust.
Identities = 92/172 (53%), Positives = 132/172 (76%), Gaps = 0/172 (0%)
Query 1 DTSYLPDKERDAKLAAERQRLIEEYRRKEEEEKRMPLTVTFSYWDGSGHRRRIQILRGLT 60
DTS+LPD++R+ + R+ L +E+ RK+E+ K + +TFSYWDGSGHR+ +++ +G T
Sbjct 165 DTSFLPDRDREEEENRLREELRQEWERKQEKIKSEEIEITFSYWDGSGHRKTVKMKKGNT 224
Query 61 VGEFLERAKRELEKEFVELRSVPTESLMYVKEDLILPHNITFFELIKSKARGKSGPLFHF 120
+ +FL+RA L K+F ELRS E LMY+KEDLI+PH+ +F++ I +KARGKSGPLF+F
Sbjct 225 IQQFLQRALEVLRKDFSELRSAGVEHLMYIKEDLIIPHHHSFYDFIVTKARGKSGPLFNF 284
Query 121 DAFDDVRVFNDVRKEKEDSHAGKIVDKKWFERNKHIFPASRWEVFNPTKTYD 172
D DD+R+ ND EK++SHAGK+V + W+E+NKHIFPASRWE ++P K +D
Sbjct 285 DVHDDIRLVNDATVEKDESHAGKVVLRSWYEKNKHIFPASRWEPYDPEKKWD 336
> mmu:108161 Fam50b, D0H6S2654E, X5L, XAP-5-like; family with
sequence similarity 50, member B; K13119 protein FAM50
Length=334
Score = 193 bits (491), Expect = 2e-49, Method: Compositional matrix adjust.
Identities = 95/172 (55%), Positives = 129/172 (75%), Gaps = 0/172 (0%)
Query 1 DTSYLPDKERDAKLAAERQRLIEEYRRKEEEEKRMPLTVTFSYWDGSGHRRRIQILRGLT 60
DTS+LPD+ER+ + R+ L +E+ K E+ K + +TFSYWDGSGHRR +++ +G T
Sbjct 158 DTSFLPDREREEEENRLREELRQEWEAKREKVKGEEVEITFSYWDGSGHRRTVRMSKGST 217
Query 61 VGEFLERAKRELEKEFVELRSVPTESLMYVKEDLILPHNITFFELIKSKARGKSGPLFHF 120
V +FL+RA + L ++F ELR+ E LMYVKEDLILPH TF++ I +KARGKSGPLF F
Sbjct 218 VQQFLKRALQGLRRDFRELRAAGVEQLMYVKEDLILPHYHTFYDFIVAKARGKSGPLFSF 277
Query 121 DAFDDVRVFNDVRKEKEDSHAGKIVDKKWFERNKHIFPASRWEVFNPTKTYD 172
D DDVR+ +D EK++SHAGK+V + W+E+NKHIFPASRWE ++P K +D
Sbjct 278 DVHDDVRLLSDATMEKDESHAGKVVLRSWYEKNKHIFPASRWEPYDPEKKWD 329
> mmu:108160 Fam50a, D0HXS9928E, XAP-5; family with sequence similarity
50, member A; K13119 protein FAM50
Length=339
Score = 191 bits (485), Expect = 1e-48, Method: Compositional matrix adjust.
Identities = 91/172 (52%), Positives = 131/172 (76%), Gaps = 0/172 (0%)
Query 1 DTSYLPDKERDAKLAAERQRLIEEYRRKEEEEKRMPLTVTFSYWDGSGHRRRIQILRGLT 60
DTS+LPD++R+ + R+ L +E+ K+E+ K + +TFSYWDGSGHRR +++ +G T
Sbjct 163 DTSFLPDRDREEEENRLREELRQEWEAKQEKIKSEEIEITFSYWDGSGHRRTVKMKKGNT 222
Query 61 VGEFLERAKRELEKEFVELRSVPTESLMYVKEDLILPHNITFFELIKSKARGKSGPLFHF 120
+ +FL++A L K+F ELRS E LMY+KEDLI+PH+ +F++ I +KARGKSGPLF+F
Sbjct 223 MQQFLQKALEILRKDFSELRSAGVEQLMYIKEDLIIPHHHSFYDFIVTKARGKSGPLFNF 282
Query 121 DAFDDVRVFNDVRKEKEDSHAGKIVDKKWFERNKHIFPASRWEVFNPTKTYD 172
D DDVR+ +D EK++SHAGK+V + W+E+NKHIFPASRWE ++P K +D
Sbjct 283 DVHDDVRLLSDATVEKDESHAGKVVLRSWYEKNKHIFPASRWEPYDPEKKWD 334
> hsa:9130 FAM50A, 9F, DXS9928E, HXC-26, XAP5; family with sequence
similarity 50, member A; K13119 protein FAM50
Length=339
Score = 191 bits (485), Expect = 1e-48, Method: Compositional matrix adjust.
Identities = 91/172 (52%), Positives = 131/172 (76%), Gaps = 0/172 (0%)
Query 1 DTSYLPDKERDAKLAAERQRLIEEYRRKEEEEKRMPLTVTFSYWDGSGHRRRIQILRGLT 60
DTS+LPD++R+ + R+ L +E+ K+E+ K + +TFSYWDGSGHRR +++ +G T
Sbjct 163 DTSFLPDRDREEEENRLREELRQEWEAKQEKIKSEEIEITFSYWDGSGHRRTVKMRKGNT 222
Query 61 VGEFLERAKRELEKEFVELRSVPTESLMYVKEDLILPHNITFFELIKSKARGKSGPLFHF 120
+ +FL++A L K+F ELRS E LMY+KEDLI+PH+ +F++ I +KARGKSGPLF+F
Sbjct 223 MQQFLQKALEILRKDFSELRSAGVEQLMYIKEDLIIPHHHSFYDFIVTKARGKSGPLFNF 282
Query 121 DAFDDVRVFNDVRKEKEDSHAGKIVDKKWFERNKHIFPASRWEVFNPTKTYD 172
D DDVR+ +D EK++SHAGK+V + W+E+NKHIFPASRWE ++P K +D
Sbjct 283 DVHDDVRLLSDATVEKDESHAGKVVLRSWYEKNKHIFPASRWEPYDPEKKWD 334
> xla:734629 fam50a-b, MGC114853, fam50-b, hxc-26, xap5; family
with sequence similarity 50, member A; K13119 protein FAM50
Length=343
Score = 191 bits (484), Expect = 1e-48, Method: Compositional matrix adjust.
Identities = 90/172 (52%), Positives = 132/172 (76%), Gaps = 0/172 (0%)
Query 1 DTSYLPDKERDAKLAAERQRLIEEYRRKEEEEKRMPLTVTFSYWDGSGHRRRIQILRGLT 60
DTS+LPD++R+ + R+ L +E+ RK+E+ K + +TFSYWDGSGHRR +++ +G +
Sbjct 167 DTSFLPDRDREEEENRLREELRQEWERKQEKIKSEEIEITFSYWDGSGHRRTVKMKKGNS 226
Query 61 VGEFLERAKRELEKEFVELRSVPTESLMYVKEDLILPHNITFFELIKSKARGKSGPLFHF 120
+ +FL++A L K+F ELRS E LMY+KEDLI+PH+ +F++ I +KARGKSGPLF+F
Sbjct 227 IQQFLQKALEILRKDFSELRSAGVEQLMYIKEDLIIPHHHSFYDFIVTKARGKSGPLFNF 286
Query 121 DAFDDVRVFNDVRKEKEDSHAGKIVDKKWFERNKHIFPASRWEVFNPTKTYD 172
D +DVR+ +D EK++SHAGK+V + W+E+NKHIFPASRWE ++P K +D
Sbjct 287 DVHEDVRLLSDASVEKDESHAGKVVLRSWYEKNKHIFPASRWEPYDPEKKWD 338
> hsa:26240 FAM50B, D6S2654E, X5L; family with sequence similarity
50, member B; K13119 protein FAM50
Length=325
Score = 190 bits (483), Expect = 2e-48, Method: Compositional matrix adjust.
Identities = 92/172 (53%), Positives = 130/172 (75%), Gaps = 0/172 (0%)
Query 1 DTSYLPDKERDAKLAAERQRLIEEYRRKEEEEKRMPLTVTFSYWDGSGHRRRIQILRGLT 60
DTS+LPD++R+ + R+ L +E+ + E+ K + VTFSYWDGSGHRR +++ +G T
Sbjct 149 DTSFLPDRDREEEENRLREELRQEWEAQREKVKDEEMEVTFSYWDGSGHRRTVRVRKGNT 208
Query 61 VGEFLERAKRELEKEFVELRSVPTESLMYVKEDLILPHNITFFELIKSKARGKSGPLFHF 120
V +FL++A + L K+F+ELRS E LM++KEDLILPH TF++ I ++ARGKSGPLF F
Sbjct 209 VQQFLKKALQGLRKDFLELRSAGVEQLMFIKEDLILPHYHTFYDFIIARARGKSGPLFSF 268
Query 121 DAFDDVRVFNDVRKEKEDSHAGKIVDKKWFERNKHIFPASRWEVFNPTKTYD 172
D DDVR+ +D EK++SHAGK+V + W+E+NKHIFPASRWE ++P K +D
Sbjct 269 DVHDDVRLLSDATMEKDESHAGKVVLRSWYEKNKHIFPASRWEAYDPEKKWD 320
> xla:379776 fam50a-a, MGC132143, MGC52848, fam50-a, fam50a, hxc-26,
mg:db03h09, xap5; family with sequence similarity 50,
member A; K13119 protein FAM50
Length=340
Score = 190 bits (482), Expect = 3e-48, Method: Compositional matrix adjust.
Identities = 89/172 (51%), Positives = 131/172 (76%), Gaps = 0/172 (0%)
Query 1 DTSYLPDKERDAKLAAERQRLIEEYRRKEEEEKRMPLTVTFSYWDGSGHRRRIQILRGLT 60
DTS+LPD++R+ + R+ L +E+ K+E+ K + +TFSYWDGSGHRR +++ +G +
Sbjct 164 DTSFLPDRDREEEENRLREELRQEWEHKQEKIKSEEIEITFSYWDGSGHRRTVKMKKGNS 223
Query 61 VGEFLERAKRELEKEFVELRSVPTESLMYVKEDLILPHNITFFELIKSKARGKSGPLFHF 120
+ +FL++A L K+F ELRS E LMY+KEDLI+PH+ +F++ I +KARGKSGPLF+F
Sbjct 224 IQQFLQKALESLRKDFSELRSAGVEQLMYIKEDLIIPHHHSFYDFIVTKARGKSGPLFNF 283
Query 121 DAFDDVRVFNDVRKEKEDSHAGKIVDKKWFERNKHIFPASRWEVFNPTKTYD 172
D +DVR+ +D EK++SHAGK+V + W+E+NKHIFPASRWE ++P K +D
Sbjct 284 DVHEDVRLLSDATVEKDESHAGKVVLRSWYEKNKHIFPASRWEPYDPEKKWD 335
> cel:C47E8.4 hypothetical protein; K13119 protein FAM50
Length=351
Score = 183 bits (465), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 87/171 (50%), Positives = 121/171 (70%), Gaps = 0/171 (0%)
Query 1 DTSYLPDKERDAKLAAERQRLIEEYRRKEEEEKRMPLTVTFSYWDGSGHRRRIQILRGLT 60
DTS+LPDKER+ L +++ L E+R K++ EK +TV ++YWDGS HR+ ++I +G T
Sbjct 171 DTSFLPDKEREEFLRKKKESLAAEWRVKQDAEKNEEITVAYAYWDGSSHRKNMKIKKGNT 230
Query 61 VGEFLERAKRELEKEFVELRSVPTESLMYVKEDLILPHNITFFELIKSKARGKSGPLFHF 120
+ + L RA L+KEF EL+S E+LM+VKEDLI+PH TF + I +KA GK+GPLF F
Sbjct 231 ISQCLGRAIEALKKEFTELKSCTAENLMFVKEDLIIPHFYTFQDFIVTKAMGKTGPLFVF 290
Query 121 DAFDDVRVFNDVRKEKEDSHAGKIVDKKWFERNKHIFPASRWEVFNPTKTY 171
D+ DVR+ D + +SH KIV + W+E+NKHI+PASRWE F P+K Y
Sbjct 291 DSASDVRIRQDAALDYGESHPAKIVLRSWYEKNKHIYPASRWEPFVPSKKY 341
> cpv:cgd1_2950 hypothetical protein ; K13119 protein FAM50
Length=216
Score = 68.2 bits (165), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 39/79 (49%), Positives = 56/79 (70%), Gaps = 0/79 (0%)
Query 1 DTSYLPDKERDAKLAAERQRLIEEYRRKEEEEKRMPLTVTFSYWDGSGHRRRIQILRGLT 60
DTS+LPDK+R+ + A R++L +E KEE+ K+ + V FSYWDGSGHRR + + R T
Sbjct 133 DTSFLPDKKRELEEAKAREKLKQEEFEKEEKLKQEIIEVVFSYWDGSGHRRSVHVPRNTT 192
Query 61 VGEFLERAKRELEKEFVEL 79
+GEFLE+ + +L+ EF E
Sbjct 193 IGEFLEKCRIKLKSEFKEF 211
> dre:100005499 hypothetical LOC100005499
Length=1336
Score = 33.9 bits (76), Expect = 0.29, Method: Composition-based stats.
Identities = 33/118 (27%), Positives = 52/118 (44%), Gaps = 23/118 (19%)
Query 1 DTSYLPD---KERDAKLAAERQRLIEEYRRKEEE----EKRMPLTVTFSYWDGSGHRRRI 53
+ +YLP E L ER L+ E R+K + EK + TFSY RR
Sbjct 1096 EVNYLPPFPFGETGESLEKERLDLLNEIRKKNNKNIIGEK---MEKTFSY-------RRT 1145
Query 54 QILRGL-TVGEFLER-----AKRELEKEFVELRSVPTESLMYVKEDLILPHNITFFEL 105
++++ V +FLER + E++ EF + ++ E K D P + FE+
Sbjct 1146 EVVKDCPAVKDFLERWPALFCESEIKNEFRRITTISLERTFLEKLDFYTPKLLALFEM 1203
> tgo:TGME49_048290 WD domain-containing protein
Length=3101
Score = 32.0 bits (71), Expect = 1.1, Method: Composition-based stats.
Identities = 16/32 (50%), Positives = 16/32 (50%), Gaps = 0/32 (0%)
Query 34 RMPLTVTFSYWDGSGHRRRIQILRGLTVGEFL 65
R PLTV W S HR I LR GEFL
Sbjct 119 RPPLTVATCVWAASEHRSEIMTLRWAPTGEFL 150
> tgo:TGME49_019800 vacuolar ATP synthase subunit B, putative
(EC:3.6.3.15); K02147 V-type H+-transporting ATPase subunit
B [EC:3.6.3.14]
Length=505
Score = 32.0 bits (71), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 15/57 (26%), Positives = 29/57 (50%), Gaps = 0/57 (0%)
Query 62 GEFLERAKRELEKEFVELRSVPTESLMYVKEDLILPHNITFFELIKSKARGKSGPLF 118
G+ ++R R L +EF+++ P V ++ I+ +++ S RG+ PLF
Sbjct 119 GKPIDRGPRVLAEEFLDINGCPINPQCRVYPKEMIQTGISAIDVMNSVVRGQKIPLF 175
> cel:K01B6.1 fozi-1; FOrm-homology and ZInc finger domains family
member (fozi-1)
Length=732
Score = 31.6 bits (70), Expect = 1.5, Method: Composition-based stats.
Identities = 18/48 (37%), Positives = 22/48 (45%), Gaps = 2/48 (4%)
Query 125 DVRVFND--VRKEKEDSHAGKIVDKKWFERNKHIFPASRWEVFNPTKT 170
D+ V +D VR EKE H+G + F N RWE F KT
Sbjct 630 DLMVLDDKTVRAEKEMEHSGSNIPLSEFVENAKTISKERWEHFKSLKT 677
> dre:560514 osbpl11, si:dkey-101k6.4; oxysterol binding protein-like
11
Length=712
Score = 30.4 bits (67), Expect = 3.0, Method: Composition-based stats.
Identities = 13/26 (50%), Positives = 17/26 (65%), Gaps = 0/26 (0%)
Query 8 KERDAKLAAERQRLIEEYRRKEEEEK 33
KERD A E +R +EE +RKEE +
Sbjct 645 KERDMDKATEHKRFLEERQRKEERHR 670
> cel:T23B5.3 hypothetical protein
Length=884
Score = 30.0 bits (66), Expect = 4.1, Method: Composition-based stats.
Identities = 15/28 (53%), Positives = 17/28 (60%), Gaps = 0/28 (0%)
Query 7 DKERDAKLAAERQRLIEEYRRKEEEEKR 34
D+ER L ERQR + E RR EEE R
Sbjct 324 DRERQEMLEVERQRRVLEDRRNSEEEIR 351
> tpv:TP02_0132 hypothetical protein
Length=642
Score = 29.3 bits (64), Expect = 6.6, Method: Composition-based stats.
Identities = 15/63 (23%), Positives = 31/63 (49%), Gaps = 0/63 (0%)
Query 80 RSVPTESLMYVKEDLILPHNITFFELIKSKARGKSGPLFHFDAFDDVRVFNDVRKEKEDS 139
R++ + LM KE +I+P N ++KS+ +S +FD + + F+ +
Sbjct 343 RNIGFQELMKAKEGVIIPKNCVNHYIVKSQILSESNEEINFDMSNYLSYFDSTVSTEASP 402
Query 140 HAG 142
++G
Sbjct 403 YSG 405
> eco:b0695 kdpD, ECK0683, JW0683, kac; fused sensory histidine
kinase in two-component regulatory system with KdpE: signal
sensing protein; K07646 two-component system, OmpR family,
sensor histidine kinase KdpD [EC:2.7.13.3]
Length=894
Score = 29.3 bits (64), Expect = 6.9, Method: Composition-based stats.
Identities = 30/120 (25%), Positives = 51/120 (42%), Gaps = 20/120 (16%)
Query 5 LPDKERDAKLAAERQRLIEEYRRKEEEEKRMPLTVTFSYW---------DGSGHRRRIQI 55
L E A++A+ER+++ + R PLTV F +GS H R+
Sbjct 647 LTASEEQARMASEREQIRNALLAALSHDLRTPLTVLFGQAEILTLDLASEGSPHARQASE 706
Query 56 LRGLT------VGEFLERAKRE-----LEKEFVELRSVPTESLMYVKEDLILPHNITFFE 104
+R V L+ A+ + L+KE++ L V +L ++ L P N++ E
Sbjct 707 IRQHVLNTTRLVNNLLDMARIQSGGFNLKKEWLTLEEVVGSALQMLEPGLSSPINLSLPE 766
> ath:AT3G20170 armadillo/beta-catenin repeat family protein
Length=475
Score = 29.3 bits (64), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 26/91 (28%), Positives = 38/91 (41%), Gaps = 5/91 (5%)
Query 3 SYLPDKERDAKLAAERQRLIEEYRRKEEEEKRMPLTVTFSYWDGSGHRRRIQILRGLTVG 62
L E +A L AE+ L+ R + E K L + WD +G+R + ++RG
Sbjct 300 CILAVAEGNAVLIAEQ--LVRILRAGDNEAK---LAASDVLWDLAGYRHSVSVIRGSGAI 354
Query 63 EFLERAKRELEKEFVELRSVPTESLMYVKED 93
L R+ EF E S L Y + D
Sbjct 355 PLLIELLRDGSLEFRERISGAISQLSYNEND 385
> cel:K03D7.8 hypothetical protein
Length=408
Score = 29.3 bits (64), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 24/73 (32%), Positives = 37/73 (50%), Gaps = 8/73 (10%)
Query 64 FLERAKRELEKEFVELRSVPTESLMYVKEDLILP----HNITFFELIKSKARGKSGPLFH 119
+L R EL + +E+ + SL+Y+KE +L +NITF L R KS P +
Sbjct 12 YLHRENVELVQGKLEVYEI---SLLYIKEMALLVLNVLNNITFISLFIYNRRRKSQPSSY 68
Query 120 FDAFDDVRVFNDV 132
FD + +FN +
Sbjct 69 RSEFDTL-IFNQM 80
> dre:567550 novel ubiquitin-protein ligase protein-like
Length=731
Score = 29.3 bits (64), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 14/49 (28%), Positives = 29/49 (59%), Gaps = 0/49 (0%)
Query 7 DKERDAKLAAERQRLIEEYRRKEEEEKRMPLTVTFSYWDGSGHRRRIQI 55
D+E + L A+++++I+E + EE + + F++ DG+ RR Q+
Sbjct 79 DQEYEQSLRADQEKIIQERLMRTNEEPADGIPLKFTFPDGTMKIRRFQV 127
> xla:100158418 mogs; mannosyl-oligosaccharide glucosidase (EC:3.2.1.106);
K01228 mannosyl-oligosaccharide glucosidase [EC:3.2.1.106]
Length=826
Score = 28.9 bits (63), Expect = 8.7, Method: Composition-based stats.
Identities = 22/85 (25%), Positives = 38/85 (44%), Gaps = 9/85 (10%)
Query 34 RMPLTVTFSYWDGSGHRRRIQILRGLTVGEFLERAKRELEKEFVELRSVPTESLMYVKED 93
++PL V + GS +R Q L GL++ + LER K +E F + + ++E
Sbjct 315 QLPLQVEVVFESGSFQQRLGQ-LSGLSLSKELERHKSNMENRF--------KGIFRLEEK 365
Query 94 LILPHNITFFELIKSKARGKSGPLF 118
+P + F + S G G +
Sbjct 366 GFIPDQVAFAQAALSNMLGGMGYFY 390
Lambda K H
0.319 0.137 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4341553636
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40