bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0084_orf2
Length=168
Score E
Sequences producing significant alignments: (Bits) Value
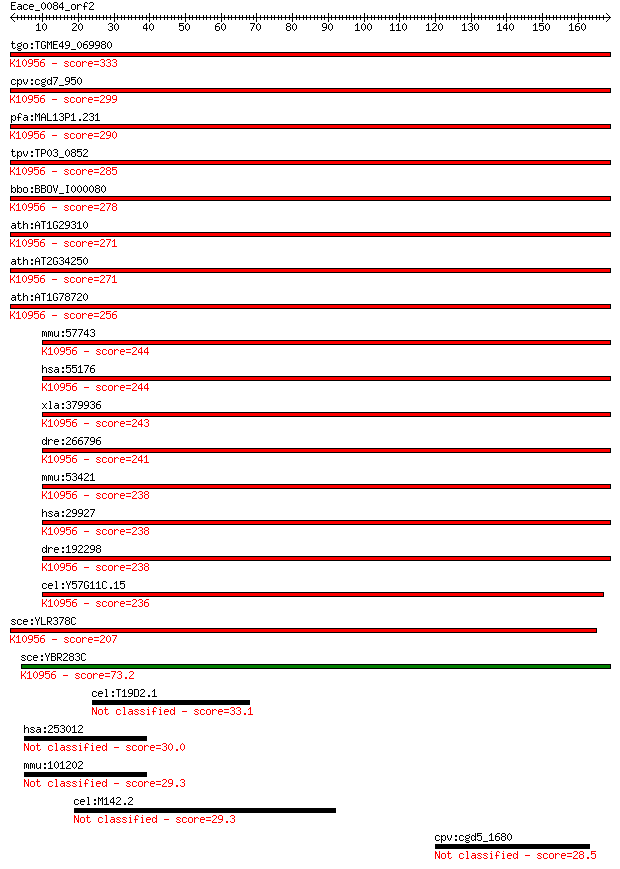
tgo:TGME49_069980 protein transport protein Sec61 alpha subuni... 333 2e-91
cpv:cgd7_950 Sec61; signal peptide plus 9 transmembrane domain... 299 3e-81
pfa:MAL13P1.231 sec61, PF13_0244; Sec61 alpha subunit, PfSec61... 290 2e-78
tpv:TP03_0852 protein transport protein Sec61; K10956 protein ... 285 6e-77
bbo:BBOV_I000080 16.m00785; protein transport protein sec61; K... 278 4e-75
ath:AT1G29310 P-P-bond-hydrolysis-driven protein transmembrane... 271 9e-73
ath:AT2G34250 protein transport protein sec61, putative; K1095... 271 9e-73
ath:AT1G78720 protein transport protein sec61, putative; K1095... 256 3e-68
mmu:57743 Sec61a2; Sec61, alpha subunit 2 (S. cerevisiae); K10... 244 7e-65
hsa:55176 SEC61A2, FLJ10578; Sec61 alpha 2 subunit (S. cerevis... 244 1e-64
xla:379936 sec61a1, MGC53497, sec61, sec61a, sec61alpha; Sec61... 243 2e-64
dre:266796 sec61al2, cb413, sec61b, wu:fb36f05, wu:fb36f12, wu... 241 8e-64
mmu:53421 Sec61a1, AA408394, AA410007, Sec61a; Sec61 alpha 1 s... 238 7e-63
hsa:29927 SEC61A1, HSEC61, SEC61, SEC61A; Sec61 alpha 1 subuni... 238 7e-63
dre:192298 sec61al1, chunp6898, fb62c11, sec61a, wu:fb62c11; S... 238 1e-62
cel:Y57G11C.15 hypothetical protein; K10956 protein transport ... 236 3e-62
sce:YLR378C SEC61; Essential subunit of Sec61 complex (Sec61p,... 207 2e-53
sce:YBR283C SSH1; Ssh1p; K10956 protein transport protein SEC6... 73.2 4e-13
cel:T19D2.1 hypothetical protein 33.1 0.47
hsa:253012 HEPACAM2; HEPACAM family member 2 30.0
mmu:101202 Hepacam2, AI987662; HEPACAM family member 2 29.3
cel:M142.2 cut-6; CUTiclin family member (cut-6) 29.3 7.0
cpv:cgd5_1680 protein with an N-terminal domain shared with th... 28.5 9.9
> tgo:TGME49_069980 protein transport protein Sec61 alpha subunit
isoform 1, putative ; K10956 protein transport protein SEC61
subunit alpha
Length=473
Score = 333 bits (854), Expect = 2e-91, Method: Compositional matrix adjust.
Identities = 164/168 (97%), Positives = 166/168 (98%), Gaps = 0/168 (0%)
Query 1 VVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCL 60
VVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCL
Sbjct 164 VVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCL 223
Query 61 FTKSTNMVALKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGNYP 120
FTKS N+VALKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQG+YP
Sbjct 224 FTKSNNIVALKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGSYP 283
Query 121 IKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKNNVLVNLLGQWQEVD 168
IKLFYTSNIPIILQTALVSNLYFLSQLLYRRFK NVLVNLLGQWQEVD
Sbjct 284 IKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKTNVLVNLLGQWQEVD 331
> cpv:cgd7_950 Sec61; signal peptide plus 9 transmembrane domain-containing
protein ; K10956 protein transport protein SEC61
subunit alpha
Length=473
Score = 299 bits (766), Expect = 3e-81, Method: Compositional matrix adjust.
Identities = 142/168 (84%), Positives = 160/168 (95%), Gaps = 0/168 (0%)
Query 1 VVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCL 60
VVI+LDEL+QKGYGLGSGISLFIATNICETIVWKAFSPTTI TG+GTEFEGA++ALFH L
Sbjct 164 VVILLDELMQKGYGLGSGISLFIATNICETIVWKAFSPTTINTGRGTEFEGAVIALFHLL 223
Query 61 FTKSTNMVALKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGNYP 120
FTK + AL+EAFYRS+A N+T+LLAT+LVFLIVIYFQGFRVDLAVKYQ+VRGQQG++P
Sbjct 224 FTKPDKISALREAFYRSHATNMTNLLATVLVFLIVIYFQGFRVDLAVKYQKVRGQQGSFP 283
Query 121 IKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKNNVLVNLLGQWQEVD 168
IKLFYTSNIPIILQTALVSNLYF SQLLYRRFK+N+LVN+LGQWQE+D
Sbjct 284 IKLFYTSNIPIILQTALVSNLYFFSQLLYRRFKSNMLVNILGQWQELD 331
> pfa:MAL13P1.231 sec61, PF13_0244; Sec61 alpha subunit, PfSec61;
K10956 protein transport protein SEC61 subunit alpha
Length=472
Score = 290 bits (742), Expect = 2e-78, Method: Compositional matrix adjust.
Identities = 137/168 (81%), Positives = 157/168 (93%), Gaps = 0/168 (0%)
Query 1 VVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCL 60
VVI+LDELLQKGYGLGSGISLFIATNICETI+WK+FSPTTI T KG EFEGA+++L +CL
Sbjct 161 VVILLDELLQKGYGLGSGISLFIATNICETIMWKSFSPTTINTDKGIEFEGAIISLIYCL 220
Query 61 FTKSTNMVALKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGNYP 120
FT+S + ALK+AFYR++APN+T+LLATILVFLIVIY QGFRVDL+VKYQ VRGQQG YP
Sbjct 221 FTESNKISALKKAFYRTHAPNVTNLLATILVFLIVIYLQGFRVDLSVKYQSVRGQQGTYP 280
Query 121 IKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKNNVLVNLLGQWQEVD 168
IKLFYTSNIPIILQTALVSNLYF SQ+LY+RFKN++LVNLLGQWQEV+
Sbjct 281 IKLFYTSNIPIILQTALVSNLYFFSQILYKRFKNSILVNLLGQWQEVE 328
> tpv:TP03_0852 protein transport protein Sec61; K10956 protein
transport protein SEC61 subunit alpha
Length=476
Score = 285 bits (728), Expect = 6e-77, Method: Compositional matrix adjust.
Identities = 132/168 (78%), Positives = 151/168 (89%), Gaps = 0/168 (0%)
Query 1 VVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCL 60
VVI+ DE+LQKGYGLGSGISLFIATNICETI+WKAFSPTTI T KGTEFEGAL++LF+C
Sbjct 164 VVILFDEMLQKGYGLGSGISLFIATNICETILWKAFSPTTISTDKGTEFEGALISLFYCF 223
Query 61 FTKSTNMVALKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGNYP 120
FTK + A KEAFYR++APN+T+LLAT L+F+IVIY QGFRVDL+VKYQ +RGQ+G YP
Sbjct 224 FTKKNKLSAFKEAFYRNHAPNVTNLLATALIFVIVIYLQGFRVDLSVKYQSMRGQRGTYP 283
Query 121 IKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKNNVLVNLLGQWQEVD 168
IKLFYTSNIPIILQTALVSNLYF SQL+YR+FKNN+ NLLGQWQE D
Sbjct 284 IKLFYTSNIPIILQTALVSNLYFFSQLVYRKFKNNLFANLLGQWQETD 331
> bbo:BBOV_I000080 16.m00785; protein transport protein sec61;
K10956 protein transport protein SEC61 subunit alpha
Length=480
Score = 278 bits (712), Expect = 4e-75, Method: Compositional matrix adjust.
Identities = 127/168 (75%), Positives = 148/168 (88%), Gaps = 0/168 (0%)
Query 1 VVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCL 60
+VI+ DE+LQKGYGLGSGISLFIATNICETI+WKAFSPTTI T KGTEFEGAL++LF+C
Sbjct 168 IVILFDEMLQKGYGLGSGISLFIATNICETILWKAFSPTTISTDKGTEFEGALISLFYCF 227
Query 61 FTKSTNMVALKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGNYP 120
FTK + A ++AFYRS+APN+T+LLAT L+F IVIY QGFRVDL +KYQ +RGQ+ YP
Sbjct 228 FTKGNKLSAFRDAFYRSHAPNVTNLLATALIFTIVIYLQGFRVDLPIKYQNMRGQRSTYP 287
Query 121 IKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKNNVLVNLLGQWQEVD 168
IKLFYTSNIPIILQTALVSNLYF SQL+YRRFKNN+ N+LGQWQE +
Sbjct 288 IKLFYTSNIPIILQTALVSNLYFFSQLIYRRFKNNLFANILGQWQETE 335
> ath:AT1G29310 P-P-bond-hydrolysis-driven protein transmembrane
transporter; K10956 protein transport protein SEC61 subunit
alpha
Length=475
Score = 271 bits (692), Expect = 9e-73, Method: Compositional matrix adjust.
Identities = 126/168 (75%), Positives = 148/168 (88%), Gaps = 0/168 (0%)
Query 1 VVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCL 60
+VI LDELLQKGYGLGSGISLFIATNICE+I+WKAFSPTTI TG+G EFEGA++ALFH L
Sbjct 164 IVICLDELLQKGYGLGSGISLFIATNICESIIWKAFSPTTINTGRGAEFEGAVIALFHML 223
Query 61 FTKSTNMVALKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGNYP 120
TKS + AL++AFYR N PN+T+LLAT+L+FLIVIYFQGFRV L V+ + RGQQG+YP
Sbjct 224 ITKSNKVAALRQAFYRQNLPNVTNLLATVLIFLIVIYFQGFRVVLPVRSKSARGQQGSYP 283
Query 121 IKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKNNVLVNLLGQWQEVD 168
IKLFYTSN+PIILQ+ALVSNLYF+SQLLYR+F N VNLLGQW+E +
Sbjct 284 IKLFYTSNMPIILQSALVSNLYFISQLLYRKFSGNFFVNLLGQWKESE 331
> ath:AT2G34250 protein transport protein sec61, putative; K10956
protein transport protein SEC61 subunit alpha
Length=475
Score = 271 bits (692), Expect = 9e-73, Method: Compositional matrix adjust.
Identities = 126/168 (75%), Positives = 148/168 (88%), Gaps = 0/168 (0%)
Query 1 VVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCL 60
+VI LDELLQKGYGLGSGISLFIATNICE+I+WKAFSPTTI TG+G EFEGA++ALFH L
Sbjct 164 IVICLDELLQKGYGLGSGISLFIATNICESIIWKAFSPTTINTGRGAEFEGAVIALFHML 223
Query 61 FTKSTNMVALKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGNYP 120
TKS + AL++AFYR N PN+T+LLAT+L+FLIVIYFQGFRV L V+ + RGQQG+YP
Sbjct 224 ITKSNKVAALRQAFYRQNLPNVTNLLATVLIFLIVIYFQGFRVVLPVRSKNARGQQGSYP 283
Query 121 IKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKNNVLVNLLGQWQEVD 168
IKLFYTSN+PIILQ+ALVSNLYF+SQLLYR+F N VNLLGQW+E +
Sbjct 284 IKLFYTSNMPIILQSALVSNLYFISQLLYRKFSGNFFVNLLGQWKESE 331
> ath:AT1G78720 protein transport protein sec61, putative; K10956
protein transport protein SEC61 subunit alpha
Length=475
Score = 256 bits (653), Expect = 3e-68, Method: Compositional matrix adjust.
Identities = 116/168 (69%), Positives = 147/168 (87%), Gaps = 0/168 (0%)
Query 1 VVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCL 60
+V+ LDELLQKGYGLGSGISLFIATNICE+I+WKAFSPTTI +G+G +FEGA++ALFH L
Sbjct 164 IVLCLDELLQKGYGLGSGISLFIATNICESIIWKAFSPTTINSGRGAQFEGAVIALFHLL 223
Query 61 FTKSTNMVALKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGNYP 120
T++ + AL+EAF+R N PN+T+L AT+L+FLIVIYFQGFRV L V+ + RGQ+G+YP
Sbjct 224 ITRTDKVRALREAFFRQNLPNVTNLHATVLIFLIVIYFQGFRVVLPVRSKNARGQRGSYP 283
Query 121 IKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKNNVLVNLLGQWQEVD 168
IKLFYTSN+PIILQ+ALVSN+YF+SQ+LYR+F N LVNL+G W+E +
Sbjct 284 IKLFYTSNMPIILQSALVSNIYFISQILYRKFGGNFLVNLIGTWKESE 331
> mmu:57743 Sec61a2; Sec61, alpha subunit 2 (S. cerevisiae); K10956
protein transport protein SEC61 subunit alpha
Length=476
Score = 244 bits (624), Expect = 7e-65, Method: Compositional matrix adjust.
Identities = 116/159 (72%), Positives = 134/159 (84%), Gaps = 0/159 (0%)
Query 10 QKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCLFTKSTNMVA 69
QKGYGLGSGISLFIATNICETIVWKAFSPTTI TG+GTEFEGA++ALFH L T++ + A
Sbjct 170 QKGYGLGSGISLFIATNICETIVWKAFSPTTINTGRGTEFEGAVIALFHLLATRTDKVRA 229
Query 70 LKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGNYPIKLFYTSNI 129
L+EAFYR N PN+ +L+AT+ VF +VIYFQGFRVDL +K R RGQ +YPIKLFYTSNI
Sbjct 230 LREAFYRQNLPNLMNLIATVFVFAVVIYFQGFRVDLPIKSARYRGQYSSYPIKLFYTSNI 289
Query 130 PIILQTALVSNLYFLSQLLYRRFKNNVLVNLLGQWQEVD 168
PIILQ+ALVSNLY +SQ+L RF N LVNLLGQW +V
Sbjct 290 PIILQSALVSNLYVISQMLSVRFSGNFLVNLLGQWADVS 328
> hsa:55176 SEC61A2, FLJ10578; Sec61 alpha 2 subunit (S. cerevisiae);
K10956 protein transport protein SEC61 subunit alpha
Length=437
Score = 244 bits (623), Expect = 1e-64, Method: Compositional matrix adjust.
Identities = 116/159 (72%), Positives = 134/159 (84%), Gaps = 0/159 (0%)
Query 10 QKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCLFTKSTNMVA 69
QKGYGLGSGISLFIATNICETIVWKAFSPTTI TG+GTEFEGA++ALFH L T++ + A
Sbjct 170 QKGYGLGSGISLFIATNICETIVWKAFSPTTINTGRGTEFEGAVIALFHLLATRTDKVRA 229
Query 70 LKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGNYPIKLFYTSNI 129
L+EAFYR N PN+ +L+AT+ VF +VIYFQGFRVDL +K R RGQ +YPIKLFYTSNI
Sbjct 230 LREAFYRQNLPNLMNLIATVFVFAVVIYFQGFRVDLPIKSARYRGQYSSYPIKLFYTSNI 289
Query 130 PIILQTALVSNLYFLSQLLYRRFKNNVLVNLLGQWQEVD 168
PIILQ+ALVSNLY +SQ+L RF N LVNLLGQW +V
Sbjct 290 PIILQSALVSNLYVISQMLSVRFSGNFLVNLLGQWADVS 328
> xla:379936 sec61a1, MGC53497, sec61, sec61a, sec61alpha; Sec61
alpha 1 subunit; K10956 protein transport protein SEC61 subunit
alpha
Length=476
Score = 243 bits (621), Expect = 2e-64, Method: Compositional matrix adjust.
Identities = 115/159 (72%), Positives = 133/159 (83%), Gaps = 0/159 (0%)
Query 10 QKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCLFTKSTNMVA 69
QKGYGLGSGISLFIATNICETIVWKAFSPTT+ TG+GTEFEGA++ALFH L T+S + A
Sbjct 170 QKGYGLGSGISLFIATNICETIVWKAFSPTTVNTGRGTEFEGAIIALFHLLATRSDKVRA 229
Query 70 LKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGNYPIKLFYTSNI 129
L+EAFYR N PN+T+L+ATI VF +VIYFQGFRVDL +K R RGQ YPIKLFYTSNI
Sbjct 230 LREAFYRQNLPNLTNLIATIFVFAVVIYFQGFRVDLPIKSARYRGQYNTYPIKLFYTSNI 289
Query 130 PIILQTALVSNLYFLSQLLYRRFKNNVLVNLLGQWQEVD 168
PIILQ+ALVSNLY +SQ+L RF N+LV+LLG W +
Sbjct 290 PIILQSALVSNLYVISQMLSARFSGNLLVSLLGTWSDTS 328
> dre:266796 sec61al2, cb413, sec61b, wu:fb36f05, wu:fb36f12,
wu:fb37d04, wu:fi26f11; Sec61 alpha like 2; K10956 protein transport
protein SEC61 subunit alpha
Length=476
Score = 241 bits (615), Expect = 8e-64, Method: Compositional matrix adjust.
Identities = 114/159 (71%), Positives = 131/159 (82%), Gaps = 0/159 (0%)
Query 10 QKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCLFTKSTNMVA 69
QKGYGLGSGISLFIATNICETIVWKAFSPTT+ TG+GTEFEGA++ALFH L T++ + A
Sbjct 170 QKGYGLGSGISLFIATNICETIVWKAFSPTTVNTGRGTEFEGAIIALFHLLATRTDKVRA 229
Query 70 LKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGNYPIKLFYTSNI 129
L+EAFYR N PN+ +L+ATI VF +VIYFQGFRVDL +K R RGQ YPIKLFYTSNI
Sbjct 230 LREAFYRQNLPNLMNLIATIFVFAVVIYFQGFRVDLPIKSARYRGQYNTYPIKLFYTSNI 289
Query 130 PIILQTALVSNLYFLSQLLYRRFKNNVLVNLLGQWQEVD 168
PIILQ+ALVSNLY +SQ+L RF N LVNLLG W +
Sbjct 290 PIILQSALVSNLYVISQMLSTRFSGNFLVNLLGTWSDTS 328
> mmu:53421 Sec61a1, AA408394, AA410007, Sec61a; Sec61 alpha 1
subunit (S. cerevisiae); K10956 protein transport protein SEC61
subunit alpha
Length=476
Score = 238 bits (607), Expect = 7e-63, Method: Compositional matrix adjust.
Identities = 112/159 (70%), Positives = 131/159 (82%), Gaps = 0/159 (0%)
Query 10 QKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCLFTKSTNMVA 69
QKGYGLGSGISLFIATNICETIVWKAFSPTT+ TG+G EFEGA++ALFH L T++ + A
Sbjct 170 QKGYGLGSGISLFIATNICETIVWKAFSPTTVNTGRGMEFEGAIIALFHLLATRTDKVRA 229
Query 70 LKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGNYPIKLFYTSNI 129
L+EAFYR N PN+ +L+ATI VF +VIYFQGFRVDL +K R RGQ YPIKLFYTSNI
Sbjct 230 LREAFYRQNLPNLMNLIATIFVFAVVIYFQGFRVDLPIKSARYRGQYNTYPIKLFYTSNI 289
Query 130 PIILQTALVSNLYFLSQLLYRRFKNNVLVNLLGQWQEVD 168
PIILQ+ALVSNLY +SQ+L RF N+LV+LLG W +
Sbjct 290 PIILQSALVSNLYVISQMLSARFSGNLLVSLLGTWSDTS 328
> hsa:29927 SEC61A1, HSEC61, SEC61, SEC61A; Sec61 alpha 1 subunit
(S. cerevisiae); K10956 protein transport protein SEC61
subunit alpha
Length=476
Score = 238 bits (607), Expect = 7e-63, Method: Compositional matrix adjust.
Identities = 112/159 (70%), Positives = 131/159 (82%), Gaps = 0/159 (0%)
Query 10 QKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCLFTKSTNMVA 69
QKGYGLGSGISLFIATNICETIVWKAFSPTT+ TG+G EFEGA++ALFH L T++ + A
Sbjct 170 QKGYGLGSGISLFIATNICETIVWKAFSPTTVNTGRGMEFEGAIIALFHLLATRTDKVRA 229
Query 70 LKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGNYPIKLFYTSNI 129
L+EAFYR N PN+ +L+ATI VF +VIYFQGFRVDL +K R RGQ YPIKLFYTSNI
Sbjct 230 LREAFYRQNLPNLMNLIATIFVFAVVIYFQGFRVDLPIKSARYRGQYNTYPIKLFYTSNI 289
Query 130 PIILQTALVSNLYFLSQLLYRRFKNNVLVNLLGQWQEVD 168
PIILQ+ALVSNLY +SQ+L RF N+LV+LLG W +
Sbjct 290 PIILQSALVSNLYVISQMLSARFSGNLLVSLLGTWSDTS 328
> dre:192298 sec61al1, chunp6898, fb62c11, sec61a, wu:fb62c11;
Sec61 alpha like 1; K10956 protein transport protein SEC61
subunit alpha
Length=476
Score = 238 bits (606), Expect = 1e-62, Method: Compositional matrix adjust.
Identities = 112/159 (70%), Positives = 130/159 (81%), Gaps = 0/159 (0%)
Query 10 QKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCLFTKSTNMVA 69
QKGYGLGSGISLFIATNICETIVWKAFSPTT+ TG+GTEFEGA++ALFH L T++ + A
Sbjct 170 QKGYGLGSGISLFIATNICETIVWKAFSPTTVNTGRGTEFEGAIIALFHLLATRTDKVRA 229
Query 70 LKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGNYPIKLFYTSNI 129
L+EAFYR N PN+ +L+AT+ VF +VIYFQGFRVDL +K R RGQ YPIKLFYTSNI
Sbjct 230 LREAFYRQNLPNLMNLIATVFVFAVVIYFQGFRVDLPIKSARYRGQYNTYPIKLFYTSNI 289
Query 130 PIILQTALVSNLYFLSQLLYRRFKNNVLVNLLGQWQEVD 168
PIILQ+ALVSNLY +SQ+L R N LVNLLG W +
Sbjct 290 PIILQSALVSNLYVISQMLSTRSSGNFLVNLLGTWSDTS 328
> cel:Y57G11C.15 hypothetical protein; K10956 protein transport
protein SEC61 subunit alpha
Length=473
Score = 236 bits (602), Expect = 3e-62, Method: Compositional matrix adjust.
Identities = 111/157 (70%), Positives = 130/157 (82%), Gaps = 0/157 (0%)
Query 10 QKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCLFTKSTNMVA 69
QKGYGLGSGISLFIATNICETIVWKAFSP T+ TG+GTEFEGA++ALFH L T+S + A
Sbjct 170 QKGYGLGSGISLFIATNICETIVWKAFSPATMNTGRGTEFEGAVIALFHLLATRSDKVRA 229
Query 70 LKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGNYPIKLFYTSNI 129
L+EAFYR N PN+ +L+AT LVF +VIYFQGFRVDL +K R RGQ +YPIKLFYTSNI
Sbjct 230 LREAFYRQNLPNLMNLMATFLVFAVVIYFQGFRVDLPIKSARYRGQYSSYPIKLFYTSNI 289
Query 130 PIILQTALVSNLYFLSQLLYRRFKNNVLVNLLGQWQE 166
PIILQ+ALVSNLY +SQ+L +F N +NLLG W +
Sbjct 290 PIILQSALVSNLYVISQMLAGKFGGNFFINLLGTWSD 326
> sce:YLR378C SEC61; Essential subunit of Sec61 complex (Sec61p,
Sbh1p, and Sss1p); forms a channel for SRP-dependent protein
import and retrograde transport of misfolded proteins out
of the ER; with Sec63 complex allows SRP-independent protein
import into ER; K10956 protein transport protein SEC61 subunit
alpha
Length=480
Score = 207 bits (526), Expect = 2e-53, Method: Compositional matrix adjust.
Identities = 93/164 (56%), Positives = 126/164 (76%), Gaps = 0/164 (0%)
Query 1 VVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCL 60
+V++LDELL KGYGLGSGISLF ATNI E I W+AF+PTT+ +G+G EFEGA++A FH L
Sbjct 163 IVMLLDELLSKGYGLGSGISLFTATNIAEQIFWRAFAPTTVNSGRGKEFEGAVIAFFHLL 222
Query 61 FTKSTNMVALKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGNYP 120
+ AL EAFYR+N PN+ +L T+ +FL V+Y QGFR +L ++ +VRGQ G YP
Sbjct 223 AVRKDKKRALVEAFYRTNLPNMFQVLMTVAIFLFVLYLQGFRYELPIRSTKVRGQIGIYP 282
Query 121 IKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKNNVLVNLLGQW 164
IKLFYTSN PI+LQ+AL SN++ +SQ+L++++ N L+ L+G W
Sbjct 283 IKLFYTSNTPIMLQSALTSNIFLISQILFQKYPTNPLIRLIGVW 326
> sce:YBR283C SSH1; Ssh1p; K10956 protein transport protein SEC61
subunit alpha
Length=490
Score = 73.2 bits (178), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 47/172 (27%), Positives = 87/172 (50%), Gaps = 7/172 (4%)
Query 4 ILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKG--TEFEGALVALFHCLF 61
+L E++ KG+G SG + I +V F + IK G+ TE +GAL+ L L
Sbjct 166 LLAEVIDKGFGFSSGAMIINTVVIATNLVADTFGVSQIKVGEDDQTEAQGALINLIQGLR 225
Query 62 TKSTNMVA-LKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGNYP 120
+K + + AF R PN+T+ + + + +IV Y Q RV+L ++ R RG YP
Sbjct 226 SKHKTFIGGIISAFNRDYLPNLTTTIIVLAIAIIVCYLQSVRVELPIRSTRARGTNNVYP 285
Query 121 IKLFYTSNIPIILQTALVSNLYFLS----QLLYRRFKNNVLVNLLGQWQEVD 168
IKL YT + ++ ++ ++ + QL+ + +++ ++G ++ +
Sbjct 286 IKLLYTGCLSVLFSYTILFYIHIFAFVLIQLVAKNEPTHIICKIMGHYENAN 337
> cel:T19D2.1 hypothetical protein
Length=872
Score = 33.1 bits (74), Expect = 0.47, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 27/44 (61%), Gaps = 1/44 (2%)
Query 24 ATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCLFTKSTNM 67
AT +C + + P TI +G+G +FE A+ ++ CL + STN+
Sbjct 516 ATRVCSRLRDENAIPNTILSGEGMQFEQAMCKIW-CLISGSTNI 558
> hsa:253012 HEPACAM2; HEPACAM family member 2
Length=462
Score = 30.0 bits (66), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 23/39 (58%), Gaps = 5/39 (12%)
Query 5 LDELLQKGYGLG-----SGISLFIATNICETIVWKAFSP 38
L++L QKG L +GISLF+ ++C +WK + P
Sbjct 339 LEKLAQKGKSLSPLASITGISLFLIISMCLLFLWKKYQP 377
> mmu:101202 Hepacam2, AI987662; HEPACAM family member 2
Length=463
Score = 29.3 bits (64), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 23/39 (58%), Gaps = 5/39 (12%)
Query 5 LDELLQKGYGLG-----SGISLFIATNICETIVWKAFSP 38
L++L Q+G L +GISLF+ ++C +WK + P
Sbjct 340 LEKLAQRGKSLSPLASITGISLFLIISMCLLFLWKKYQP 378
> cel:M142.2 cut-6; CUTiclin family member (cut-6)
Length=572
Score = 29.3 bits (64), Expect = 7.0, Method: Composition-based stats.
Identities = 17/73 (23%), Positives = 33/73 (45%), Gaps = 6/73 (8%)
Query 19 ISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCLFTKSTNMVALKEAFYRSN 78
I L + + C +++ +P KG E ++V +FH LF T+ + FY
Sbjct 277 IGLTVPFSACNVHRYRSLNP------KGIFVEVSIVFMFHSLFMTKTDQTVKVQCFYMEA 330
Query 79 APNITSLLATILV 91
++T L+ ++
Sbjct 331 DKHVTVPLSVSMI 343
> cpv:cgd5_1680 protein with an N-terminal domain shared with
the TOG/ALp4p ike microtubule associated proteins
Length=1359
Score = 28.5 bits (62), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 17/43 (39%), Positives = 23/43 (53%), Gaps = 3/43 (6%)
Query 120 PIKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKNNVLVNLLG 162
PI +Y SNI L + SNL F++ L Y K L N++G
Sbjct 1017 PINKYYNSNISESLNDDINSNLIFVNTLEYETIK---LFNIIG 1056
Lambda K H
0.327 0.142 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4157683456
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40