bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0131_orf4
Length=54
Score E
Sequences producing significant alignments: (Bits) Value
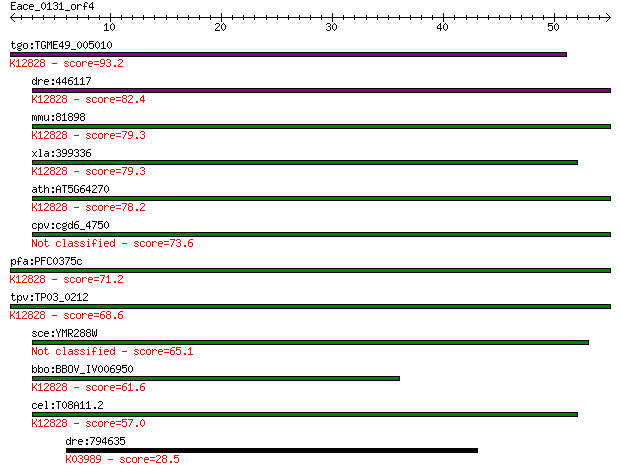
tgo:TGME49_005010 splicing factor 3B subunit 1, putative (EC:5... 93.2 2e-19
dre:446117 sf3b1, wu:fb99f09; splicing factor 3b, subunit 1; K... 82.4 3e-16
mmu:81898 Sf3b1, 155kDa, 2810001M05Rik, AA409119, Prp10, SAP15... 79.3 3e-15
xla:399336 sf3b1, MGC115390, hsh155, prp10, prpf10, sap155; sp... 79.3 3e-15
ath:AT5G64270 splicing factor, putative; K12828 splicing facto... 78.2 7e-15
cpv:cgd6_4750 splicing factor 3B subunit1-like HEAT repeat con... 73.6 1e-13
pfa:PFC0375c U2 snRNP spliceosome subunit, putative; K12828 sp... 71.2 8e-13
tpv:TP03_0212 splicing factor 3B subunit 1; K12828 splicing fa... 68.6 5e-12
sce:YMR288W HSH155; Hsh155p 65.1 5e-11
bbo:BBOV_IV006950 23.m06336; splicing factor; K12828 splicing ... 61.6 6e-10
cel:T08A11.2 hypothetical protein; K12828 splicing factor 3B s... 57.0 2e-08
dre:794635 complement C4-2-like; K03989 complement component 4 28.5
> tgo:TGME49_005010 splicing factor 3B subunit 1, putative (EC:5.5.1.4);
K12828 splicing factor 3B subunit 1
Length=1386
Score = 93.2 bits (230), Expect = 2e-19, Method: Composition-based stats.
Identities = 38/50 (76%), Positives = 44/50 (88%), Gaps = 0/50 (0%)
Query 1 ILGLFHPARKVREVYWRVYNTVYIGHQDAMVAFYPKLPDDGKHSFARHEL 50
+LGLFHPA+KVREVYWRVYN +YIGHQD+MVAFYP LPDD K ++R EL
Sbjct 1333 LLGLFHPAKKVREVYWRVYNNLYIGHQDSMVAFYPPLPDDEKGCYSRDEL 1382
> dre:446117 sf3b1, wu:fb99f09; splicing factor 3b, subunit 1;
K12828 splicing factor 3B subunit 1
Length=1315
Score = 82.4 bits (202), Expect = 3e-16, Method: Composition-based stats.
Identities = 32/52 (61%), Positives = 45/52 (86%), Gaps = 0/52 (0%)
Query 3 GLFHPARKVREVYWRVYNTVYIGHQDAMVAFYPKLPDDGKHSFARHELELFI 54
GLFHPARKVR+VYW++YN++YIG QDA++A YP + +D K+S+ R+ELE F+
Sbjct 1264 GLFHPARKVRDVYWKIYNSIYIGSQDALIAHYPLIFNDEKNSYVRYELEYFL 1315
> mmu:81898 Sf3b1, 155kDa, 2810001M05Rik, AA409119, Prp10, SAP155,
SF3b155, TA-8, Targ4; splicing factor 3b, subunit 1; K12828
splicing factor 3B subunit 1
Length=1304
Score = 79.3 bits (194), Expect = 3e-15, Method: Composition-based stats.
Identities = 29/52 (55%), Positives = 45/52 (86%), Gaps = 0/52 (0%)
Query 3 GLFHPARKVREVYWRVYNTVYIGHQDAMVAFYPKLPDDGKHSFARHELELFI 54
GLFHPARKVR+VYW++YN++YIG QDA++A YP++ +D K+++ R+EL+ +
Sbjct 1253 GLFHPARKVRDVYWKIYNSIYIGSQDALIAHYPRIYNDDKNTYIRYELDYIL 1304
> xla:399336 sf3b1, MGC115390, hsh155, prp10, prpf10, sap155;
splicing factor 3b, subunit 1, 155kDa; K12828 splicing factor
3B subunit 1
Length=1307
Score = 79.3 bits (194), Expect = 3e-15, Method: Composition-based stats.
Identities = 29/49 (59%), Positives = 44/49 (89%), Gaps = 0/49 (0%)
Query 3 GLFHPARKVREVYWRVYNTVYIGHQDAMVAFYPKLPDDGKHSFARHELE 51
GLFHPARKVR+VYW++YN++YIG QDA++A YP++ +D K+++ R+EL+
Sbjct 1256 GLFHPARKVRDVYWKIYNSIYIGSQDALIAHYPRIYNDEKNTYIRYELD 1304
> ath:AT5G64270 splicing factor, putative; K12828 splicing factor
3B subunit 1
Length=1269
Score = 78.2 bits (191), Expect = 7e-15, Method: Composition-based stats.
Identities = 31/52 (59%), Positives = 42/52 (80%), Gaps = 0/52 (0%)
Query 3 GLFHPARKVREVYWRVYNTVYIGHQDAMVAFYPKLPDDGKHSFARHELELFI 54
GLFHPARKVREVYW++YN++YIG QD +VA YP L D+ + ++R EL +F+
Sbjct 1218 GLFHPARKVREVYWKIYNSLYIGAQDTLVAAYPVLEDEQNNVYSRPELTMFV 1269
> cpv:cgd6_4750 splicing factor 3B subunit1-like HEAT repeat containing
protein
Length=1031
Score = 73.6 bits (179), Expect = 1e-13, Method: Composition-based stats.
Identities = 29/52 (55%), Positives = 38/52 (73%), Gaps = 0/52 (0%)
Query 3 GLFHPARKVREVYWRVYNTVYIGHQDAMVAFYPKLPDDGKHSFARHELELFI 54
GLFHPA+KVR VYWR+YN +YIG QD++V F+P +P G +F +E FI
Sbjct 980 GLFHPAKKVRSVYWRIYNNLYIGSQDSLVPFFPPIPQIGNRNFDINEFYYFI 1031
> pfa:PFC0375c U2 snRNP spliceosome subunit, putative; K12828
splicing factor 3B subunit 1
Length=1386
Score = 71.2 bits (173), Expect = 8e-13, Method: Composition-based stats.
Identities = 28/54 (51%), Positives = 37/54 (68%), Gaps = 0/54 (0%)
Query 1 ILGLFHPARKVREVYWRVYNTVYIGHQDAMVAFYPKLPDDGKHSFARHELELFI 54
+ G+FHP+RKVRE+YW++YN VYIGHQD++V YP +F R EL I
Sbjct 1333 VQGIFHPSRKVREIYWKIYNNVYIGHQDSLVPIYPPFELLNDSTFVRDELRYTI 1386
> tpv:TP03_0212 splicing factor 3B subunit 1; K12828 splicing
factor 3B subunit 1
Length=1107
Score = 68.6 bits (166), Expect = 5e-12, Method: Composition-based stats.
Identities = 29/54 (53%), Positives = 38/54 (70%), Gaps = 0/54 (0%)
Query 1 ILGLFHPARKVREVYWRVYNTVYIGHQDAMVAFYPKLPDDGKHSFARHELELFI 54
I GLFHPAR+VRE YWRVYN +Y+GHQDA+V YP + + ++ +EL I
Sbjct 1054 IQGLFHPARRVREAYWRVYNNLYLGHQDALVPLYPLITEGVENKHQANELLYMI 1107
> sce:YMR288W HSH155; Hsh155p
Length=971
Score = 65.1 bits (157), Expect = 5e-11, Method: Composition-based stats.
Identities = 26/50 (52%), Positives = 35/50 (70%), Gaps = 0/50 (0%)
Query 3 GLFHPARKVREVYWRVYNTVYIGHQDAMVAFYPKLPDDGKHSFARHELEL 52
GLFHPA+ VR+ +WRVYN +Y+ +QDAMV FYP PD+ + +L L
Sbjct 922 GLFHPAKNVRKAFWRVYNNMYVMYQDAMVPFYPVTPDNNEEYIEELDLVL 971
> bbo:BBOV_IV006950 23.m06336; splicing factor; K12828 splicing
factor 3B subunit 1
Length=1147
Score = 61.6 bits (148), Expect = 6e-10, Method: Composition-based stats.
Identities = 23/33 (69%), Positives = 29/33 (87%), Gaps = 0/33 (0%)
Query 3 GLFHPARKVREVYWRVYNTVYIGHQDAMVAFYP 35
GLFHPA KVREVYWR+YN +Y+G+QDA+V +P
Sbjct 1096 GLFHPATKVREVYWRLYNNLYVGNQDALVPLFP 1128
> cel:T08A11.2 hypothetical protein; K12828 splicing factor 3B
subunit 1
Length=1322
Score = 57.0 bits (136), Expect = 2e-08, Method: Composition-based stats.
Identities = 21/49 (42%), Positives = 35/49 (71%), Gaps = 0/49 (0%)
Query 3 GLFHPARKVREVYWRVYNTVYIGHQDAMVAFYPKLPDDGKHSFARHELE 51
L+HPARKVRE W+V+N + +G DA++A YP++ + + + R+EL+
Sbjct 1271 ALWHPARKVREPVWKVFNNLILGSADALIAAYPRIENTPTNQYVRYELD 1319
> dre:794635 complement C4-2-like; K03989 complement component
4
Length=1717
Score = 28.5 bits (62), Expect = 5.0, Method: Composition-based stats.
Identities = 15/38 (39%), Positives = 20/38 (52%), Gaps = 1/38 (2%)
Query 6 HPARKVREVYWRVYNTVYIGHQDAMVAFYPK-LPDDGK 42
HP + +Y V N V Q V+FYPK P+DG+
Sbjct 444 HPMKSTSFLYMSVTNKVLSSGQFLEVSFYPKGNPEDGQ 481
Lambda K H
0.329 0.146 0.476
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2014103000
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40