bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
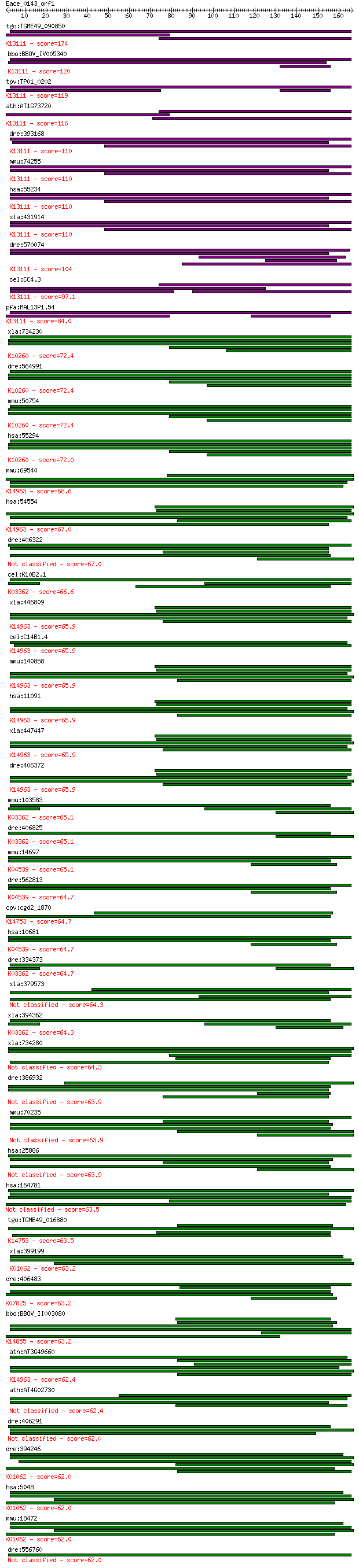
Query= Eace_0143_orf1
Length=166
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_090850 WD-40 repeat protein, putative ; K13111 WD40... 174 1e-43
bbo:BBOV_IV005340 23.m06217; WD domain, G-beta repeat containi... 120 3e-27
tpv:TP01_0202 hypothetical protein; K13111 WD40 repeat-contain... 119 5e-27
ath:AT1G73720 transducin family protein / WD-40 repeat family ... 116 4e-26
dre:393168 smu1, MGC56147, zgc:56147; smu-1 suppressor of mec-... 110 2e-24
mmu:74255 Smu1, 2600001O03Rik, 2610203K23Rik, AB044414, AI8450... 110 2e-24
hsa:55234 SMU1, BWD, DKFZp761L0916, FLJ10805, FLJ10870, FLJ119... 110 2e-24
xla:431914 smu1, MGC81475; smu-1 suppressor of mec-8 and unc-5... 110 3e-24
dre:570074 WD40 repeat-containing protein SMU1-like; K13111 WD... 104 1e-22
cel:CC4.3 smu-1; Suppressor of Mec and Unc defects family memb... 97.1 2e-20
pfa:MAL13P1.54 conserved Plasmodium protein, unknown function;... 84.0 2e-16
xla:734230 fbxw7; F-box and WD repeat domain containing 7; K10... 72.4 6e-13
dre:564991 fbxw7, si:ch211-208n2.1; F-box and WD repeat domain... 72.4 7e-13
mmu:50754 Fbxw7, 1110001A17Rik, AGO, Cdc4, Fbw7, Fbwd6, Fbx30,... 72.4 7e-13
hsa:55294 FBXW7, AGO, CDC4, DKFZp686F23254, FBW6, FBW7, FBX30,... 72.0 8e-13
mmu:69544 Wdr5b, 2310009C03Rik, AI606931; WD repeat domain 5B;... 68.6 1e-11
hsa:54554 WDR5B, FLJ11287, MGC49879; WD repeat domain 5B; K149... 67.0 3e-11
dre:406322 poc1a, wdr51a, wu:fl82d01, zgc:56055; POC1 centriol... 67.0 3e-11
cel:K10B2.1 lin-23; abnormal cell LINeage family member (lin-2... 66.6 3e-11
xla:446809 wdr5-a, MGC80538, big-3, swd3, xwdr5; WD repeat dom... 65.9 5e-11
cel:C14B1.4 tag-125; Temporarily Assigned Gene name family mem... 65.9 6e-11
mmu:140858 Wdr5, 2410008O07Rik, AA408785, AA960360, Big, Big-3... 65.9 6e-11
hsa:11091 WDR5, BIG-3, SWD3; WD repeat domain 5; K14963 COMPAS... 65.9 6e-11
xla:447447 wdr5-b, MGC81485, big-3, swd3, wdr5, xwdr5; WD repe... 65.9 6e-11
dre:406372 wdr5, wu:fk47f04, zgc:56591, zgc:76895; WD repeat d... 65.9 6e-11
mmu:103583 Fbxw11, 2310065A07Rik, AA536858, BTRC2, BTRCP2, Fbx... 65.1 1e-10
dre:406825 fbxw11b, btrc2, fbxw11a, wu:fa12e12, wu:fb11f03, zg... 65.1 1e-10
mmu:14697 Gnb5, GBS, Gbeta5, flr; guanine nucleotide binding p... 65.1 1e-10
dre:562813 gnb5a, MGC158678, zgc:158678; guanine nucleotide bi... 64.7 1e-10
cpv:cgd2_1870 guanine nucleotide-binding protein ; K14753 guan... 64.7 1e-10
hsa:10681 GNB5, FLJ37457, FLJ43714, GB5; guanine nucleotide bi... 64.7 1e-10
dre:334373 fbxw11a, btrc2, fbxw11, fbxw11b, fbxw1b, wu:fd14d12... 64.7 1e-10
xla:379573 poc1a, MGC69111, pix2, wdr51a; POC1 centriolar prot... 64.3 2e-10
xla:394362 btrc, MGC83554, beta-TrCP, betaTrCP, btrc-a, btrcp,... 64.3 2e-10
xla:734280 wdr69, MGC85213; WD repeat domain 69 64.3
dre:386932 poc1b, TUWD12, fl36w17, wdr51b, wu:fl36w17, zgc:635... 63.9 2e-10
mmu:70235 Poc1a, 2510040D07Rik, Wdr51a; POC1 centriolar protei... 63.9 2e-10
hsa:25886 POC1A, DKFZp434C245, MGC131902, PIX2, WDR51A; POC1 c... 63.9 3e-10
hsa:164781 WDR69, FLJ25955; WD repeat domain 69 63.5
tgo:TGME49_016880 receptor for activated C kinase, RACK protei... 63.5 3e-10
xla:399199 pafah1b1, lis2, mdcr, mds, pafah, pafah1b1-b; plate... 63.2 4e-10
dre:406483 gnb3b, gnb3, wu:fk54b04, zgc:73058, zgc:77780; guan... 63.2 4e-10
bbo:BBOV_II003080 18.m06257; WD-repeat protein; K14855 ribosom... 63.2 4e-10
ath:AT3G49660 transducin family protein / WD-40 repeat family ... 62.4 6e-10
ath:AT4G02730 transducin family protein / WD-40 repeat family ... 62.4 7e-10
dre:406291 katnb1, wu:fj32f02, wu:fj65h01, zgc:56071; katanin ... 62.0 8e-10
dre:394246 pafah1b1a; platelet-activating factor acetylhydrola... 62.0 8e-10
hsa:5048 PAFAH1B1, LIS1, LIS2, MDCR, MDS, PAFAH; platelet-acti... 62.0 9e-10
mmu:18472 Pafah1b1, LIS-1, Lis1, MGC25297, MMS10-U, Mdsh, Ms10... 62.0 9e-10
dre:556760 wdr69, si:dkey-223n17.5, zgc:153626, zgc:153780; WD... 62.0 1e-09
> tgo:TGME49_090850 WD-40 repeat protein, putative ; K13111 WD40
repeat-containing protein SMU1
Length=521
Score = 174 bits (440), Expect = 1e-43, Method: Composition-based stats.
Identities = 83/164 (50%), Positives = 116/164 (70%), Gaps = 6/164 (3%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
+++GS DG +++W+ T + A+ + + +++ + ++++ +
Sbjct 230 LVSGSIDGFVEVWEWTTGQLNKEL-----AYQKEDALMMHESAVVAVEFSRDSEVLATGS 284
Query 63 KSNSITLMNFA-GKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
+ + + A G+ +K D+AHD AI SI+FSKDNTH+LT SFDTTARIHGLK+GKTLK
Sbjct 285 QDGQLKVWIVATGQCARKFDRAHDGAITSISFSKDNTHLLTSSFDTTARIHGLKSGKTLK 344
Query 122 EFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
EFRGHLTFVNCALYLPDNTR++TGSADGK+KIWD+KTQDCL TF
Sbjct 345 EFRGHLTFVNCALYLPDNTRVVTGSADGKVKIWDAKTQDCLHTF 388
Score = 103 bits (256), Expect = 3e-22, Method: Composition-based stats.
Identities = 44/79 (55%), Positives = 62/79 (78%), Gaps = 1/79 (1%)
Query 1 TRIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVG-ADLMY 59
TR++TGSADGK+KIWD+KTQDCL TF PP+P +M +Q LP++ +IILAP++ D++Y
Sbjct 363 TRVVTGSADGKVKIWDAKTQDCLHTFAPPLPPYMNASQHLPAINNIILAPKHGSEKDMIY 422
Query 60 VCTKSNSITLMNFAGKTIK 78
VC+K+++I LM G IK
Sbjct 423 VCSKTSTIMLMTLDGHAIK 441
Score = 41.6 bits (96), Expect = 0.001, Method: Composition-based stats.
Identities = 30/100 (30%), Positives = 44/100 (44%), Gaps = 11/100 (11%)
Query 74 GKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKE--------FRG 125
KTIK K+H A FS D H+++GS D + G+ KE
Sbjct 207 AKTIKFGSKSHPECAA---FSPDGHHLVSGSIDGFVEVWEWTTGQLNKELAYQKEDALMM 263
Query 126 HLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
H + V + D+ + TGS DG++K+W T C + F
Sbjct 264 HESAVVAVEFSRDSEVLATGSQDGQLKVWIVATGQCARKF 303
> bbo:BBOV_IV005340 23.m06217; WD domain, G-beta repeat containing
protein; K13111 WD40 repeat-containing protein SMU1
Length=533
Score = 120 bits (300), Expect = 3e-27, Method: Composition-based stats.
Identities = 60/164 (36%), Positives = 104/164 (63%), Gaps = 6/164 (3%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
+I+GS+DG I++W+ + + + + + I+ + ++++
Sbjct 240 LISGSSDGFIEVWNWHSGQLDLDLE-----YQKNDRFMLHDTLIVSLAVSRDSEILASGD 294
Query 63 KSNSITLMNFA-GKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
K +I + A G+ ++K+D AHD A+ +TFS+++ +LTGSFD TA+IHGLK+G+++K
Sbjct 295 KDGNIKIWKIATGECMRKMDNAHDGAVTCMTFSRNSMSLLTGSFDKTAKIHGLKSGRSIK 354
Query 122 EFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
EF+GH + VN A+Y D ++ITGS+DG IK+WDS+T D L++F
Sbjct 355 EFKGHHSIVNAAIYSYDGNKVITGSSDGYIKVWDSRTGDLLKSF 398
Score = 59.3 bits (142), Expect = 5e-09, Method: Composition-based stats.
Identities = 48/158 (30%), Positives = 77/158 (48%), Gaps = 8/158 (5%)
Query 2 RIITGSADGKIKIWDSKTQDCLQTF---TPPVPAHMQTAQCLPSVMSIILAPRNVGAD-L 57
++ITGS+DG IK+WDS+T D L++F T P + Q P ++ I+A G D L
Sbjct 374 KVITGSSDGYIKVWDSRTGDLLKSFLAYTGP-GSGTQIPNDSPRAVNCIIALPYTGTDEL 432
Query 58 MYVCTKSNSITLMNFAGKTIKK--IDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLK 115
+ VCT+S S+ + G +++ I++A ++ + S+ + T K
Sbjct 433 ILVCTRSTSLVIYKLNGISVRNYSIEEAPNSNFLDVAVSRKMNWVYTVGEGNQLYCFN-K 491
Query 116 AGKTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKI 153
AG FR H V + P + + T S DG IK+
Sbjct 492 AGPLEYTFRVHEKDVIGLAHHPQDAVLATWSLDGTIKL 529
Score = 29.3 bits (64), Expect = 5.7, Method: Composition-based stats.
Identities = 9/24 (37%), Positives = 17/24 (70%), Gaps = 0/24 (0%)
Query 132 CALYLPDNTRIITGSADGKIKIWD 155
C ++ P +I+GS+DG I++W+
Sbjct 230 CVVFTPSGQYLISGSSDGFIEVWN 253
> tpv:TP01_0202 hypothetical protein; K13111 WD40 repeat-containing
protein SMU1
Length=526
Score = 119 bits (298), Expect = 5e-27, Method: Composition-based stats.
Identities = 59/164 (35%), Positives = 102/164 (62%), Gaps = 6/164 (3%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
+I+GS+DG I++W+ + H + L + +++ + ++++
Sbjct 238 LISGSSDGFIEVWNWSLGVLDTELSYQANDHFMLHETLITCLAV-----SRDSEVLASGD 292
Query 63 KSNSITLMNF-AGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
+ +I + G+ +K ++ +H A+ TFS+D++ +LTGSFD+ ARIHGLK+GK LK
Sbjct 293 QKGNIKIWKIDTGECLKTMNNSHKGAVTCATFSRDSSCLLTGSFDSLARIHGLKSGKPLK 352
Query 122 EFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
EFRGH + VN +Y D TR+ITGS+DG +K+WD++T +CL++F
Sbjct 353 EFRGHTSIVNTVVYSMDGTRVITGSSDGFVKVWDTRTCECLKSF 396
Score = 48.1 bits (113), Expect = 1e-05, Method: Composition-based stats.
Identities = 26/77 (33%), Positives = 43/77 (55%), Gaps = 3/77 (3%)
Query 1 TRIITGSADGKIKIWDSKTQDCLQTFTPPVPAHM--QTAQCLPSVMSIILAPRNVGADLM 58
TR+ITGS+DG +K+WD++T +CL++F + + + ++ IL G M
Sbjct 371 TRVITGSSDGFVKVWDTRTCECLKSFAAFIQRENDDRDGPLISKSVNTILNLSTAGQSEM 430
Query 59 Y-VCTKSNSITLMNFAG 74
+ VC+KS + L N G
Sbjct 431 FLVCSKSPVLKLFNMNG 447
Score = 30.0 bits (66), Expect = 4.0, Method: Composition-based stats.
Identities = 9/24 (37%), Positives = 18/24 (75%), Gaps = 0/24 (0%)
Query 132 CALYLPDNTRIITGSADGKIKIWD 155
C ++ P+ +I+GS+DG I++W+
Sbjct 228 CVVFTPNGQYLISGSSDGFIEVWN 251
> ath:AT1G73720 transducin family protein / WD-40 repeat family
protein; K13111 WD40 repeat-containing protein SMU1
Length=511
Score = 116 bits (290), Expect = 4e-26, Method: Composition-based stats.
Identities = 52/92 (56%), Positives = 70/92 (76%), Gaps = 1/92 (1%)
Query 74 GKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCA 133
G I++ D AH + S++FS+D + +L+ SFD TARIHGLK+GK LKEFRGH ++VN A
Sbjct 295 GVCIRRFD-AHSQGVTSLSFSRDGSQLLSTSFDQTARIHGLKSGKLLKEFRGHTSYVNHA 353
Query 134 LYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
++ D +RIIT S+D +K+WDSKT DCLQTF
Sbjct 354 IFTSDGSRIITASSDCTVKVWDSKTTDCLQTF 385
Score = 70.5 bits (171), Expect = 2e-12, Method: Composition-based stats.
Identities = 37/78 (47%), Positives = 49/78 (62%), Gaps = 6/78 (7%)
Query 1 TRIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYV 60
+RIIT S+D +K+WDSKT DCLQTF PP P A SV SI L P+N + + V
Sbjct 360 SRIITASSDCTVKVWDSKTTDCLQTFKPPPPLRGTDA----SVNSIHLFPKN--TEHIVV 413
Query 61 CTKSNSITLMNFAGKTIK 78
C K++SI +M G+ +K
Sbjct 414 CNKTSSIYIMTLQGQVVK 431
Score = 37.4 bits (85), Expect = 0.024, Method: Composition-based stats.
Identities = 33/103 (32%), Positives = 46/103 (44%), Gaps = 11/103 (10%)
Query 71 NFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKE-------- 122
N TIK K+H A A FS D + + S D + +GK K+
Sbjct 202 NVLTHTIKFGKKSH-AECAR--FSPDGQFLASSSVDGFIEVWDYISGKLKKDLQYQADES 258
Query 123 FRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
F H V C + D+ + +GS DGKIKIW +T C++ F
Sbjct 259 FMMHDDPVLCIDFSRDSEMLASGSQDGKIKIWRIRTGVCIRRF 301
> dre:393168 smu1, MGC56147, zgc:56147; smu-1 suppressor of mec-8
and unc-52 homolog (C. elegans); K13111 WD40 repeat-containing
protein SMU1
Length=513
Score = 110 bits (275), Expect = 2e-24, Method: Composition-based stats.
Identities = 57/164 (34%), Positives = 90/164 (54%), Gaps = 6/164 (3%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
++TGS DG I++W+ T + + MS + +++
Sbjct 229 LVTGSVDGFIEVWNFTTGKIRKDLKYQAQDNFMMMDDAVLCMSF-----SRDTEMLATGA 283
Query 63 KSNSITLMNF-AGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
+ I + +G+ +++ ++AH + ++FSKD+T IL+ SFD T RIHGLK+GK LK
Sbjct 284 QDGKIKVWKIQSGQCLRRYERAHSKGVTCLSFSKDSTQILSASFDQTIRIHGLKSGKCLK 343
Query 122 EFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
EFRGH +FVN A PD I+ S+DG +K+W+ KT +C TF
Sbjct 344 EFRGHSSFVNEATLTPDGHHAISASSDGTVKVWNMKTTECTSTF 387
Score = 53.9 bits (128), Expect = 2e-07, Method: Composition-based stats.
Identities = 41/153 (26%), Positives = 65/153 (42%), Gaps = 8/153 (5%)
Query 4 ITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCTK 63
I+ S+DG +K+W+ KT +C TF + +A +V ++IL P+N + VC +
Sbjct 365 ISASSDGTVKVWNMKTTECTSTFK----SLGTSAGTDITVNNVILLPKN--PEHFVVCNR 418
Query 64 SNSITLMNFAGKTIKKID--KAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
SN++ +MN G+ ++ K T S I D GK +
Sbjct 419 SNTVVIMNMQGQIVRSFSSGKREGGDFVCCTLSPRGEWIYCVGEDYVLYCFSTVTGKLER 478
Query 122 EFRGHLTFVNCALYLPDNTRIITGSADGKIKIW 154
H V + P I T S DG +K+W
Sbjct 479 TLTVHEKDVIGIAHHPHQNLIGTYSEDGLLKLW 511
Score = 43.1 bits (100), Expect = 4e-04, Method: Composition-based stats.
Identities = 30/126 (23%), Positives = 51/126 (40%), Gaps = 8/126 (6%)
Query 48 LAPRNVGADLMYVCTKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDT 107
L P + DL + F + + I + + FS D +++TGS D
Sbjct 177 LLPPGMTIDLFRGKAAVKDVEEERFPTQLARHIKFGQKSHVECARFSPDGQYLVTGSVDG 236
Query 108 TARIHGLKAGKTLKE--FRGHLTF------VNCALYLPDNTRIITGSADGKIKIWDSKTQ 159
+ GK K+ ++ F V C + D + TG+ DGKIK+W ++
Sbjct 237 FIEVWNFTTGKIRKDLKYQAQDNFMMMDDAVLCMSFSRDTEMLATGAQDGKIKVWKIQSG 296
Query 160 DCLQTF 165
CL+ +
Sbjct 297 QCLRRY 302
> mmu:74255 Smu1, 2600001O03Rik, 2610203K23Rik, AB044414, AI845086,
AW556129, Bwd; smu-1 suppressor of mec-8 and unc-52 homolog
(C. elegans); K13111 WD40 repeat-containing protein SMU1
Length=513
Score = 110 bits (275), Expect = 2e-24, Method: Composition-based stats.
Identities = 57/164 (34%), Positives = 94/164 (57%), Gaps = 6/164 (3%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
++TGS DG I++W+ T + + +V+ + + +++
Sbjct 229 LVTGSVDGFIEVWNFTTGKIRKDLKYQAQDNFMMMD--DAVLCMCFSR---DTEMLATGA 283
Query 63 KSNSITLMNF-AGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
+ I + +G+ +++ ++AH + ++FSKD++ IL+ SFD T RIHGLK+GKTLK
Sbjct 284 QDGKIKVWKIQSGQCLRRFERAHSKGVTCLSFSKDSSQILSASFDQTIRIHGLKSGKTLK 343
Query 122 EFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
EFRGH +FVN A + D II+ S+DG +KIW+ KT +C TF
Sbjct 344 EFRGHSSFVNEATFTQDGHYIISASSDGTVKIWNMKTTECSNTF 387
Score = 57.4 bits (137), Expect = 2e-08, Method: Composition-based stats.
Identities = 44/154 (28%), Positives = 65/154 (42%), Gaps = 8/154 (5%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
II+ S+DG +KIW+ KT +C TF + TA +V S+IL P+N + VC
Sbjct 364 IISASSDGTVKIWNMKTTECSNTFK----SLGSTAGTDITVNSVILLPKN--PEHFVVCN 417
Query 63 KSNSITLMNFAGKTIKKID--KAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTL 120
+SN++ +MN G+ ++ K S I D GK
Sbjct 418 RSNTVVIMNMQGQIVRSFSSGKREGGDFVCCALSPRGEWIYCVGEDFVLYCFSTVTGKLE 477
Query 121 KEFRGHLTFVNCALYLPDNTRIITGSADGKIKIW 154
+ H V + P I T S DG +K+W
Sbjct 478 RTLTVHEKDVIGIAHHPHQNLIATYSEDGLLKLW 511
Score = 44.7 bits (104), Expect = 2e-04, Method: Composition-based stats.
Identities = 31/126 (24%), Positives = 51/126 (40%), Gaps = 8/126 (6%)
Query 48 LAPRNVGADLMYVCTKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDT 107
L P + DL + F + + I + + FS D +++TGS D
Sbjct 177 LLPPGMTIDLFRGKAAVKDVEEEKFPTQLSRHIKFGQKSHVECARFSPDGQYLVTGSVDG 236
Query 108 TARIHGLKAGKTLKE--FRGHLTF------VNCALYLPDNTRIITGSADGKIKIWDSKTQ 159
+ GK K+ ++ F V C + D + TG+ DGKIK+W ++
Sbjct 237 FIEVWNFTTGKIRKDLKYQAQDNFMMMDDAVLCMCFSRDTEMLATGAQDGKIKVWKIQSG 296
Query 160 DCLQTF 165
CL+ F
Sbjct 297 QCLRRF 302
> hsa:55234 SMU1, BWD, DKFZp761L0916, FLJ10805, FLJ10870, FLJ11970,
MGC117363, RP11-54K16.3, SMU-1, fSAP57; smu-1 suppressor
of mec-8 and unc-52 homolog (C. elegans); K13111 WD40 repeat-containing
protein SMU1
Length=513
Score = 110 bits (275), Expect = 2e-24, Method: Composition-based stats.
Identities = 57/164 (34%), Positives = 94/164 (57%), Gaps = 6/164 (3%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
++TGS DG I++W+ T + + +V+ + + +++
Sbjct 229 LVTGSVDGFIEVWNFTTGKIRKDLKYQAQDNFMMMD--DAVLCMCFSR---DTEMLATGA 283
Query 63 KSNSITLMNF-AGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
+ I + +G+ +++ ++AH + ++FSKD++ IL+ SFD T RIHGLK+GKTLK
Sbjct 284 QDGKIKVWKIQSGQCLRRFERAHSKGVTCLSFSKDSSQILSASFDQTIRIHGLKSGKTLK 343
Query 122 EFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
EFRGH +FVN A + D II+ S+DG +KIW+ KT +C TF
Sbjct 344 EFRGHSSFVNEATFTQDGHYIISASSDGTVKIWNMKTTECSNTF 387
Score = 57.4 bits (137), Expect = 2e-08, Method: Composition-based stats.
Identities = 44/154 (28%), Positives = 65/154 (42%), Gaps = 8/154 (5%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
II+ S+DG +KIW+ KT +C TF + TA +V S+IL P+N + VC
Sbjct 364 IISASSDGTVKIWNMKTTECSNTFK----SLGSTAGTDITVNSVILLPKN--PEHFVVCN 417
Query 63 KSNSITLMNFAGKTIKKID--KAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTL 120
+SN++ +MN G+ ++ K S I D GK
Sbjct 418 RSNTVVIMNMQGQIVRSFSSGKREGGDFVCCALSPRGEWIYCVGEDFVLYCFSTVTGKLE 477
Query 121 KEFRGHLTFVNCALYLPDNTRIITGSADGKIKIW 154
+ H V + P I T S DG +K+W
Sbjct 478 RTLTVHEKDVIGIAHHPHQNLIATYSEDGLLKLW 511
Score = 44.7 bits (104), Expect = 2e-04, Method: Composition-based stats.
Identities = 31/126 (24%), Positives = 51/126 (40%), Gaps = 8/126 (6%)
Query 48 LAPRNVGADLMYVCTKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDT 107
L P + DL + F + + I + + FS D +++TGS D
Sbjct 177 LLPPGMTIDLFRGKAAVKDVEEEKFPTQLSRHIKFGQKSHVECARFSPDGQYLVTGSVDG 236
Query 108 TARIHGLKAGKTLKE--FRGHLTF------VNCALYLPDNTRIITGSADGKIKIWDSKTQ 159
+ GK K+ ++ F V C + D + TG+ DGKIK+W ++
Sbjct 237 FIEVWNFTTGKIRKDLKYQAQDNFMMMDDAVLCMCFSRDTEMLATGAQDGKIKVWKIQSG 296
Query 160 DCLQTF 165
CL+ F
Sbjct 297 QCLRRF 302
> xla:431914 smu1, MGC81475; smu-1 suppressor of mec-8 and unc-52
homolog; K13111 WD40 repeat-containing protein SMU1
Length=513
Score = 110 bits (274), Expect = 3e-24, Method: Composition-based stats.
Identities = 57/164 (34%), Positives = 94/164 (57%), Gaps = 6/164 (3%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
++TGS DG I++W+ T + + +V+ + + +++
Sbjct 229 LVTGSVDGFIEVWNFTTGKIRKDLKYQAQDNFMMMD--DAVLCMCFSR---DTEMLATGA 283
Query 63 KSNSITLMNF-AGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
+ I + +G+ +++ ++AH + ++FSKD++ IL+ SFD T RIHGLK+GKTLK
Sbjct 284 QDGKIKVWKIQSGQCLRRFERAHSKGVTCLSFSKDSSQILSASFDQTIRIHGLKSGKTLK 343
Query 122 EFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
EFRGH +FVN A + D II+ S+DG +KIW+ KT +C TF
Sbjct 344 EFRGHSSFVNEATFTQDGHYIISASSDGTVKIWNMKTTECSNTF 387
Score = 58.2 bits (139), Expect = 1e-08, Method: Composition-based stats.
Identities = 45/154 (29%), Positives = 66/154 (42%), Gaps = 8/154 (5%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
II+ S+DG +KIW+ KT +C TF + TA +V S+IL P+N + VC
Sbjct 364 IISASSDGTVKIWNMKTTECSNTFK----SLGSTAGTDITVNSVILLPKN--PEHFVVCN 417
Query 63 KSNSITLMNFAGKTIKKID--KAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTL 120
+SN++ +MN G+ ++ K T S I D GK
Sbjct 418 RSNTVVIMNMQGQIVRSFSSGKREGGDFVCCTLSPRGEWIYCVGEDFVLYCFSTVTGKLE 477
Query 121 KEFRGHLTFVNCALYLPDNTRIITGSADGKIKIW 154
+ H V + P I T S DG +K+W
Sbjct 478 RTLTVHEKDVIGIAHHPHQNLIGTYSEDGLLKLW 511
Score = 44.3 bits (103), Expect = 2e-04, Method: Composition-based stats.
Identities = 31/126 (24%), Positives = 51/126 (40%), Gaps = 8/126 (6%)
Query 48 LAPRNVGADLMYVCTKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDT 107
L P + DL + F + + I + + FS D +++TGS D
Sbjct 177 LLPPGMTIDLFRGKAAVKDVEEEKFPTQLSRHIKFGQKSHVECARFSPDGQYLVTGSVDG 236
Query 108 TARIHGLKAGKTLKE--FRGHLTF------VNCALYLPDNTRIITGSADGKIKIWDSKTQ 159
+ GK K+ ++ F V C + D + TG+ DGKIK+W ++
Sbjct 237 FIEVWNFTTGKIRKDLKYQAQDNFMMMDDAVLCMCFSRDTEMLATGAQDGKIKVWKIQSG 296
Query 160 DCLQTF 165
CL+ F
Sbjct 297 QCLRRF 302
> dre:570074 WD40 repeat-containing protein SMU1-like; K13111
WD40 repeat-containing protein SMU1
Length=513
Score = 104 bits (260), Expect = 1e-22, Method: Composition-based stats.
Identities = 58/165 (35%), Positives = 88/165 (53%), Gaps = 10/165 (6%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
+ITGS DG I++W+ T + AQ +M + V D + +
Sbjct 229 LITGSVDGFIEVWNFTTGKICKDLK-------YQAQDNFMMMDEAVLCLAVSHDSHVIAS 281
Query 63 KSNSITLMNFA---GKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKT 119
+ + + G + ++++AH A+ S+ F++D+T +L+ SFD T RIHGLK+GK
Sbjct 282 GAQDGKIQVWRILNGTCLNRLERAHSKAVTSVCFNRDSTQLLSSSFDQTIRIHGLKSGKC 341
Query 120 LKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQT 164
LKEFRGH VN +Y D +I+ SADG +K+W KT DC T
Sbjct 342 LKEFRGHTAHVNDVIYSHDGQHVISASADGTVKVWTVKTMDCTHT 386
Score = 48.9 bits (115), Expect = 8e-06, Method: Composition-based stats.
Identities = 39/155 (25%), Positives = 62/155 (40%), Gaps = 10/155 (6%)
Query 3 IITGSADGKIKIWDSKTQDCLQTF-TPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVC 61
+I+ SADG +K+W KT DC T TP +P +V +++L P+ + V
Sbjct 364 VISASADGTVKVWTVKTMDCTHTIKTPDIPEGTDI-----TVNNVVLVPKT--PEHFVVS 416
Query 62 TKSNSITLMNFAGKTIKKIDKA--HDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKT 119
++N++ + N G+ ++ A T S I D GK
Sbjct 417 NRTNTVVVTNIHGQVMRSFCSGVREGADFVCCTLSPRGEWIYCVGEDYVLYCFSTITGKL 476
Query 120 LKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIW 154
+ H V + P I T S DG +K+W
Sbjct 477 ERTLTVHEKDVIGIAHHPHQNLIATYSEDGLLKLW 511
Score = 35.4 bits (80), Expect = 0.100, Method: Composition-based stats.
Identities = 23/78 (29%), Positives = 35/78 (44%), Gaps = 8/78 (10%)
Query 93 FSKDNTHILTGSFDTTARIHGLKAGKTLKE--FRGHLTF------VNCALYLPDNTRIIT 144
FS D +++TGS D + GK K+ ++ F V C D+ I +
Sbjct 222 FSPDGQYLITGSVDGFIEVWNFTTGKICKDLKYQAQDNFMMMDEAVLCLAVSHDSHVIAS 281
Query 145 GSADGKIKIWDSKTQDCL 162
G+ DGKI++W CL
Sbjct 282 GAQDGKIQVWRILNGTCL 299
Score = 35.0 bits (79), Expect = 0.10, Method: Composition-based stats.
Identities = 15/34 (44%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 125 GHLTFVNCALYLPDNTRIITGSADGKIKIWDSKT 158
G + V CA + PD +ITGS DG I++W+ T
Sbjct 212 GPKSHVECARFSPDGQYLITGSVDGFIEVWNFTT 245
Score = 33.9 bits (76), Expect = 0.24, Method: Composition-based stats.
Identities = 22/82 (26%), Positives = 39/82 (47%), Gaps = 1/82 (1%)
Query 85 DAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEF-RGHLTFVNCALYLPDNTRII 143
D A+ + S D+ I +G+ D ++ + G L R H V + D+T+++
Sbjct 264 DEAVLCLAVSHDSHVIASGAQDGKIQVWRILNGTCLNRLERAHSKAVTSVCFNRDSTQLL 323
Query 144 TGSADGKIKIWDSKTQDCLQTF 165
+ S D I+I K+ CL+ F
Sbjct 324 SSSFDQTIRIHGLKSGKCLKEF 345
> cel:CC4.3 smu-1; Suppressor of Mec and Unc defects family member
(smu-1); K13111 WD40 repeat-containing protein SMU1
Length=510
Score = 97.1 bits (240), Expect = 2e-20, Method: Composition-based stats.
Identities = 39/92 (42%), Positives = 60/92 (65%), Gaps = 0/92 (0%)
Query 74 GKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCA 133
G +++ D+AH + ++ FSKDN+HIL+G D R+HG+K+GK LKE RGH +++
Sbjct 296 GDCLRRFDRAHTKGVCAVRFSKDNSHILSGGNDHVVRVHGMKSGKCLKEMRGHSSYITDV 355
Query 134 LYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
Y + II+ S DG I++W K+ +CL TF
Sbjct 356 RYSDEGNHIISCSTDGSIRVWHGKSGECLSTF 387
Score = 48.5 bits (114), Expect = 9e-06, Method: Composition-based stats.
Identities = 36/125 (28%), Positives = 61/125 (48%), Gaps = 18/125 (14%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
+ TGS DGKIK+W +T DCL+ F AH + + A R D ++ +
Sbjct 279 LATGSIDGKIKVWKVETGDCLRRFD---RAHTKG----------VCAVR-FSKDNSHILS 324
Query 63 KSNSITLMNF---AGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKT 119
N + +GK +K++ + H + I + +S + HI++ S D + R+ K+G+
Sbjct 325 GGNDHVVRVHGMKSGKCLKEM-RGHSSYITDVRYSDEGNHIISCSTDGSIRVWHGKSGEC 383
Query 120 LKEFR 124
L FR
Sbjct 384 LSTFR 388
Score = 47.0 bits (110), Expect = 3e-05, Method: Composition-based stats.
Identities = 27/84 (32%), Positives = 40/84 (47%), Gaps = 8/84 (9%)
Query 90 SITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHL--------TFVNCALYLPDNTR 141
S FS D ++++GS D + GK K+ + V C + D+
Sbjct 219 SAVFSPDANYLVSGSKDGFIEVWNYMNGKLRKDLKYQAQDNLMMMDAAVRCISFSRDSEM 278
Query 142 IITGSADGKIKIWDSKTQDCLQTF 165
+ TGS DGKIK+W +T DCL+ F
Sbjct 279 LATGSIDGKIKVWKVETGDCLRRF 302
Score = 42.7 bits (99), Expect = 6e-04, Method: Composition-based stats.
Identities = 21/78 (26%), Positives = 43/78 (55%), Gaps = 9/78 (11%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
II+ S DG I++W K+ +CL TF + ++++I P++ M VC
Sbjct 364 IISCSTDGSIRVWHGKSGECLSTFRVGSEDY--------PILNVIPIPKS-DPPQMIVCN 414
Query 63 KSNSITLMNFAGKTIKKI 80
+SN++ ++N +G+ ++ +
Sbjct 415 RSNTLYVVNISGQVVRTM 432
> pfa:MAL13P1.54 conserved Plasmodium protein, unknown function;
K13111 WD40 repeat-containing protein SMU1
Length=527
Score = 84.0 bits (206), Expect = 2e-16, Method: Composition-based stats.
Identities = 60/175 (34%), Positives = 91/175 (52%), Gaps = 28/175 (16%)
Query 3 IITGSADGKIKIWDSKT-----------QDCLQTFTPPVPAHMQTAQCLPSVMSIILAPR 51
+ITGS+DG I+IW+ T Q+ L P+ C+ I+L+
Sbjct 234 LITGSSDGFIEIWNWITGELNLDLEYQKQNNLMIHDNPI-----VTLCISKDDEILLSGD 288
Query 52 NVGADLMYVCTKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARI 111
+ G L+ + I L KI AH+ ++ SI F+ D T ILT S+D + +I
Sbjct 289 SKG--LIKIWRIKTGICL---------KIINAHNDSLISIQFNNDQTQILTSSYDKSVKI 337
Query 112 HGLKAGKTLKEFRGHL-TFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
GLK+ K LKEFR H + V+ A+Y DN++II + GKI I++ KT +C+ +F
Sbjct 338 FGLKSLKCLKEFRKHEDSVVHSAIYTLDNSKIICATDQGKIFIYNQKTLECITSF 392
Score = 44.3 bits (103), Expect = 2e-04, Method: Composition-based stats.
Identities = 28/78 (35%), Positives = 44/78 (56%), Gaps = 3/78 (3%)
Query 1 TRIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYV 60
++II + GKI I++ KT +C+ +F V + PS+ +IIL +N+ D + V
Sbjct 367 SKIICATDQGKIFIYNQKTLECITSFY--VYFNKSENLIFPSLNNIILINKNL-EDHILV 423
Query 61 CTKSNSITLMNFAGKTIK 78
C+KS +MN GK IK
Sbjct 424 CSKSPYCYIMNMKGKIIK 441
Score = 30.4 bits (67), Expect = 2.7, Method: Composition-based stats.
Identities = 16/39 (41%), Positives = 24/39 (61%), Gaps = 1/39 (2%)
Query 118 KTLKEFR-GHLTFVNCALYLPDNTRIITGSADGKIKIWD 155
K LK + G + V C + +N +ITGS+DG I+IW+
Sbjct 209 KILKSIKFGKESNVECCISSYNNDYLITGSSDGFIEIWN 247
> xla:734230 fbxw7; F-box and WD repeat domain containing 7; K10260
F-box and WD-40 domain protein 7
Length=706
Score = 72.4 bits (176), Expect = 6e-13, Method: Composition-based stats.
Identities = 52/188 (27%), Positives = 89/188 (47%), Gaps = 29/188 (15%)
Query 3 IITGSADGKIKIWDSKTQDCLQTF---TPPVPA--------------------HMQTAQC 39
II+GS D +K+W+++T +C+ T T V ++T QC
Sbjct 433 IISGSTDRTLKVWNAETGECIHTLYGHTSTVRCMHLHEKRVVSGSRDATLRVWDIETGQC 492
Query 40 LPSVMSIILAPRNVGADLMYVCTKSNS--ITLMNFAGKTIKKIDKAHDAAIASITFSKDN 97
L +M + A R V D V + + + + + +T + H + S+ F D
Sbjct 493 LHVLMGHVAAVRCVQYDGRRVVSGAYDFMVKVWDPETETCLHTLQGHTNRVYSLQF--DG 550
Query 98 THILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSK 157
H+++GS DT+ R+ ++ G + GH + + + L DN +++G+AD +KIWD K
Sbjct 551 IHVVSGSLDTSIRVWDVETGNCIHTLTGHQSLTS-GMELKDNI-LVSGNADSTVKIWDIK 608
Query 158 TQDCLQTF 165
T CLQT
Sbjct 609 TGQCLQTL 616
Score = 61.2 bits (147), Expect = 2e-09, Method: Composition-based stats.
Identities = 43/164 (26%), Positives = 72/164 (43%), Gaps = 19/164 (11%)
Query 2 RIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVC 61
RI++GS D +K+W + T CL+T T S M N+ +
Sbjct 392 RIVSGSDDNTLKVWSAVTGKCLRTLVG------HTGGVWSSQMR-----DNI------II 434
Query 62 TKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
+ S TL + +T + I + +++GS D T R+ ++ G+ L
Sbjct 435 SGSTDRTLKVWNAETGECIHTLYGHTSTVRCMHLHEKRVVSGSRDATLRVWDIETGQCLH 494
Query 122 EFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
GH+ V C Y D R+++G+ D +K+WD +T+ CL T
Sbjct 495 VLMGHVAAVRCVQY--DGRRVVSGAYDFMVKVWDPETETCLHTL 536
Score = 48.9 bits (115), Expect = 7e-06, Method: Composition-based stats.
Identities = 41/169 (24%), Positives = 76/169 (44%), Gaps = 26/169 (15%)
Query 2 RIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVC 61
R+++G+ D +K+WD +T+ CL T H L D ++V
Sbjct 512 RVVSGAYDFMVKVWDPETETCLHTLQ----GHTNRVYSLQ-------------FDGIHVV 554
Query 62 TKS--NSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKT 119
+ S SI + + H + + + KDN +++G+ D+T +I +K G+
Sbjct 555 SGSLDTSIRVWDVETGNCIHTLTGHQSLTSGMEL-KDNI-LVSGNADSTVKIWDIKTGQC 612
Query 120 LKEFRG---HLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
L+ +G H + V C + + +IT S DG +K+W KT + ++
Sbjct 613 LQTLQGPNKHQSAVTCLQF--NKNFVITSSDDGTVKLWGLKTGEFIRNL 659
Score = 43.1 bits (100), Expect = 4e-04, Method: Composition-based stats.
Identities = 29/88 (32%), Positives = 47/88 (53%), Gaps = 5/88 (5%)
Query 79 KIDKAHD-AAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLP 137
K+ K HD I + F + I++GS D T ++ GK L+ GH V + +
Sbjct 373 KVLKGHDDHVITCLQFCGN--RIVSGSDDNTLKVWSAVTGKCLRTLVGHTGGVWSS-QMR 429
Query 138 DNTRIITGSADGKIKIWDSKTQDCLQTF 165
DN II+GS D +K+W+++T +C+ T
Sbjct 430 DNI-IISGSTDRTLKVWNAETGECIHTL 456
Score = 36.2 bits (82), Expect = 0.049, Method: Composition-based stats.
Identities = 24/60 (40%), Positives = 31/60 (51%), Gaps = 4/60 (6%)
Query 106 DTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
DT R LK+ K LK GH V L N RI++GS D +K+W + T CL+T
Sbjct 361 DTNWRRGDLKSPKVLK---GHDDHVITCLQFCGN-RIVSGSDDNTLKVWSAVTGKCLRTL 416
> dre:564991 fbxw7, si:ch211-208n2.1; F-box and WD repeat domain
containing 7; K10260 F-box and WD-40 domain protein 7
Length=605
Score = 72.4 bits (176), Expect = 7e-13, Method: Composition-based stats.
Identities = 52/188 (27%), Positives = 89/188 (47%), Gaps = 29/188 (15%)
Query 3 IITGSADGKIKIWDSKTQDCLQTF---TPPVPA--------------------HMQTAQC 39
II+GS D +K+W+++T +C+ T T V ++T QC
Sbjct 332 IISGSTDRTLKVWNAETGECIHTLYGHTSTVRCMHLHEKRVVSGSRDATLRVWDIETGQC 391
Query 40 LPSVMSIILAPRNVGADLMYVCTKSNS--ITLMNFAGKTIKKIDKAHDAAIASITFSKDN 97
L +M + A R V D V + + + + + +T + H + S+ F D
Sbjct 392 LHVLMGHVAAVRCVQYDGRRVVSGAYDFMVKVWDPETETCLHTLQGHTNRVYSLQF--DG 449
Query 98 THILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSK 157
H+++GS DT+ R+ ++ G + GH + + + L DN +++G+AD +KIWD K
Sbjct 450 IHVVSGSLDTSIRVWDVETGNCIHTLTGHQSLTS-GMELKDNI-LVSGNADSTVKIWDIK 507
Query 158 TQDCLQTF 165
T CLQT
Sbjct 508 TGQCLQTL 515
Score = 60.8 bits (146), Expect = 2e-09, Method: Composition-based stats.
Identities = 43/164 (26%), Positives = 72/164 (43%), Gaps = 19/164 (11%)
Query 2 RIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVC 61
RI++GS D +K+W + T CL+T T S M N+ +
Sbjct 291 RIVSGSDDNTLKVWSAVTGKCLRTLVG------HTGGVWSSQMR-----DNI------II 333
Query 62 TKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
+ S TL + +T + I + +++GS D T R+ ++ G+ L
Sbjct 334 SGSTDRTLKVWNAETGECIHTLYGHTSTVRCMHLHEKRVVSGSRDATLRVWDIETGQCLH 393
Query 122 EFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
GH+ V C Y D R+++G+ D +K+WD +T+ CL T
Sbjct 394 VLMGHVAAVRCVQY--DGRRVVSGAYDFMVKVWDPETETCLHTL 435
Score = 50.8 bits (120), Expect = 2e-06, Method: Composition-based stats.
Identities = 41/169 (24%), Positives = 77/169 (45%), Gaps = 26/169 (15%)
Query 2 RIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVC 61
R+++G+ D +K+WD +T+ CL T H L D ++V
Sbjct 411 RVVSGAYDFMVKVWDPETETCLHTLQ----GHTNRVYSLQ-------------FDGIHVV 453
Query 62 TKS--NSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKT 119
+ S SI + + H + + + KDN +++G+ D+T +I +K G+
Sbjct 454 SGSLDTSIRVWDVETGNCIHTLTGHQSLTSGMEL-KDNI-LVSGNADSTVKIWDIKTGQC 511
Query 120 LKEFRG---HLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
L+ +G H + V C + + +IT S DG +K+WD +T + ++
Sbjct 512 LQTLQGPHKHQSAVTCLQF--NKNFVITSSDDGTVKLWDLRTGEFIRNL 558
Score = 42.7 bits (99), Expect = 5e-04, Method: Composition-based stats.
Identities = 29/88 (32%), Positives = 47/88 (53%), Gaps = 5/88 (5%)
Query 79 KIDKAHD-AAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLP 137
K+ K HD I + F + I++GS D T ++ GK L+ GH V + +
Sbjct 272 KVLKGHDDHVITCLQFCGN--RIVSGSDDNTLKVWSAVTGKCLRTLVGHTGGVWSS-QMR 328
Query 138 DNTRIITGSADGKIKIWDSKTQDCLQTF 165
DN II+GS D +K+W+++T +C+ T
Sbjct 329 DNI-IISGSTDRTLKVWNAETGECIHTL 355
Score = 36.2 bits (82), Expect = 0.045, Method: Composition-based stats.
Identities = 25/69 (36%), Positives = 34/69 (49%), Gaps = 4/69 (5%)
Query 97 NTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDS 156
+ +I DT R LK+ K LK GH V L N RI++GS D +K+W +
Sbjct 251 SAYIRQHRIDTNWRRGDLKSPKVLK---GHDDHVITCLQFCGN-RIVSGSDDNTLKVWSA 306
Query 157 KTQDCLQTF 165
T CL+T
Sbjct 307 VTGKCLRTL 315
> mmu:50754 Fbxw7, 1110001A17Rik, AGO, Cdc4, Fbw7, Fbwd6, Fbx30,
Fbxo30, Fbxw6, SEL-10; F-box and WD-40 domain protein 7;
K10260 F-box and WD-40 domain protein 7
Length=710
Score = 72.4 bits (176), Expect = 7e-13, Method: Composition-based stats.
Identities = 52/188 (27%), Positives = 89/188 (47%), Gaps = 29/188 (15%)
Query 3 IITGSADGKIKIWDSKTQDCLQTF---TPPVPA--------------------HMQTAQC 39
II+GS D +K+W+++T +C+ T T V ++T QC
Sbjct 437 IISGSTDRTLKVWNAETGECIHTLYGHTSTVRCMHLHEKRVVSGSRDATLRVWDIETGQC 496
Query 40 LPSVMSIILAPRNVGADLMYVCTKSNS--ITLMNFAGKTIKKIDKAHDAAIASITFSKDN 97
L +M + A R V D V + + + + + +T + H + S+ F D
Sbjct 497 LHVLMGHVAAVRCVQYDGRRVVSGAYDFMVKVWDPETETCLHTLQGHTNRVYSLQF--DG 554
Query 98 THILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSK 157
H+++GS DT+ R+ ++ G + GH + + + L DN +++G+AD +KIWD K
Sbjct 555 IHVVSGSLDTSIRVWDVETGNCIHTLTGHQSLTS-GMELKDNI-LVSGNADSTVKIWDIK 612
Query 158 TQDCLQTF 165
T CLQT
Sbjct 613 TGQCLQTL 620
Score = 60.8 bits (146), Expect = 2e-09, Method: Composition-based stats.
Identities = 43/164 (26%), Positives = 72/164 (43%), Gaps = 19/164 (11%)
Query 2 RIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVC 61
RI++GS D +K+W + T CL+T T S M N+ +
Sbjct 396 RIVSGSDDNTLKVWSAVTGKCLRTLVG------HTGGVWSSQMR-----DNI------II 438
Query 62 TKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
+ S TL + +T + I + +++GS D T R+ ++ G+ L
Sbjct 439 SGSTDRTLKVWNAETGECIHTLYGHTSTVRCMHLHEKRVVSGSRDATLRVWDIETGQCLH 498
Query 122 EFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
GH+ V C Y D R+++G+ D +K+WD +T+ CL T
Sbjct 499 VLMGHVAAVRCVQY--DGRRVVSGAYDFMVKVWDPETETCLHTL 540
Score = 51.6 bits (122), Expect = 1e-06, Method: Composition-based stats.
Identities = 42/169 (24%), Positives = 77/169 (45%), Gaps = 26/169 (15%)
Query 2 RIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVC 61
R+++G+ D +K+WD +T+ CL T H L D ++V
Sbjct 516 RVVSGAYDFMVKVWDPETETCLHTLQ----GHTNRVYSLQ-------------FDGIHVV 558
Query 62 TKS--NSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKT 119
+ S SI + + H + + + KDN +++G+ D+T +I +K G+
Sbjct 559 SGSLDTSIRVWDVETGNCIHTLTGHQSLTSGMEL-KDNI-LVSGNADSTVKIWDIKTGQC 616
Query 120 LKEFRG---HLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
L+ +G H + V C + + +IT S DG +K+WD KT + ++
Sbjct 617 LQTLQGPSKHQSAVTCLQF--NKNFVITSSDDGTVKLWDLKTGEFIRNL 663
Score = 42.7 bits (99), Expect = 5e-04, Method: Composition-based stats.
Identities = 29/88 (32%), Positives = 47/88 (53%), Gaps = 5/88 (5%)
Query 79 KIDKAHD-AAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLP 137
K+ K HD I + F + I++GS D T ++ GK L+ GH V + +
Sbjct 377 KVLKGHDDHVITCLQFCGN--RIVSGSDDNTLKVWSAVTGKCLRTLVGHTGGVWSS-QMR 433
Query 138 DNTRIITGSADGKIKIWDSKTQDCLQTF 165
DN II+GS D +K+W+++T +C+ T
Sbjct 434 DNI-IISGSTDRTLKVWNAETGECIHTL 460
Score = 36.2 bits (82), Expect = 0.058, Method: Composition-based stats.
Identities = 25/69 (36%), Positives = 34/69 (49%), Gaps = 4/69 (5%)
Query 97 NTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDS 156
+ +I DT R LK+ K LK GH V L N RI++GS D +K+W +
Sbjct 356 SAYIRQHRIDTNWRRGELKSPKVLK---GHDDHVITCLQFCGN-RIVSGSDDNTLKVWSA 411
Query 157 KTQDCLQTF 165
T CL+T
Sbjct 412 VTGKCLRTL 420
> hsa:55294 FBXW7, AGO, CDC4, DKFZp686F23254, FBW6, FBW7, FBX30,
FBXO30, FBXW6, FLJ16457, SEL-10, SEL10; F-box and WD repeat
domain containing 7; K10260 F-box and WD-40 domain protein
7
Length=589
Score = 72.0 bits (175), Expect = 8e-13, Method: Composition-based stats.
Identities = 52/188 (27%), Positives = 89/188 (47%), Gaps = 29/188 (15%)
Query 3 IITGSADGKIKIWDSKTQDCLQTF---TPPVPA--------------------HMQTAQC 39
II+GS D +K+W+++T +C+ T T V ++T QC
Sbjct 316 IISGSTDRTLKVWNAETGECIHTLYGHTSTVRCMHLHEKRVVSGSRDATLRVWDIETGQC 375
Query 40 LPSVMSIILAPRNVGADLMYVCTKSNS--ITLMNFAGKTIKKIDKAHDAAIASITFSKDN 97
L +M + A R V D V + + + + + +T + H + S+ F D
Sbjct 376 LHVLMGHVAAVRCVQYDGRRVVSGAYDFMVKVWDPETETCLHTLQGHTNRVYSLQF--DG 433
Query 98 THILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSK 157
H+++GS DT+ R+ ++ G + GH + + + L DN +++G+AD +KIWD K
Sbjct 434 IHVVSGSLDTSIRVWDVETGNCIHTLTGHQSLTS-GMELKDNI-LVSGNADSTVKIWDIK 491
Query 158 TQDCLQTF 165
T CLQT
Sbjct 492 TGQCLQTL 499
Score = 60.8 bits (146), Expect = 2e-09, Method: Composition-based stats.
Identities = 43/164 (26%), Positives = 72/164 (43%), Gaps = 19/164 (11%)
Query 2 RIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVC 61
RI++GS D +K+W + T CL+T T S M N+ +
Sbjct 275 RIVSGSDDNTLKVWSAVTGKCLRTLVG------HTGGVWSSQMR-----DNI------II 317
Query 62 TKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
+ S TL + +T + I + +++GS D T R+ ++ G+ L
Sbjct 318 SGSTDRTLKVWNAETGECIHTLYGHTSTVRCMHLHEKRVVSGSRDATLRVWDIETGQCLH 377
Query 122 EFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
GH+ V C Y D R+++G+ D +K+WD +T+ CL T
Sbjct 378 VLMGHVAAVRCVQY--DGRRVVSGAYDFMVKVWDPETETCLHTL 419
Score = 51.6 bits (122), Expect = 1e-06, Method: Composition-based stats.
Identities = 42/169 (24%), Positives = 77/169 (45%), Gaps = 26/169 (15%)
Query 2 RIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVC 61
R+++G+ D +K+WD +T+ CL T H L D ++V
Sbjct 395 RVVSGAYDFMVKVWDPETETCLHTLQ----GHTNRVYSLQ-------------FDGIHVV 437
Query 62 TKS--NSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKT 119
+ S SI + + H + + + KDN +++G+ D+T +I +K G+
Sbjct 438 SGSLDTSIRVWDVETGNCIHTLTGHQSLTSGMEL-KDNI-LVSGNADSTVKIWDIKTGQC 495
Query 120 LKEFRG---HLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
L+ +G H + V C + + +IT S DG +K+WD KT + ++
Sbjct 496 LQTLQGPNKHQSAVTCLQF--NKNFVITSSDDGTVKLWDLKTGEFIRNL 542
Score = 42.7 bits (99), Expect = 6e-04, Method: Composition-based stats.
Identities = 29/88 (32%), Positives = 47/88 (53%), Gaps = 5/88 (5%)
Query 79 KIDKAHD-AAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLP 137
K+ K HD I + F + I++GS D T ++ GK L+ GH V + +
Sbjct 256 KVLKGHDDHVITCLQFCGN--RIVSGSDDNTLKVWSAVTGKCLRTLVGHTGGVWSS-QMR 312
Query 138 DNTRIITGSADGKIKIWDSKTQDCLQTF 165
DN II+GS D +K+W+++T +C+ T
Sbjct 313 DNI-IISGSTDRTLKVWNAETGECIHTL 339
Score = 35.8 bits (81), Expect = 0.062, Method: Composition-based stats.
Identities = 25/69 (36%), Positives = 34/69 (49%), Gaps = 4/69 (5%)
Query 97 NTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDS 156
+ +I DT R LK+ K LK GH V L N RI++GS D +K+W +
Sbjct 235 SAYIRQHRIDTNWRRGELKSPKVLK---GHDDHVITCLQFCGN-RIVSGSDDNTLKVWSA 290
Query 157 KTQDCLQTF 165
T CL+T
Sbjct 291 VTGKCLRTL 299
> mmu:69544 Wdr5b, 2310009C03Rik, AI606931; WD repeat domain 5B;
K14963 COMPASS component SWD3
Length=328
Score = 68.6 bits (166), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 31/89 (34%), Positives = 53/89 (59%), Gaps = 0/89 (0%)
Query 78 KKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLP 137
KK H I+ + +S D++ +++ S D T ++ +++GK LK +GH FV C + P
Sbjct 74 KKTLYGHSLEISDVAWSSDSSRLVSASDDKTLKVWDMRSGKCLKTLKGHSDFVFCCDFNP 133
Query 138 DNTRIITGSADGKIKIWDSKTQDCLQTFT 166
+ I++GS D +KIW+ KT CL+T +
Sbjct 134 PSNLIVSGSFDESVKIWEVKTGKCLKTLS 162
Score = 60.1 bits (144), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 43/171 (25%), Positives = 82/171 (47%), Gaps = 22/171 (12%)
Query 1 TRIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYV 60
+R+++ S D +K+WD ++ CL+T H C N ++L+
Sbjct 94 SRLVSASDDKTLKVWDMRSGKCLKTLK----GHSDFVFC---------CDFNPPSNLIVS 140
Query 61 CTKSNSITLMNF-AGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKT 119
+ S+ + GK +K + AH I+++ F+ + + I++GS+D RI +G+
Sbjct 141 GSFDESVKIWEVKTGKCLKTL-SAHSDPISAVNFNCNGSLIVSGSYDGLCRIWDAASGQC 199
Query 120 LK----EFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTFT 166
L+ E ++FV + P+ I+T + D +K+WD CL+T+T
Sbjct 200 LRTLADEGNPPVSFVK---FSPNGKYILTATLDNTLKLWDYSRGRCLKTYT 247
Score = 56.2 bits (134), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 39/165 (23%), Positives = 75/165 (45%), Gaps = 17/165 (10%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
I++GS D +KIW+ KT CL+T + AH I N L+ +
Sbjct 138 IVSGSFDESVKIWEVKTGKCLKTLS----AHSDP---------ISAVNFNCNGSLIVSGS 184
Query 63 KSNSITLMNFA-GKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
+ + A G+ ++ + + ++ + FS + +ILT + D T ++ G+ LK
Sbjct 185 YDGLCRIWDAASGQCLRTLADEGNPPVSFVKFSPNGKYILTATLDNTLKLWDYSRGRCLK 244
Query 122 EFRGHLTFVNC---ALYLPDNTRIITGSADGKIKIWDSKTQDCLQ 163
+ GH C + + +++GS D + IW+ +T++ +Q
Sbjct 245 TYTGHKNEKYCLFASFSVTGRKWVVSGSEDNMVYIWNLQTKEIVQ 289
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 46/169 (27%), Positives = 75/169 (44%), Gaps = 30/169 (17%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
I++GS DG +IWD+ + CL+T + P V + +P + T
Sbjct 180 IVSGSYDGLCRIWDAASGQCLRTLAD---------EGNPPVSFVKFSPN---GKYILTAT 227
Query 63 KSNSITLMNFA-GKTIK-----KIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKA 116
N++ L +++ G+ +K K +K A S+T K +++GS D I L+
Sbjct 228 LDNTLKLWDYSRGRCLKTYTGHKNEKYCLFASFSVTGRK---WVVSGSEDNMVYIWNLQT 284
Query 117 GKTLKEFRGHLTFVNCALYLPDNTRIITGSA----DGKIKIWDSKTQDC 161
+ ++ +GH V A P T+ I SA D IK+W S DC
Sbjct 285 KEIVQRLQGHTDVVISAACHP--TKNIIASAALENDKTIKVWSS---DC 328
> hsa:54554 WDR5B, FLJ11287, MGC49879; WD repeat domain 5B; K14963
COMPASS component SWD3
Length=330
Score = 67.0 bits (162), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 32/95 (33%), Positives = 57/95 (60%), Gaps = 1/95 (1%)
Query 72 FAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVN 131
+ GK K + H+ I+ + +S D++ +++ S D T ++ +++GK LK +GH +V
Sbjct 71 YDGKYEKTL-YGHNLEISDVAWSSDSSRLVSASDDKTLKLWDVRSGKCLKTLKGHSNYVF 129
Query 132 CALYLPDNTRIITGSADGKIKIWDSKTQDCLQTFT 166
C + P + II+GS D +KIW+ KT CL+T +
Sbjct 130 CCNFNPPSNLIISGSFDETVKIWEVKTGKCLKTLS 164
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 31/93 (33%), Positives = 50/93 (53%), Gaps = 1/93 (1%)
Query 73 AGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNC 132
+GK +K + K H + F+ + I++GSFD T +I +K GK LK H V+
Sbjct 114 SGKCLKTL-KGHSNYVFCCNFNPPSNLIISGSFDETVKIWEVKTGKCLKTLSAHSDPVSA 172
Query 133 ALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
+ + I++GS DG +IWD+ + CL+T
Sbjct 173 VHFNCSGSLIVSGSYDGLCRIWDAASGQCLKTL 205
Score = 55.5 bits (132), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 44/173 (25%), Positives = 79/173 (45%), Gaps = 26/173 (15%)
Query 1 TRIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCL---PSVMSIILAPRNVGADL 57
+R+++ S D +K+WD ++ CL+T + H C P II + +
Sbjct 96 SRLVSASDDKTLKLWDVRSGKCLKT----LKGHSNYVFCCNFNPPSNLIISGSFDETVKI 151
Query 58 MYVCTKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAG 117
V T GK +K + AH ++++ F+ + I++GS+D RI +G
Sbjct 152 WEVKT-----------GKCLKTL-SAHSDPVSAVHFNCSGSLIVSGSYDGLCRIWDAASG 199
Query 118 KTLKEF----RGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTFT 166
+ LK ++FV + P+ I+T + D +K+WD CL+T+T
Sbjct 200 QCLKTLVDDDNPPVSFVK---FSPNGKYILTATLDNTLKLWDYSRGRCLKTYT 249
Score = 55.5 bits (132), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 43/168 (25%), Positives = 78/168 (46%), Gaps = 23/168 (13%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTP---PVPA-HMQTAQCLPSVMSIILAPRNVGADLM 58
II+GS D +KIW+ KT CL+T + PV A H + S+I++ G +
Sbjct 140 IISGSFDETVKIWEVKTGKCLKTLSAHSDPVSAVHFNCSG------SLIVSGSYDGLCRI 193
Query 59 YVCTKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGK 118
+ +G+ +K + + ++ + FS + +ILT + D T ++ G+
Sbjct 194 WDAA----------SGQCLKTLVDDDNPPVSFVKFSPNGKYILTATLDNTLKLWDYSRGR 243
Query 119 TLKEFRGHLTFVNC---ALYLPDNTRIITGSADGKIKIWDSKTQDCLQ 163
LK + GH C + I++GS D + IW+ +T++ +Q
Sbjct 244 CLKTYTGHKNEKYCIFANFSVTGGKWIVSGSEDNLVYIWNLQTKEIVQ 291
Score = 48.9 bits (115), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 24/83 (28%), Positives = 43/83 (51%), Gaps = 0/83 (0%)
Query 83 AHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRI 142
H A++S+ FS + + + S D I G GK K GH ++ + D++R+
Sbjct 39 GHTEAVSSVKFSPNGEWLASSSADRLIIIWGAYDGKYEKTLYGHNLEISDVAWSSDSSRL 98
Query 143 ITGSADGKIKIWDSKTQDCLQTF 165
++ S D +K+WD ++ CL+T
Sbjct 99 VSASDDKTLKLWDVRSGKCLKTL 121
Score = 39.3 bits (90), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 41/160 (25%), Positives = 70/160 (43%), Gaps = 23/160 (14%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
I++GS DG +IWD+ + CL+T P V + +P + T
Sbjct 182 IVSGSYDGLCRIWDAASGQCLKTLVDDDN---------PPVSFVKFSPN---GKYILTAT 229
Query 63 KSNSITLMNFA-GKTIK-----KIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKA 116
N++ L +++ G+ +K K +K A S+T K I++GS D I L+
Sbjct 230 LDNTLKLWDYSRGRCLKTYTGHKNEKYCIFANFSVTGGK---WIVSGSEDNLVYIWNLQT 286
Query 117 GKTLKEFRGHLTFVNCALYLPDNTRIITGS--ADGKIKIW 154
+ +++ +GH V A P I + + D IK+W
Sbjct 287 KEIVQKLQGHTDVVISAACHPTENLIASAALENDKTIKLW 326
> dre:406322 poc1a, wdr51a, wu:fl82d01, zgc:56055; POC1 centriolar
protein homolog A (Chlamydomonas)
Length=416
Score = 67.0 bits (162), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 41/164 (25%), Positives = 76/164 (46%), Gaps = 13/164 (7%)
Query 2 RIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVC 61
+I +GS D + +W+ K Q FT H C + +P A L+
Sbjct 32 QIASGSVDASVMVWNMKPQSRAYRFT----GHKDAVTC------VQFSP---SAHLLASS 78
Query 62 TKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
++ ++ L + K + +AH ++ S+ FS D +LT S D + ++ + K +
Sbjct 79 SRDKTVRLWVPSVKGESVLFRAHTGSVRSVCFSADGQSLLTASDDQSIKLWSVHRQKIIC 138
Query 122 EFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
R H +V CA + PD +++ S D +K+WD+ ++ + TF
Sbjct 139 TLREHNNWVRCARFSPDGQLMVSVSDDRTVKLWDASSRQLIHTF 182
Score = 55.1 bits (131), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 33/152 (21%), Positives = 67/152 (44%), Gaps = 13/152 (8%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
+++ S D +K+WD+ ++ + TF P + PS I A +
Sbjct 159 MVSVSDDRTVKLWDASSRQLIHTFCEP-GGYSSYVDFHPSSTCIATA------------S 205
Query 63 KSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKE 122
N++ + + T+ + + H AA+ +++F H+LT S D+T +I L G+ L
Sbjct 206 SDNTVRVWDIRTHTLLQHYQVHSAAVNALSFHPSGNHLLTASSDSTLKILDLLEGRLLYT 265
Query 123 FRGHLTFVNCALYLPDNTRIITGSADGKIKIW 154
GH +C + + + +D ++ +W
Sbjct 266 LHGHQGSASCVSFSRSGDQFASAGSDQQVMVW 297
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 22/79 (27%), Positives = 36/79 (45%), Gaps = 0/79 (0%)
Query 76 TIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALY 135
T+++ K H AI S+ FS I +GS D + + +K F GH V C +
Sbjct 9 TLERNFKGHRDAITSLDFSPSGKQIASGSVDASVMVWNMKPQSRAYRFTGHKDAVTCVQF 68
Query 136 LPDNTRIITGSADGKIKIW 154
P + + S D +++W
Sbjct 69 SPSAHLLASSSRDKTVRLW 87
Score = 39.3 bits (90), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 33/153 (21%), Positives = 60/153 (39%), Gaps = 13/153 (8%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
++T S D IK+W Q + T H +C A + LM +
Sbjct 117 LLTASDDQSIKLWSVHRQKIICTLR----EHNNWVRC---------ARFSPDGQLMVSVS 163
Query 63 KSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKE 122
++ L + + + + + + F +T I T S D T R+ ++ L+
Sbjct 164 DDRTVKLWDASSRQLIHTFCEPGGYSSYVDFHPSSTCIATASSDNTVRVWDIRTHTLLQH 223
Query 123 FRGHLTFVNCALYLPDNTRIITGSADGKIKIWD 155
++ H VN + P ++T S+D +KI D
Sbjct 224 YQVHSAAVNALSFHPSGNHLLTASSDSTLKILD 256
Score = 32.3 bits (72), Expect = 0.69, Method: Compositional matrix adjust.
Identities = 13/46 (28%), Positives = 22/46 (47%), Gaps = 0/46 (0%)
Query 121 KEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTFT 166
+ F+GH + + P +I +GS D + +W+ K Q FT
Sbjct 12 RNFKGHRDAITSLDFSPSGKQIASGSVDASVMVWNMKPQSRAYRFT 57
> cel:K10B2.1 lin-23; abnormal cell LINeage family member (lin-23);
K03362 F-box and WD-40 domain protein 1/11
Length=665
Score = 66.6 bits (161), Expect = 3e-11, Method: Composition-based stats.
Identities = 41/168 (24%), Positives = 80/168 (47%), Gaps = 26/168 (15%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPA--HMQTAQCLPSVMSIILAPRNVGADLMYV 60
II+GS+D +++WD +T +C++T A H++ A +M
Sbjct 275 IISGSSDATVRVWDVETGECIKTLIHHCEAVLHLRFAN-----------------GIMVT 317
Query 61 CTKSNSITLMNFAGK---TIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAG 117
C+K SI + + TI+++ H AA+ + F D+ +I++ S D T ++ +
Sbjct 318 CSKDRSIAVWDMVSPRDITIRRVLVGHRAAVNVVDF--DDRYIVSASGDRTIKVWSMDTL 375
Query 118 KTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
+ ++ GH + C Y +++GS+D I++WD + CL+
Sbjct 376 EFVRTLAGHRRGIACLQY--RGRLVVSGSSDNTIRLWDIHSGVCLRVL 421
Score = 46.6 bits (109), Expect = 4e-05, Method: Composition-based stats.
Identities = 24/70 (34%), Positives = 38/70 (54%), Gaps = 2/70 (2%)
Query 96 DNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWD 155
D+ I++G D T +I K + GH V C Y DN II+GS+D +++WD
Sbjct 231 DDDKIVSGLRDNTIKIWDRKDYSCSRILSGHTGSVLCLQY--DNRVIISGSSDATVRVWD 288
Query 156 SKTQDCLQTF 165
+T +C++T
Sbjct 289 VETGECIKTL 298
Score = 36.6 bits (83), Expect = 0.037, Method: Composition-based stats.
Identities = 25/94 (26%), Positives = 46/94 (48%), Gaps = 6/94 (6%)
Query 63 KSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKE 122
+ N+I + + + +I H ++ + + DN I++GS D T R+ ++ G+ +K
Sbjct 240 RDNTIKIWDRKDYSCSRILSGHTGSVLCLQY--DNRVIISGSSDATVRVWDVETGECIKT 297
Query 123 FRGHLTFVNCALYLP-DNTRIITGSADGKIKIWD 155
H V L+L N ++T S D I +WD
Sbjct 298 LIHHCEAV---LHLRFANGIMVTCSKDRSIAVWD 328
Score = 30.4 bits (67), Expect = 2.7, Method: Composition-based stats.
Identities = 10/15 (66%), Positives = 14/15 (93%), Gaps = 0/15 (0%)
Query 2 RIITGSADGKIKIWD 16
RI++G+ DGKIK+WD
Sbjct 437 RIVSGAYDGKIKVWD 451
> xla:446809 wdr5-a, MGC80538, big-3, swd3, xwdr5; WD repeat domain
5; K14963 COMPASS component SWD3
Length=334
Score = 65.9 bits (159), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 33/94 (35%), Positives = 53/94 (56%), Gaps = 1/94 (1%)
Query 72 FAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVN 131
+ GK K I H I+ + +S D+ +++ S D T +I + +GK LK +GH +V
Sbjct 75 YDGKFEKTIS-GHKLGISDVAWSSDSNLLVSASDDKTLKIWDISSGKCLKTLKGHSNYVF 133
Query 132 CALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
C + P + I++GS D ++IWD KT CL+T
Sbjct 134 CCNFNPQSNLIVSGSFDESVRIWDVKTGKCLKTL 167
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 31/93 (33%), Positives = 50/93 (53%), Gaps = 1/93 (1%)
Query 73 AGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNC 132
+GK +K + K H + F+ + I++GSFD + RI +K GK LK H V+
Sbjct 118 SGKCLKTL-KGHSNYVFCCNFNPQSNLIVSGSFDESVRIWDVKTGKCLKTLPAHSDPVSA 176
Query 133 ALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
+ D + I++ S DG +IWD+ + CL+T
Sbjct 177 VHFNRDGSLIVSSSYDGLCRIWDTASGQCLKTL 209
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 41/169 (24%), Positives = 79/169 (46%), Gaps = 22/169 (13%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
+++ S D +KIWD + CL+T + H C N ++L+ +
Sbjct 102 LVSASDDKTLKIWDISSGKCLKT----LKGHSNYVFC---------CNFNPQSNLIVSGS 148
Query 63 KSNSITLMNF-AGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
S+ + + GK +K + AH ++++ F++D + I++ S+D RI +G+ LK
Sbjct 149 FDESVRIWDVKTGKCLKTL-PAHSDPVSAVHFNRDGSLIVSSSYDGLCRIWDTASGQCLK 207
Query 122 EF----RGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTFT 166
++FV + P+ I+ + D +K+WD CL+T+T
Sbjct 208 TLIDDDNPPVSFVK---FSPNGKYILAATLDNTLKLWDYSKGKCLKTYT 253
Score = 53.1 bits (126), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 41/166 (24%), Positives = 76/166 (45%), Gaps = 19/166 (11%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQ--TAQCLPSVMSIILAPRNVGADLMYV 60
I++GS D ++IWD KT CL+T +PAH +A S+I++ G ++
Sbjct 144 IVSGSFDESVRIWDVKTGKCLKT----LPAHSDPVSAVHFNRDGSLIVSSSYDGLCRIWD 199
Query 61 CTKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTL 120
+G+ +K + + ++ + FS + +IL + D T ++ GK L
Sbjct 200 TA----------SGQCLKTLIDDDNPPVSFVKFSPNGKYILAATLDNTLKLWDYSKGKCL 249
Query 121 KEFRGHLTFVNC---ALYLPDNTRIITGSADGKIKIWDSKTQDCLQ 163
K + H C + I++GS D + IW+ +T++ +Q
Sbjct 250 KTYTCHKNEKYCIFANFSVTGGKWIVSGSEDNLVYIWNLQTKEVVQ 295
Score = 46.6 bits (109), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 26/90 (28%), Positives = 44/90 (48%), Gaps = 0/90 (0%)
Query 76 TIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALY 135
T+K H A++S+ FS + + + S D +I G GK K GH ++ +
Sbjct 36 TLKFTLAGHTKAVSSVKFSPNGEWLASSSADKLIKIWGAYDGKFEKTISGHKLGISDVAW 95
Query 136 LPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
D+ +++ S D +KIWD + CL+T
Sbjct 96 SSDSNLLVSASDDKTLKIWDISSGKCLKTL 125
> cel:C14B1.4 tag-125; Temporarily Assigned Gene name family member
(tag-125); K14963 COMPASS component SWD3
Length=376
Score = 65.9 bits (159), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 47/165 (28%), Positives = 81/165 (49%), Gaps = 17/165 (10%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
+++GS D ++IWD KT C++T +PAH S +S N L+ +
Sbjct 186 VVSGSFDESVRIWDVKTGMCIKT----LPAHSDPV----SAVSF-----NRDGSLIASGS 232
Query 63 KSNSITLMNFA-GKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
+ + + A G+ IK + + +A + FS + +IL + D+T ++ GKTLK
Sbjct 233 YDGLVRIWDTANGQCIKTLVDDENPPVAFVKFSPNGKYILASNLDSTLKLWDFSKGKTLK 292
Query 122 EFRGHLTFVNC---ALYLPDNTRIITGSADGKIKIWDSKTQDCLQ 163
++ GH C + II+GS D KI IW+ +T++ +Q
Sbjct 293 QYTGHENSKYCIFANFSVTGGKWIISGSEDCKIYIWNLQTREIVQ 337
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 46/163 (28%), Positives = 74/163 (45%), Gaps = 17/163 (10%)
Query 5 TGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCTKS 64
T SAD +KIW+ C +T T H V I + +D V + S
Sbjct 104 TSSADKTVKIWNMDHMICERTLT----GHKL------GVNDIAWS-----SDSRCVVSAS 148
Query 65 NSITLMNFAGKTIK--KIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKE 122
+ TL F T + K K H+ + F+ ++ +++GSFD + RI +K G +K
Sbjct 149 DDKTLKIFEIVTSRMTKTLKGHNNYVFCCNFNPQSSLVVSGSFDESVRIWDVKTGMCIKT 208
Query 123 FRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
H V+ + D + I +GS DG ++IWD+ C++T
Sbjct 209 LPAHSDPVSAVSFNRDGSLIASGSYDGLVRIWDTANGQCIKTL 251
> mmu:140858 Wdr5, 2410008O07Rik, AA408785, AA960360, Big, Big-3;
WD repeat domain 5; K14963 COMPASS component SWD3
Length=334
Score = 65.9 bits (159), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 33/94 (35%), Positives = 53/94 (56%), Gaps = 1/94 (1%)
Query 72 FAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVN 131
+ GK K I H I+ + +S D+ +++ S D T +I + +GK LK +GH +V
Sbjct 75 YDGKFEKTIS-GHKLGISDVAWSSDSNLLVSASDDKTLKIWDVSSGKCLKTLKGHSNYVF 133
Query 132 CALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
C + P + I++GS D ++IWD KT CL+T
Sbjct 134 CCNFNPQSNLIVSGSFDESVRIWDVKTGKCLKTL 167
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 31/93 (33%), Positives = 50/93 (53%), Gaps = 1/93 (1%)
Query 73 AGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNC 132
+GK +K + K H + F+ + I++GSFD + RI +K GK LK H V+
Sbjct 118 SGKCLKTL-KGHSNYVFCCNFNPQSNLIVSGSFDESVRIWDVKTGKCLKTLPAHSDPVSA 176
Query 133 ALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
+ D + I++ S DG +IWD+ + CL+T
Sbjct 177 VHFNRDGSLIVSSSYDGLCRIWDTASGQCLKTL 209
Score = 56.2 bits (134), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 41/165 (24%), Positives = 74/165 (44%), Gaps = 17/165 (10%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
I++GS D ++IWD KT CL+T +PAH + N L+ +
Sbjct 144 IVSGSFDESVRIWDVKTGKCLKT----LPAHSDPVSAVH---------FNRDGSLIVSSS 190
Query 63 KSNSITLMNFA-GKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
+ + A G+ +K + + ++ + FS + +IL + D T ++ GK LK
Sbjct 191 YDGLCRIWDTASGQCLKTLIDDDNPPVSFVKFSPNGKYILAATLDNTLKLWDYSKGKCLK 250
Query 122 EFRGHLTFVNC---ALYLPDNTRIITGSADGKIKIWDSKTQDCLQ 163
+ GH C + I++GS D + IW+ +T++ +Q
Sbjct 251 TYTGHKNEKYCIFANFSVTGGKWIVSGSEDNLVYIWNLQTKEIVQ 295
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 41/169 (24%), Positives = 78/169 (46%), Gaps = 22/169 (13%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
+++ S D +KIWD + CL+T H C N ++L+ +
Sbjct 102 LVSASDDKTLKIWDVSSGKCLKTLK----GHSNYVFC---------CNFNPQSNLIVSGS 148
Query 63 KSNSITLMNF-AGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
S+ + + GK +K + AH ++++ F++D + I++ S+D RI +G+ LK
Sbjct 149 FDESVRIWDVKTGKCLKTL-PAHSDPVSAVHFNRDGSLIVSSSYDGLCRIWDTASGQCLK 207
Query 122 EF----RGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTFT 166
++FV + P+ I+ + D +K+WD CL+T+T
Sbjct 208 TLIDDDNPPVSFVK---FSPNGKYILAATLDNTLKLWDYSKGKCLKTYT 253
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 24/83 (28%), Positives = 41/83 (49%), Gaps = 0/83 (0%)
Query 83 AHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRI 142
H A++S+ FS + + + S D +I G GK K GH ++ + D+ +
Sbjct 43 GHTKAVSSVKFSPNGEWLASSSADKLIKIWGAYDGKFEKTISGHKLGISDVAWSSDSNLL 102
Query 143 ITGSADGKIKIWDSKTQDCLQTF 165
++ S D +KIWD + CL+T
Sbjct 103 VSASDDKTLKIWDVSSGKCLKTL 125
> hsa:11091 WDR5, BIG-3, SWD3; WD repeat domain 5; K14963 COMPASS
component SWD3
Length=334
Score = 65.9 bits (159), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 33/94 (35%), Positives = 53/94 (56%), Gaps = 1/94 (1%)
Query 72 FAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVN 131
+ GK K I H I+ + +S D+ +++ S D T +I + +GK LK +GH +V
Sbjct 75 YDGKFEKTIS-GHKLGISDVAWSSDSNLLVSASDDKTLKIWDVSSGKCLKTLKGHSNYVF 133
Query 132 CALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
C + P + I++GS D ++IWD KT CL+T
Sbjct 134 CCNFNPQSNLIVSGSFDESVRIWDVKTGKCLKTL 167
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 31/93 (33%), Positives = 50/93 (53%), Gaps = 1/93 (1%)
Query 73 AGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNC 132
+GK +K + K H + F+ + I++GSFD + RI +K GK LK H V+
Sbjct 118 SGKCLKTL-KGHSNYVFCCNFNPQSNLIVSGSFDESVRIWDVKTGKCLKTLPAHSDPVSA 176
Query 133 ALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
+ D + I++ S DG +IWD+ + CL+T
Sbjct 177 VHFNRDGSLIVSSSYDGLCRIWDTASGQCLKTL 209
Score = 56.2 bits (134), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 41/165 (24%), Positives = 74/165 (44%), Gaps = 17/165 (10%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
I++GS D ++IWD KT CL+T +PAH + N L+ +
Sbjct 144 IVSGSFDESVRIWDVKTGKCLKT----LPAHSDPVSAVH---------FNRDGSLIVSSS 190
Query 63 KSNSITLMNFA-GKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
+ + A G+ +K + + ++ + FS + +IL + D T ++ GK LK
Sbjct 191 YDGLCRIWDTASGQCLKTLIDDDNPPVSFVKFSPNGKYILAATLDNTLKLWDYSKGKCLK 250
Query 122 EFRGHLTFVNC---ALYLPDNTRIITGSADGKIKIWDSKTQDCLQ 163
+ GH C + I++GS D + IW+ +T++ +Q
Sbjct 251 TYTGHKNEKYCIFANFSVTGGKWIVSGSEDNLVYIWNLQTKEIVQ 295
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 41/169 (24%), Positives = 78/169 (46%), Gaps = 22/169 (13%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
+++ S D +KIWD + CL+T H C N ++L+ +
Sbjct 102 LVSASDDKTLKIWDVSSGKCLKTLK----GHSNYVFC---------CNFNPQSNLIVSGS 148
Query 63 KSNSITLMNF-AGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
S+ + + GK +K + AH ++++ F++D + I++ S+D RI +G+ LK
Sbjct 149 FDESVRIWDVKTGKCLKTL-PAHSDPVSAVHFNRDGSLIVSSSYDGLCRIWDTASGQCLK 207
Query 122 EF----RGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTFT 166
++FV + P+ I+ + D +K+WD CL+T+T
Sbjct 208 TLIDDDNPPVSFVK---FSPNGKYILAATLDNTLKLWDYSKGKCLKTYT 253
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 24/83 (28%), Positives = 41/83 (49%), Gaps = 0/83 (0%)
Query 83 AHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRI 142
H A++S+ FS + + + S D +I G GK K GH ++ + D+ +
Sbjct 43 GHTKAVSSVKFSPNGEWLASSSADKLIKIWGAYDGKFEKTISGHKLGISDVAWSSDSNLL 102
Query 143 ITGSADGKIKIWDSKTQDCLQTF 165
++ S D +KIWD + CL+T
Sbjct 103 VSASDDKTLKIWDVSSGKCLKTL 125
> xla:447447 wdr5-b, MGC81485, big-3, swd3, wdr5, xwdr5; WD repeat
domain 5; K14963 COMPASS component SWD3
Length=334
Score = 65.9 bits (159), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 33/94 (35%), Positives = 53/94 (56%), Gaps = 1/94 (1%)
Query 72 FAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVN 131
+ GK K I H I+ + +S D+ +++ S D T +I + +GK LK +GH +V
Sbjct 75 YDGKFEKTIS-GHKLGISDVAWSSDSNLLVSASDDKTLKIWDVSSGKCLKTLKGHSNYVF 133
Query 132 CALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
C + P + I++GS D ++IWD KT CL+T
Sbjct 134 CCNFNPQSNLIVSGSFDESVRIWDVKTGKCLKTL 167
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 31/93 (33%), Positives = 50/93 (53%), Gaps = 1/93 (1%)
Query 73 AGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNC 132
+GK +K + K H + F+ + I++GSFD + RI +K GK LK H V+
Sbjct 118 SGKCLKTL-KGHSNYVFCCNFNPQSNLIVSGSFDESVRIWDVKTGKCLKTLPAHSDPVSA 176
Query 133 ALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
+ D + I++ S DG +IWD+ + CL+T
Sbjct 177 VHFNRDGSLIVSSSYDGLCRIWDTASGQCLKTL 209
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 41/169 (24%), Positives = 79/169 (46%), Gaps = 22/169 (13%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
+++ S D +KIWD + CL+T + H C N ++L+ +
Sbjct 102 LVSASDDKTLKIWDVSSGKCLKT----LKGHSNYVFC---------CNFNPQSNLIVSGS 148
Query 63 KSNSITLMNF-AGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
S+ + + GK +K + AH ++++ F++D + I++ S+D RI +G+ LK
Sbjct 149 FDESVRIWDVKTGKCLKTL-PAHSDPVSAVHFNRDGSLIVSSSYDGLCRIWDTASGQCLK 207
Query 122 EF----RGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTFT 166
++FV + P+ I+ + D +K+WD CL+T+T
Sbjct 208 TLIDDDNPPVSFVK---FSPNGKYILAATLDNTLKLWDYSKGKCLKTYT 253
Score = 53.1 bits (126), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 41/166 (24%), Positives = 76/166 (45%), Gaps = 19/166 (11%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQ--TAQCLPSVMSIILAPRNVGADLMYV 60
I++GS D ++IWD KT CL+T +PAH +A S+I++ G ++
Sbjct 144 IVSGSFDESVRIWDVKTGKCLKT----LPAHSDPVSAVHFNRDGSLIVSSSYDGLCRIWD 199
Query 61 CTKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTL 120
+G+ +K + + ++ + FS + +IL + D T ++ GK L
Sbjct 200 TA----------SGQCLKTLIDDDNPPVSFVKFSPNGKYILAATLDNTLKLWDYSKGKCL 249
Query 121 KEFRGHLTFVNC---ALYLPDNTRIITGSADGKIKIWDSKTQDCLQ 163
K + H C + I++GS D + IW+ +T++ +Q
Sbjct 250 KTYTCHKNEKYCIFANFSVTGGKWIVSGSEDNLVYIWNLQTKEVVQ 295
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 26/90 (28%), Positives = 44/90 (48%), Gaps = 0/90 (0%)
Query 76 TIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALY 135
T+K H A++S+ FS + + + S D +I G GK K GH ++ +
Sbjct 36 TLKFTLAGHTKAVSSVKFSPNGEWLASSSADKLIKIWGAYDGKFEKTISGHKLGISDVAW 95
Query 136 LPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
D+ +++ S D +KIWD + CL+T
Sbjct 96 SSDSNLLVSASDDKTLKIWDVSSGKCLKTL 125
> dre:406372 wdr5, wu:fk47f04, zgc:56591, zgc:76895; WD repeat
domain 5; K14963 COMPASS component SWD3
Length=334
Score = 65.9 bits (159), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 33/94 (35%), Positives = 53/94 (56%), Gaps = 1/94 (1%)
Query 72 FAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVN 131
+ GK K I H I+ + +S D+ +++ S D T +I + +GK LK +GH +V
Sbjct 75 YDGKFEKTI-SGHKLGISDVAWSSDSNLLVSASDDKTLKIWDVSSGKCLKTLKGHSNYVF 133
Query 132 CALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
C + P + I++GS D ++IWD KT CL+T
Sbjct 134 CCNFNPQSNLIVSGSFDESVRIWDVKTGKCLKTL 167
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 31/93 (33%), Positives = 50/93 (53%), Gaps = 1/93 (1%)
Query 73 AGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNC 132
+GK +K + K H + F+ + I++GSFD + RI +K GK LK H V+
Sbjct 118 SGKCLKTL-KGHSNYVFCCNFNPQSNLIVSGSFDESVRIWDVKTGKCLKTLPAHSDPVSA 176
Query 133 ALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
+ D + I++ S DG +IWD+ + CL+T
Sbjct 177 VHFNRDGSLIVSSSYDGLCRIWDTASGQCLKTL 209
Score = 57.0 bits (136), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 43/166 (25%), Positives = 79/166 (47%), Gaps = 19/166 (11%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQ--TAQCLPSVMSIILAPRNVGADLMYV 60
I++GS D ++IWD KT CL+T +PAH +A S+I++ G +
Sbjct 144 IVSGSFDESVRIWDVKTGKCLKT----LPAHSDPVSAVHFNRDGSLIVSSSYDG-----L 194
Query 61 CTKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTL 120
C ++ + G+ +K + + ++ + FS + +IL + D T ++ GK L
Sbjct 195 CRIWDTAS-----GQCLKTLIDDDNPPVSFVKFSPNGKYILAATLDNTLKLWDYSKGKCL 249
Query 121 KEFRGHLTFVNC---ALYLPDNTRIITGSADGKIKIWDSKTQDCLQ 163
K + GH C + I++GS D + IW+ +T++ +Q
Sbjct 250 KTYTGHKNEKYCIFANFSVTGGKWIVSGSEDNMVYIWNLQTKEIVQ 295
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 41/169 (24%), Positives = 79/169 (46%), Gaps = 22/169 (13%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
+++ S D +KIWD + CL+T + H C N ++L+ +
Sbjct 102 LVSASDDKTLKIWDVSSGKCLKT----LKGHSNYVFC---------CNFNPQSNLIVSGS 148
Query 63 KSNSITLMNF-AGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
S+ + + GK +K + AH ++++ F++D + I++ S+D RI +G+ LK
Sbjct 149 FDESVRIWDVKTGKCLKTL-PAHSDPVSAVHFNRDGSLIVSSSYDGLCRIWDTASGQCLK 207
Query 122 EF----RGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTFT 166
++FV + P+ I+ + D +K+WD CL+T+T
Sbjct 208 TLIDDDNPPVSFVK---FSPNGKYILAATLDNTLKLWDYSKGKCLKTYT 253
Score = 46.2 bits (108), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 26/90 (28%), Positives = 43/90 (47%), Gaps = 0/90 (0%)
Query 76 TIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALY 135
T+K H A++S+ FS + + S D +I G GK K GH ++ +
Sbjct 36 TLKFTLAGHTKAVSSVKFSPSGEWLASSSADKLIKIWGAYDGKFEKTISGHKLGISDVAW 95
Query 136 LPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
D+ +++ S D +KIWD + CL+T
Sbjct 96 SSDSNLLVSASDDKTLKIWDVSSGKCLKTL 125
> mmu:103583 Fbxw11, 2310065A07Rik, AA536858, BTRC2, BTRCP2, Fbxw1b,
HOS, mKIAA0696; F-box and WD-40 domain protein 11; K03362
F-box and WD-40 domain protein 1/11
Length=563
Score = 65.1 bits (157), Expect = 1e-10, Method: Composition-based stats.
Identities = 51/194 (26%), Positives = 79/194 (40%), Gaps = 58/194 (29%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
I+TGS+D +++WD T + L T H A +L R LM C+
Sbjct 314 IVTGSSDSTVRVWDVNTGEVLNTLI-----HHNEA---------VLHLR-FSNGLMVTCS 358
Query 63 KSNSITLMNFAGKT---IKKIDKAHDAAIASITFSKD----------------------- 96
K SI + + A T ++++ H AA+ + F
Sbjct 359 KDRSIAVWDMASATDITLRRVLVGHRAAVNVVDFDDKYIVSASGDRTIKVWSTSTCEFVR 418
Query 97 --NTH-------------ILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTR 141
N H +++GS D T R+ ++ G L+ GH V C + DN R
Sbjct 419 TLNGHKRGIACLQYRDRLVVSGSSDNTIRLWDIECGACLRVLEGHEELVRCIRF--DNKR 476
Query 142 IITGSADGKIKIWD 155
I++G+ DGKIK+WD
Sbjct 477 IVSGAYDGKIKVWD 490
Score = 42.4 bits (98), Expect = 7e-04, Method: Composition-based stats.
Identities = 23/70 (32%), Positives = 36/70 (51%), Gaps = 2/70 (2%)
Query 96 DNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWD 155
D+ I++G D + +I + + LK GH V C Y D I+TGS+D +++WD
Sbjct 270 DDDKIISGLRDNSIKIWDKSSLECLKVLTGHTGSVLCLQY--DERVIVTGSSDSTVRVWD 327
Query 156 SKTQDCLQTF 165
T + L T
Sbjct 328 VNTGEVLNTL 337
Score = 35.0 bits (79), Expect = 0.11, Method: Composition-based stats.
Identities = 16/37 (43%), Positives = 22/37 (59%), Gaps = 2/37 (5%)
Query 130 VNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTFT 166
V C Y D+ +II+G D IKIWD + +CL+ T
Sbjct 264 VYCLQY--DDDKIISGLRDNSIKIWDKSSLECLKVLT 298
Score = 30.0 bits (66), Expect = 3.3, Method: Composition-based stats.
Identities = 10/15 (66%), Positives = 14/15 (93%), Gaps = 0/15 (0%)
Query 2 RIITGSADGKIKIWD 16
RI++G+ DGKIK+WD
Sbjct 476 RIVSGAYDGKIKVWD 490
> dre:406825 fbxw11b, btrc2, fbxw11a, wu:fa12e12, wu:fb11f03,
zgc:63728; F-box and WD-40 domain protein 11b; K03362 F-box
and WD-40 domain protein 1/11
Length=527
Score = 65.1 bits (157), Expect = 1e-10, Method: Composition-based stats.
Identities = 56/224 (25%), Positives = 85/224 (37%), Gaps = 76/224 (33%)
Query 2 RIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPR---------- 51
+II+G D IKIWD +T +CL+ T H + CL +I+
Sbjct 237 KIISGLRDNSIKIWDKQTLECLKILT----GHTGSVLCLQYDERVIVTGSSDSTVRVWDV 292
Query 52 NVGA-------------------DLMYVCTKSNSITLMNFAGKT---IKKIDKAHDAAIA 89
N G LM C+K SI + + A T ++++ H AA+
Sbjct 293 NSGEVLNTLIHHNEAVLHLRFCNGLMVTCSKDRSIAVWDMASPTDISLRRVLVGHRAAVN 352
Query 90 SITFSKD-------------------------NTH-------------ILTGSFDTTARI 111
+ F N H +++GS D T R+
Sbjct 353 VVDFDDKYIVSASGDRTIKVWSTSTCEFVRTLNGHKRGIACLQYRDRLVVSGSSDNTIRL 412
Query 112 HGLKAGKTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWD 155
++ G L+ GH V C + DN RI++G+ DGKIK+WD
Sbjct 413 WDIECGACLRVLEGHEELVRCIRF--DNKRIVSGAYDGKIKVWD 454
Score = 37.0 bits (84), Expect = 0.034, Method: Composition-based stats.
Identities = 17/37 (45%), Positives = 23/37 (62%), Gaps = 2/37 (5%)
Query 130 VNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTFT 166
V C Y D+ +II+G D IKIWD +T +CL+ T
Sbjct 228 VYCLQY--DDEKIISGLRDNSIKIWDKQTLECLKILT 262
> mmu:14697 Gnb5, GBS, Gbeta5, flr; guanine nucleotide binding
protein (G protein), beta 5; K04539 guanine nucleotide binding
protein (G protein), beta 5
Length=395
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 48/166 (28%), Positives = 75/166 (45%), Gaps = 9/166 (5%)
Query 2 RIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVC 61
RI++ S DGK+ +WDS T + T P M A PS +I + + +
Sbjct 119 RIVSSSQDGKVIVWDSFTTNKEHAVTMPCTWVMACAY-APSGCAIACGGLDNKCSVYPLT 177
Query 62 TKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
N N A K KK H +++ +F+ + ILT S D T + +++G+ L+
Sbjct 178 FDKNE----NMAAK--KKSVAMHTNYLSACSFTNSDMQILTASGDGTCALWDVESGQLLQ 231
Query 122 EFRGHLTFVNCALYLPDNT--RIITGSADGKIKIWDSKTQDCLQTF 165
F GH V C P T ++G D K +WD ++ C+Q F
Sbjct 232 SFHGHGADVLCLDLAPSETGNTFVSGGCDKKAMVWDMRSGQCVQAF 277
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 34/156 (21%), Positives = 70/156 (44%), Gaps = 13/156 (8%)
Query 2 RIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVC 61
+I+T S DG +WD ++ LQ+F H A V+ + LAP G +
Sbjct 209 QILTASGDGTCALWDVESGQLLQSF------HGHGAD----VLCLDLAPSETGNTFVSGG 258
Query 62 TKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
++ +G+ ++ + H++ + S+ + +GS D T R++ L+A + +
Sbjct 259 CDKKAMVWDMRSGQCVQAFE-THESDVNSVRYYPSGDAFASGSDDATCRLYDLRADREVA 317
Query 122 EF-RGHLTFVNCALYLPDNTRII-TGSADGKIKIWD 155
+ + + F ++ + R++ G D I +WD
Sbjct 318 IYSKESIIFGASSVDFSLSGRLLFAGYNDYTINVWD 353
Score = 38.1 bits (87), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 17/41 (41%), Positives = 24/41 (58%), Gaps = 0/41 (0%)
Query 118 KTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKT 158
KT + +GH V C + D RI++ S DGK+ +WDS T
Sbjct 96 KTRRTLKGHGNKVLCMDWCKDKRRIVSSSQDGKVIVWDSFT 136
> dre:562813 gnb5a, MGC158678, zgc:158678; guanine nucleotide
binding protein (G protein), beta 5a; K04539 guanine nucleotide
binding protein (G protein), beta 5
Length=355
Score = 64.7 bits (156), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 47/166 (28%), Positives = 75/166 (45%), Gaps = 9/166 (5%)
Query 2 RIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVC 61
RI++ S DGK+ +WD+ T + T P M A PS ++ + + +
Sbjct 79 RIVSSSQDGKVIVWDAFTTNKEHAVTMPCTWVMACAY-APSGCAVACGGLDNKCSVYPLS 137
Query 62 TKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
N N A K KK H +++ F+ + ILT S D T + +++G+ L+
Sbjct 138 LDKNE----NLAAK--KKSVAMHTNYLSACCFTNSDMQILTSSGDGTCALWDVESGQMLQ 191
Query 122 EFRGHLTFVNCALYLPDNT--RIITGSADGKIKIWDSKTQDCLQTF 165
F GH V C P T ++G D K +WD +T C+Q+F
Sbjct 192 SFHGHAADVLCLDLAPSETGNTFVSGGCDKKACVWDMRTGQCVQSF 237
Score = 45.8 bits (107), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 39/157 (24%), Positives = 72/157 (45%), Gaps = 15/157 (9%)
Query 2 RIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYV- 60
+I+T S DG +WD ++ LQ+F H A V+ + LAP G +
Sbjct 169 QILTSSGDGTCALWDVESGQMLQSF------HGHAAD----VLCLDLAPSETGNTFVSGG 218
Query 61 CTKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTL 120
C K + M G+ ++ + +HD+ I S+ + +GS D T R++ L+A + +
Sbjct 219 CDKKACVWDMR-TGQCVQSFE-SHDSDINSVRYYPSGDAFASGSDDATCRLYDLRADREV 276
Query 121 KEF-RGHLTFVNCALYLPDNTRIITGSA-DGKIKIWD 155
+ + + F ++ + R++ G D I +WD
Sbjct 277 AIYSKESIIFGASSVDFSLSGRLLFGGYNDYTINVWD 313
Score = 37.0 bits (84), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 16/41 (39%), Positives = 24/41 (58%), Gaps = 0/41 (0%)
Query 118 KTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKT 158
KT + +GH V C + D RI++ S DGK+ +WD+ T
Sbjct 56 KTRRTLKGHGNKVLCMDWCKDKRRIVSSSQDGKVIVWDAFT 96
> cpv:cgd2_1870 guanine nucleotide-binding protein ; K14753 guanine
nucleotide-binding protein subunit beta-2-like 1 protein
Length=313
Score = 64.7 bits (156), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 39/122 (31%), Positives = 65/122 (53%), Gaps = 10/122 (8%)
Query 43 VMSIILAPRNVGADLMYVCTKSNSITLMNF--AGKTIKKIDKA------HDAAIASITFS 94
V SI P N +D++ ++ S+ + N A K I A H+ A+ + S
Sbjct 18 VTSIAAPPDN--SDIVVSASRDKSVLVWNLSDANKDQHSIGSAKTRLTGHNQAVNDVAVS 75
Query 95 KDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIW 154
D +++++GS D T R+ + AGK+++ F GH + V PDN +II+GS D IK+W
Sbjct 76 SDGSYVVSGSCDKTLRLFDVNAGKSVRNFVGHTSDVFSVALSPDNRQIISGSRDHTIKVW 135
Query 155 DS 156
++
Sbjct 136 NA 137
Score = 31.2 bits (69), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 30/158 (18%), Positives = 66/158 (41%), Gaps = 17/158 (10%)
Query 1 TRIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYV 60
+ +++GS D ++++D ++ F H V S+ L+P N +
Sbjct 79 SYVVSGSCDKTLRLFDVNAGKSVRNFV----GHTS------DVFSVALSPDN---RQIIS 125
Query 61 CTKSNSITLMNFAGKTIKKI-DKAHDAAIASITFSKDNTHIL--TGSFDTTARIHGLKAG 117
++ ++I + N G + + D H+ ++ + FS + L + +D ++ +
Sbjct 126 GSRDHTIKVWNAMGVCMYTLLDGQHNDWVSCVRFSPSTSQPLFVSCGWDKIVKVWSHEFK 185
Query 118 KTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWD 155
T GH + + PD + +G DG + +WD
Sbjct 186 PTCN-LVGHSSVLYTVTIAPDGSLCASGGKDGVVMLWD 222
> hsa:10681 GNB5, FLJ37457, FLJ43714, GB5; guanine nucleotide
binding protein (G protein), beta 5; K04539 guanine nucleotide
binding protein (G protein), beta 5
Length=353
Score = 64.7 bits (156), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 48/166 (28%), Positives = 75/166 (45%), Gaps = 9/166 (5%)
Query 2 RIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVC 61
RI++ S DGK+ +WDS T + T P M A PS +I + + +
Sbjct 77 RIVSSSQDGKVIVWDSFTTNKEHAVTMPCTWVMACAYA-PSGCAIACGGLDNKCSVYPLT 135
Query 62 TKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
N N A K KK H +++ +F+ + ILT S D T + +++G+ L+
Sbjct 136 FDKNE----NMAAK--KKSVAMHTNYLSACSFTNSDMQILTASGDGTCALWDVESGQLLQ 189
Query 122 EFRGHLTFVNCALYLPDNT--RIITGSADGKIKIWDSKTQDCLQTF 165
F GH V C P T ++G D K +WD ++ C+Q F
Sbjct 190 SFHGHGADVLCLDLAPSETGNTFVSGGCDKKAMVWDMRSGQCVQAF 235
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 35/156 (22%), Positives = 70/156 (44%), Gaps = 13/156 (8%)
Query 2 RIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVC 61
+I+T S DG +WD ++ LQ+F H A V+ + LAP G +
Sbjct 167 QILTASGDGTCALWDVESGQLLQSF------HGHGAD----VLCLDLAPSETGNTFVSGG 216
Query 62 TKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
++ +G+ ++ + H++ I S+ + +GS D T R++ L+A + +
Sbjct 217 CDKKAMVWDMRSGQCVQAFE-THESDINSVRYYPSGDAFASGSDDATCRLYDLRADREVA 275
Query 122 EF-RGHLTFVNCALYLPDNTRII-TGSADGKIKIWD 155
+ + + F ++ + R++ G D I +WD
Sbjct 276 IYSKESIIFGASSVDFSLSGRLLFAGYNDYTINVWD 311
Score = 38.1 bits (87), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 17/41 (41%), Positives = 24/41 (58%), Gaps = 0/41 (0%)
Query 118 KTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKT 158
KT + +GH V C + D RI++ S DGK+ +WDS T
Sbjct 54 KTRRTLKGHGNKVLCMDWCKDKRRIVSSSQDGKVIVWDSFT 94
> dre:334373 fbxw11a, btrc2, fbxw11, fbxw11b, fbxw1b, wu:fd14d12,
wu:fi43f07; F-box and WD-40 domain protein 11a; K03362 F-box
and WD-40 domain protein 1/11
Length=527
Score = 64.7 bits (156), Expect = 1e-10, Method: Composition-based stats.
Identities = 48/194 (24%), Positives = 77/194 (39%), Gaps = 58/194 (29%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
I+TGS+D +++WD + + L T A + C LM C+
Sbjct 278 IVTGSSDSTVRVWDVSSGEVLNTLIHHNEAVLHLRFC---------------NGLMVTCS 322
Query 63 KSNSITLMNFAGKT---IKKIDKAHDAAIASITFSKD----------------------- 96
K SI + + A T ++++ H AA+ + F
Sbjct 323 KDRSIAVWDMASATDISLRRVLVGHRAAVNVVDFDDKYIVSASGDRTIKVWSTSTCEFVR 382
Query 97 --NTH-------------ILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTR 141
N H +++GS D T R+ ++ G L+ GH V C + DN R
Sbjct 383 TLNGHKRGIACLQYRDRLVVSGSSDNTIRLWDIECGACLRVLEGHEELVRCIRF--DNKR 440
Query 142 IITGSADGKIKIWD 155
I++G+ DGKIK+WD
Sbjct 441 IVSGAYDGKIKVWD 454
Score = 35.8 bits (81), Expect = 0.075, Method: Composition-based stats.
Identities = 16/37 (43%), Positives = 23/37 (62%), Gaps = 2/37 (5%)
Query 130 VNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTFT 166
V C Y D+ +II+G D IKIWD ++ +CL+ T
Sbjct 228 VYCLQY--DDDKIISGLRDNSIKIWDKQSLECLKVLT 262
Score = 30.0 bits (66), Expect = 3.4, Method: Composition-based stats.
Identities = 10/15 (66%), Positives = 14/15 (93%), Gaps = 0/15 (0%)
Query 2 RIITGSADGKIKIWD 16
RI++G+ DGKIK+WD
Sbjct 440 RIVSGAYDGKIKVWD 454
> xla:379573 poc1a, MGC69111, pix2, wdr51a; POC1 centriolar protein
homolog A (Chlamydomonas)
Length=399
Score = 64.3 bits (155), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 34/124 (27%), Positives = 64/124 (51%), Gaps = 3/124 (2%)
Query 42 SVMSIILAPRNVGADLMYVCTKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHIL 101
+++S+ +P L+ ++ ++ L + K KAH + S++FS D ++
Sbjct 20 AILSVDFSP---SGHLIASASRDKTVRLWVPSVKGESTAFKAHTGTVRSVSFSGDGQSLV 76
Query 102 TGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDC 161
T S D T ++ + K L H+ +V CA + PD I++ S D IK+WD +++C
Sbjct 77 TASDDKTIKVWTVHRQKFLFSLNQHINWVRCAKFSPDGRLIVSASDDKTIKLWDKTSREC 136
Query 162 LQTF 165
+Q+F
Sbjct 137 IQSF 140
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 32/152 (21%), Positives = 66/152 (43%), Gaps = 13/152 (8%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
I++ S D IK+WD +++C+Q+F + PS I A +
Sbjct 117 IVSASDDKTIKLWDKTSRECIQSFCEH-GGFVNFVDFHPSGTCIAAAATD---------- 165
Query 63 KSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKE 122
N++ + + + + + H + S++F +++T S D+T ++ L G+ L
Sbjct 166 --NTVKVWDIRMNKLIQHYQVHSGVVNSLSFHPSGNYLITASNDSTLKVLDLLEGRLLYT 223
Query 123 FRGHLTFVNCALYLPDNTRIITGSADGKIKIW 154
GH V C + + +G +D ++ +W
Sbjct 224 LHGHQGPVTCVKFSREGDFFASGGSDEQVMVW 255
Score = 37.4 bits (85), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 20/73 (27%), Positives = 35/73 (47%), Gaps = 0/73 (0%)
Query 93 FSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIK 152
FS D I++ S D T ++ + + ++ F H FVN + P T I + D +K
Sbjct 110 FSPDGRLIVSASDDKTIKLWDKTSRECIQSFCEHGGFVNFVDFHPSGTCIAAAATDNTVK 169
Query 153 IWDSKTQDCLQTF 165
+WD + +Q +
Sbjct 170 VWDIRMNKLIQHY 182
Score = 37.0 bits (84), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 31/153 (20%), Positives = 59/153 (38%), Gaps = 13/153 (8%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
++T S D IK+W Q L + H+ +C A + L+ +
Sbjct 75 LVTASDDKTIKVWTVHRQKFLFSLN----QHINWVRC---------AKFSPDGRLIVSAS 121
Query 63 KSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKE 122
+I L + + + H + + F T I + D T ++ ++ K ++
Sbjct 122 DDKTIKLWDKTSRECIQSFCEHGGFVNFVDFHPSGTCIAAAATDNTVKVWDIRMNKLIQH 181
Query 123 FRGHLTFVNCALYLPDNTRIITGSADGKIKIWD 155
++ H VN + P +IT S D +K+ D
Sbjct 182 YQVHSGVVNSLSFHPSGNYLITASNDSTLKVLD 214
> xla:394362 btrc, MGC83554, beta-TrCP, betaTrCP, btrc-a, btrcp,
btrcp1, fbw1a, fbxw1, fbxw1a, fwd1; beta-transducin repeat
containing; K03362 F-box and WD-40 domain protein 1/11
Length=518
Score = 64.3 bits (155), Expect = 2e-10, Method: Composition-based stats.
Identities = 48/194 (24%), Positives = 76/194 (39%), Gaps = 58/194 (29%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
IITGS+D +++WD T + L T A + +M C+
Sbjct 285 IITGSSDSTVRVWDVNTGEMLNTLIHHCEAVLHL---------------RFNNGMMVTCS 329
Query 63 KSNSITLMNFAGK---TIKKIDKAHDAAIASITFSKD----------------------- 96
K SI + + A T++++ H AA+ + F
Sbjct 330 KDRSIAVWDMASATDITLRRVLVGHRAAVNVVDFDDKYIVSASGDRTIKVWNTSTCEFVR 389
Query 97 --NTH-------------ILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTR 141
N H +++GS D T R+ ++ G L+ GH V C + DN R
Sbjct 390 TLNGHKRGIACLQYRDRLVVSGSSDNTIRLWDIECGACLRVLEGHEELVRCIRF--DNKR 447
Query 142 IITGSADGKIKIWD 155
I++G+ DGKIK+WD
Sbjct 448 IVSGAYDGKIKVWD 461
Score = 39.7 bits (91), Expect = 0.005, Method: Composition-based stats.
Identities = 23/70 (32%), Positives = 34/70 (48%), Gaps = 2/70 (2%)
Query 96 DNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWD 155
D+ I++G D T +I + + GH V C Y D IITGS+D +++WD
Sbjct 241 DDQKIVSGLRDNTIKIWDKNTLECKRVLMGHTGSVLCLQY--DERVIITGSSDSTVRVWD 298
Query 156 SKTQDCLQTF 165
T + L T
Sbjct 299 VNTGEMLNTL 308
Score = 31.6 bits (70), Expect = 1.3, Method: Composition-based stats.
Identities = 14/32 (43%), Positives = 19/32 (59%), Gaps = 2/32 (6%)
Query 130 VNCALYLPDNTRIITGSADGKIKIWDSKTQDC 161
V C Y D+ +I++G D IKIWD T +C
Sbjct 235 VYCLQY--DDQKIVSGLRDNTIKIWDKNTLEC 264
Score = 30.0 bits (66), Expect = 3.3, Method: Composition-based stats.
Identities = 10/15 (66%), Positives = 14/15 (93%), Gaps = 0/15 (0%)
Query 2 RIITGSADGKIKIWD 16
RI++G+ DGKIK+WD
Sbjct 447 RIVSGAYDGKIKVWD 461
> xla:734280 wdr69, MGC85213; WD repeat domain 69
Length=415
Score = 64.3 bits (155), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 47/168 (27%), Positives = 72/168 (42%), Gaps = 19/168 (11%)
Query 2 RIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCL---PSVMSIILAPRNVGADLM 58
+I TGS D K+W ++T C TF H CL P I + A L
Sbjct 149 KIATGSFDKTCKLWSAETGKCYHTFR----GHTAEIVCLVFNPQSTLIATGSMDTTAKLW 204
Query 59 YVCTKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGK 118
+ + ++TL H A I S++F+ ++TGSFD T + + +G+
Sbjct 205 DIQSGEEALTL------------SGHAAEIISLSFNTTGDRLITGSFDHTVSVWEIPSGR 252
Query 119 TLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTFT 166
+ GH ++ A + D + I T S D K+WDS C+ T T
Sbjct 253 RIHTLIGHRGEISSAQFNWDCSLIATASMDKSCKLWDSLNGKCVATLT 300
Score = 58.2 bits (139), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 41/165 (24%), Positives = 69/165 (41%), Gaps = 15/165 (9%)
Query 2 RIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVC 61
R+ITGS D + +W+ +P+ + + I A N L+
Sbjct 233 RLITGSFDHTVSVWE-------------IPSGRRIHTLIGHRGEISSAQFNWDCSLIATA 279
Query 62 TKSNSITLMN-FAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTL 120
+ S L + GK + + HD + +TF + T S D TAR++ + K L
Sbjct 280 SMDKSCKLWDSLNGKCVATL-TGHDDEVLDVTFDSTGQLVATASADGTARVYSASSRKCL 338
Query 121 KEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
+ GH ++ + RI+T S+D ++WD T +CLQ
Sbjct 339 AKLEGHEGEISKICFNAQGNRIVTASSDKTSRLWDPHTGECLQVL 383
Score = 52.8 bits (125), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 27/88 (30%), Positives = 45/88 (51%), Gaps = 1/88 (1%)
Query 79 KIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALY-LP 137
K+ +AH + ++ F+K + +TGS+D T ++ +G+ L GH V + P
Sbjct 86 KVLRAHILPLTNVAFNKSGSSFITGSYDRTCKVWDTASGEELHTLEGHRNVVYAIQFNNP 145
Query 138 DNTRIITGSADGKIKIWDSKTQDCLQTF 165
+I TGS D K+W ++T C TF
Sbjct 146 YGDKIATGSFDKTCKLWSAETGKCYHTF 173
Score = 51.6 bits (122), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 27/75 (36%), Positives = 39/75 (52%), Gaps = 1/75 (1%)
Query 82 KAHDAAIASITFSKD-NTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNT 140
+ H + +I F+ I TGSFD T ++ + GK FRGH + C ++ P +T
Sbjct 131 EGHRNVVYAIQFNNPYGDKIATGSFDKTCKLWSAETGKCYHTFRGHTAEIVCLVFNPQST 190
Query 141 RIITGSADGKIKIWD 155
I TGS D K+WD
Sbjct 191 LIATGSMDTTAKLWD 205
Score = 50.4 bits (119), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 42/153 (27%), Positives = 63/153 (41%), Gaps = 15/153 (9%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
I T S D K+WDS C+ T T + S ++ G +Y +
Sbjct 276 IATASMDKSCKLWDSLNGKCVATLTGHDDEVLDVT--FDSTGQLVATASADGTARVYSAS 333
Query 63 KSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKE 122
K + K++ H+ I+ I F+ I+T S D T+R+ G+ L+
Sbjct 334 SR----------KCLAKLE-GHEGEISKICFNAQGNRIVTASSDKTSRLWDPHTGECLQV 382
Query 123 FRGHL-TFVNCALYLPDNTRIITGSADGKIKIW 154
+GH +CA NT IITGS D +IW
Sbjct 383 LKGHTDEIFSCAFNYEGNT-IITGSKDNTCRIW 414
> dre:386932 poc1b, TUWD12, fl36w17, wdr51b, wu:fl36w17, zgc:63538;
POC1 centriolar protein homolog B (Chlamydomonas)
Length=490
Score = 63.9 bits (154), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 41/156 (26%), Positives = 69/156 (44%), Gaps = 18/156 (11%)
Query 29 PVPAHMQTAQCLPSVMSIILAP-----RNVG-------------ADLMYVCTKSNSITLM 70
P + T C S+M LAP R VG L+ ++ ++ L
Sbjct 28 PNNKQLATGSCDKSLMIWNLAPKARAFRFVGHTDVITGVNFAPSGSLVASSSRDQTVRLW 87
Query 71 NFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFV 130
+ K + KAH A++ S+ FS+D ++T S D + ++ G++ K L H +V
Sbjct 88 TPSIKGESTVFKAHTASVRSVNFSRDGQRLVTASDDKSVKVWGVERKKFLYSLNRHTNWV 147
Query 131 NCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTFT 166
CA + PD I + D +++WD+ + C FT
Sbjct 148 RCARFSPDGRLIASCGDDRTVRLWDTSSHQCTNIFT 183
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 34/154 (22%), Positives = 64/154 (41%), Gaps = 13/154 (8%)
Query 2 RIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVC 61
R++T S D +K+W + + L + H +C A + L+ C
Sbjct 116 RLVTASDDKSVKVWGVERKKFLYSLN----RHTNWVRC---------ARFSPDGRLIASC 162
Query 62 TKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
++ L + + I + + + F+ T I + D T +I ++ K ++
Sbjct 163 GDDRTVRLWDTSSHQCTNIFTDYGGSATFVDFNSSGTCIASSGADNTIKIWDIRTNKLIQ 222
Query 122 EFRGHLTFVNCALYLPDNTRIITGSADGKIKIWD 155
++ H VNC + P +I+GS+D IKI D
Sbjct 223 HYKVHNAGVNCFSFHPSGNYLISGSSDSTIKILD 256
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 34/159 (21%), Positives = 64/159 (40%), Gaps = 24/159 (15%)
Query 2 RIITGSADGK-IKIWDSKTQDCLQTFT-----PPVPAHMQTAQCLPSVMSIILAPRNVGA 55
R+I D + +++WD+ + C FT + C+ S GA
Sbjct 157 RLIASCGDDRTVRLWDTSSHQCTNIFTDYGGSATFVDFNSSGTCIASS----------GA 206
Query 56 DLMYVCTKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLK 115
D N+I + + + + K H+A + +F ++++GS D+T +I L
Sbjct 207 D--------NTIKIWDIRTNKLIQHYKVHNAGVNCFSFHPSGNYLISGSSDSTIKILDLL 258
Query 116 AGKTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIW 154
G+ + GH V + D +G AD ++ +W
Sbjct 259 EGRLIYTLHGHKGPVLTVTFSRDGDLFASGGADSQVLMW 297
Score = 37.0 bits (84), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 13/35 (37%), Positives = 23/35 (65%), Gaps = 0/35 (0%)
Query 121 KEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWD 155
+ F+GH ++CA + P+N ++ TGS D + IW+
Sbjct 12 RHFKGHKDVISCADFNPNNKQLATGSCDKSLMIWN 46
Score = 36.2 bits (82), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 19/79 (24%), Positives = 35/79 (44%), Gaps = 0/79 (0%)
Query 76 TIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALY 135
T+++ K H I+ F+ +N + TGS D + I L F GH + +
Sbjct 9 TLERHFKGHKDVISCADFNPNNKQLATGSCDKSLMIWNLAPKARAFRFVGHTDVITGVNF 68
Query 136 LPDNTRIITGSADGKIKIW 154
P + + + S D +++W
Sbjct 69 APSGSLVASSSRDQTVRLW 87
> mmu:70235 Poc1a, 2510040D07Rik, Wdr51a; POC1 centriolar protein
homolog A (Chlamydomonas)
Length=405
Score = 63.9 bits (154), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 42/166 (25%), Positives = 74/166 (44%), Gaps = 19/166 (11%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCL---PSVMSIILAPRNVGADLMY 59
+ +GS D + IW K Q FT H C+ PS + R+
Sbjct 34 LASGSMDSTLMIWHMKPQSRAYRFT----GHKDAVTCVNFSPSGHLLASGSRD------- 82
Query 60 VCTKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKT 119
K+ I + N G++ + +AH A + S+ F D ++T S D T ++ +
Sbjct 83 ---KTVRIWVPNVKGEST--VFRAHTATVRSVHFCSDGQSLVTASDDKTVKVWSTHRQRF 137
Query 120 LKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
L H+ +V CA + PD I++ S D +K+WD +++C+ ++
Sbjct 138 LFSLTQHINWVRCAKFSPDGRLIVSASDDKTVKLWDKTSRECIHSY 183
Score = 48.1 bits (113), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 39/79 (49%), Gaps = 0/79 (0%)
Query 76 TIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALY 135
++++ K H A+ + FS + H+ +GS D+T I +K F GH V C +
Sbjct 10 SLERHFKGHRDAVTCVDFSLNTKHLASGSMDSTLMIWHMKPQSRAYRFTGHKDAVTCVNF 69
Query 136 LPDNTRIITGSADGKIKIW 154
P + +GS D ++IW
Sbjct 70 SPSGHLLASGSRDKTVRIW 88
Score = 45.8 bits (107), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 32/154 (20%), Positives = 66/154 (42%), Gaps = 13/154 (8%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
I++ S D +K+WD +++C+ ++ + PS I A G D
Sbjct 160 IVSASDDKTVKLWDKTSRECIHSYCEH-GGFVTYVDFHPSGTCIAAA----GMD------ 208
Query 63 KSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKE 122
N++ + + + + + H AA+ +++F +++T S D+T +I L G+ L
Sbjct 209 --NTVKVWDARTHRLLQHYQLHSAAVNALSFHPSGNYLITASSDSTLKILDLMEGRLLYT 266
Query 123 FRGHLTFVNCALYLPDNTRIITGSADGKIKIWDS 156
GH + +G +D ++ +W S
Sbjct 267 LHGHQGPATTVAFSRTGEYFASGGSDEQVMVWKS 300
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 31/153 (20%), Positives = 59/153 (38%), Gaps = 13/153 (8%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
++T S D +K+W + Q L + T H+ +C A + L+ +
Sbjct 118 LVTASDDKTVKVWSTHRQRFLFSLT----QHINWVRC---------AKFSPDGRLIVSAS 164
Query 63 KSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKE 122
++ L + + H + + F T I D T ++ + + L+
Sbjct 165 DDKTVKLWDKTSRECIHSYCEHGGFVTYVDFHPSGTCIAAAGMDNTVKVWDARTHRLLQH 224
Query 123 FRGHLTFVNCALYLPDNTRIITGSADGKIKIWD 155
++ H VN + P +IT S+D +KI D
Sbjct 225 YQLHSAAVNALSFHPSGNYLITASSDSTLKILD 257
Score = 39.3 bits (90), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 23/84 (27%), Positives = 34/84 (40%), Gaps = 0/84 (0%)
Query 83 AHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRI 142
H A+ + FS + +GS D T RI FR H V + D +
Sbjct 59 GHKDAVTCVNFSPSGHLLASGSRDKTVRIWVPNVKGESTVFRAHTATVRSVHFCSDGQSL 118
Query 143 ITGSADGKIKIWDSKTQDCLQTFT 166
+T S D +K+W + Q L + T
Sbjct 119 VTASDDKTVKVWSTHRQRFLFSLT 142
Score = 30.4 bits (67), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 14/46 (30%), Positives = 21/46 (45%), Gaps = 0/46 (0%)
Query 121 KEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTFT 166
+ F+GH V C + + + +GS D + IW K Q FT
Sbjct 13 RHFKGHRDAVTCVDFSLNTKHLASGSMDSTLMIWHMKPQSRAYRFT 58
> hsa:25886 POC1A, DKFZp434C245, MGC131902, PIX2, WDR51A; POC1
centriolar protein homolog A (Chlamydomonas)
Length=359
Score = 63.9 bits (154), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 42/167 (25%), Positives = 74/167 (44%), Gaps = 19/167 (11%)
Query 2 RIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCL---PSVMSIILAPRNVGADLM 58
++ +GS D + +W K Q FT H C+ PS + R+
Sbjct 33 QLASGSMDSCLMVWHMKPQSRAYRFT----GHKDAVTCVNFSPSGHLLASGSRD------ 82
Query 59 YVCTKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGK 118
K+ I + N G++ + +AH A + S+ F D +T S D T ++ K
Sbjct 83 ----KTVRIWVPNVKGEST--VFRAHTATVRSVHFCSDGQSFVTASDDKTVKVWATHRQK 136
Query 119 TLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
L H+ +V CA + PD I++ S D +K+WD +++C+ ++
Sbjct 137 FLFSLSQHINWVRCAKFSPDGRLIVSASDDKTVKLWDKSSRECVHSY 183
Score = 46.2 bits (108), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 32/154 (20%), Positives = 65/154 (42%), Gaps = 13/154 (8%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
I++ S D +K+WD +++C+ ++ + PS I A G D
Sbjct 160 IVSASDDKTVKLWDKSSRECVHSYCEH-GGFVTYVDFHPSGTCIAAA----GMD------ 208
Query 63 KSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKE 122
N++ + + + + + H AA+ ++F +++T S D+T +I L G+ L
Sbjct 209 --NTVKVWDVRTHRLLQHYQLHSAAVNGLSFHPSGNYLITASSDSTLKILDLMEGRLLYT 266
Query 123 FRGHLTFVNCALYLPDNTRIITGSADGKIKIWDS 156
GH + +G +D ++ +W S
Sbjct 267 LHGHQGPATTVAFSRTGEYFASGGSDEQVMVWKS 300
Score = 42.0 bits (97), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 20/79 (25%), Positives = 37/79 (46%), Gaps = 0/79 (0%)
Query 76 TIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALY 135
++++ K H A+ + FS + + +GS D+ + +K F GH V C +
Sbjct 10 SLERHFKGHRDAVTCVDFSINTKQLASGSMDSCLMVWHMKPQSRAYRFTGHKDAVTCVNF 69
Query 136 LPDNTRIITGSADGKIKIW 154
P + +GS D ++IW
Sbjct 70 SPSGHLLASGSRDKTVRIW 88
Score = 39.3 bits (90), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 30/153 (19%), Positives = 60/153 (39%), Gaps = 13/153 (8%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
+T S D +K+W + Q L + + H+ +C A + L+ +
Sbjct 118 FVTASDDKTVKVWATHRQKFLFSLS----QHINWVRC---------AKFSPDGRLIVSAS 164
Query 63 KSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKE 122
++ L + + + H + + F T I D T ++ ++ + L+
Sbjct 165 DDKTVKLWDKSSRECVHSYCEHGGFVTYVDFHPSGTCIAAAGMDNTVKVWDVRTHRLLQH 224
Query 123 FRGHLTFVNCALYLPDNTRIITGSADGKIKIWD 155
++ H VN + P +IT S+D +KI D
Sbjct 225 YQLHSAAVNGLSFHPSGNYLITASSDSTLKILD 257
Score = 29.6 bits (65), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 13/46 (28%), Positives = 22/46 (47%), Gaps = 0/46 (0%)
Query 121 KEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTFT 166
+ F+GH V C + + ++ +GS D + +W K Q FT
Sbjct 13 RHFKGHRDAVTCVDFSINTKQLASGSMDSCLMVWHMKPQSRAYRFT 58
> hsa:164781 WDR69, FLJ25955; WD repeat domain 69
Length=415
Score = 63.5 bits (153), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 47/168 (27%), Positives = 69/168 (41%), Gaps = 19/168 (11%)
Query 2 RIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCL---PSVMSIILAPRNVGADLM 58
+I TGS D K+W +T C TF H CL P + + A L
Sbjct 149 KIATGSFDKTCKLWSVETGKCYHTF----RGHTAEIVCLSFNPQSTLVATGSMDTTAKLW 204
Query 59 YVCTKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGK 118
+ TL + H A I S++F+ I+TGSFD T + G+
Sbjct 205 DIQNGEEVYTL------------RGHSAEIISLSFNTSGDRIITGSFDHTVVVWDADTGR 252
Query 119 TLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTFT 166
+ GH ++ A + D + I+TGS D K+WD+ C+ T T
Sbjct 253 KVNILIGHCAEISSASFNWDCSLILTGSMDKTCKLWDATNGKCVATLT 300
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 49/155 (31%), Positives = 68/155 (43%), Gaps = 19/155 (12%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
I+TGS D K+WD+ C+ T T + + C +I T
Sbjct 276 ILTGSMDKTCKLWDATNGKCVATLTGHDDEILDS--CFDYTGKLI-------------AT 320
Query 63 KSNSITLMNFAGKTIKKIDK--AHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTL 120
S T F+ T K I K H+ I+ I+F+ H+LTGS D TARI + G+ L
Sbjct 321 ASADGTARIFSAATRKCIAKLEGHEGEISKISFNPQGNHLLTGSSDKTARIWDAQTGQCL 380
Query 121 KEFRGHL-TFVNCALYLPDNTRIITGSADGKIKIW 154
+ GH +CA N +ITGS D +IW
Sbjct 381 QVLEGHTDEIFSCAFNYKGNI-VITGSKDNTCRIW 414
Score = 53.5 bits (127), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 45/165 (27%), Positives = 67/165 (40%), Gaps = 15/165 (9%)
Query 2 RIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVC 61
RIITGS D + +WD+ T ++ C I A N L+
Sbjct 233 RIITGSFDHTVVVWDADTGR---------KVNILIGHC----AEISSASFNWDCSLILTG 279
Query 62 TKSNSITLMNFA-GKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTL 120
+ + L + GK + + HD I F I T S D TARI K +
Sbjct 280 SMDKTCKLWDATNGKCVATL-TGHDDEILDSCFDYTGKLIATASADGTARIFSAATRKCI 338
Query 121 KEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
+ GH ++ + P ++TGS+D +IWD++T CLQ
Sbjct 339 AKLEGHEGEISKISFNPQGNHLLTGSSDKTARIWDAQTGQCLQVL 383
Score = 50.1 bits (118), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 28/88 (31%), Positives = 43/88 (48%), Gaps = 1/88 (1%)
Query 79 KIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNC-ALYLP 137
K+ KAH + ++ +K + +TGS+D T ++ +G+ L GH V A P
Sbjct 86 KVLKAHILPLTNVALNKSGSCFITGSYDRTCKLWDTASGEELNTLEGHRNVVYAIAFNNP 145
Query 138 DNTRIITGSADGKIKIWDSKTQDCLQTF 165
+I TGS D K+W +T C TF
Sbjct 146 YGDKIATGSFDKTCKLWSVETGKCYHTF 173
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 46/164 (28%), Positives = 74/164 (45%), Gaps = 17/164 (10%)
Query 1 TRIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYV 60
T + TGS D K+WD Q+ + +T + H II N D +
Sbjct 190 TLVATGSMDTTAKLWD--IQNGEEVYT--LRGHS---------AEIISLSFNTSGD--RI 234
Query 61 CTKSNSITLMNFAGKTIKKIDK--AHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGK 118
T S T++ + T +K++ H A I+S +F+ D + ILTGS D T ++ GK
Sbjct 235 ITGSFDHTVVVWDADTGRKVNILIGHCAEISSASFNWDCSLILTGSMDKTCKLWDATNGK 294
Query 119 TLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCL 162
+ GH + + + I T SADG +I+ + T+ C+
Sbjct 295 CVATLTGHDDEILDSCFDYTGKLIATASADGTARIFSAATRKCI 338
> tgo:TGME49_016880 receptor for activated C kinase, RACK protein,
putative (EC:2.7.11.7); K14753 guanine nucleotide-binding
protein subunit beta-2-like 1 protein
Length=321
Score = 63.5 bits (153), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 27/74 (36%), Positives = 44/74 (59%), Gaps = 0/74 (0%)
Query 83 AHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRI 142
H + + + D L+GS+D T R+ L AG T++ F+GH + VN + PDN +I
Sbjct 66 GHSQCVQDVVINSDGQFALSGSWDKTLRLWDLNAGVTVRSFQGHTSDVNSVAFSPDNRQI 125
Query 143 ITGSADGKIKIWDS 156
++GS D IK+W++
Sbjct 126 VSGSRDRTIKLWNT 139
Score = 39.7 bits (91), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 36/165 (21%), Positives = 69/165 (41%), Gaps = 12/165 (7%)
Query 2 RIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVC 61
+I++GS D IK+W++ + C T H C + +P + L+ C
Sbjct 124 QIVSGSRDRTIKLWNTLAE-CKYTIVD--DQHNDWVSC------VRFSP-SANKPLIVSC 173
Query 62 TKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
+ + N + ++ H + + ++T S D + +G D A + + GK L
Sbjct 174 GWDKLVKVWNLSNCKLRTNLVGHTSVLYTVTISPDGSLCASGGKDGVAMLWDVNEGKHLY 233
Query 122 EFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTFT 166
+ T +N + P N + + D +KIWD + ++ L T
Sbjct 234 SLDSNST-INALCFSPCN-YWLCAATDKSVKIWDLENKNVLSEIT 276
Score = 38.1 bits (87), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 26/91 (28%), Positives = 45/91 (49%), Gaps = 14/91 (15%)
Query 73 AGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFR------GH 126
AG T++ + H + + S+ FS DN I++GS D T +K TL E + H
Sbjct 99 AGVTVRSF-QGHTSDVNSVAFSPDNRQIVSGSRDRT-----IKLWNTLAECKYTIVDDQH 152
Query 127 LTFVNCALYLPDNTR--IITGSADGKIKIWD 155
+V+C + P + I++ D +K+W+
Sbjct 153 NDWVSCVRFSPSANKPLIVSCGWDKLVKVWN 183
Score = 33.9 bits (76), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 32/155 (20%), Positives = 62/155 (40%), Gaps = 16/155 (10%)
Query 4 ITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCTK 63
++GS D +++WD +++F H V S+ +P N + ++
Sbjct 84 LSGSWDKTLRLWDLNAGVTVRSFQ----GHTS------DVNSVAFSPDN---RQIVSGSR 130
Query 64 SNSITLMN-FAGKTIKKIDKAHDAAIASITFS--KDNTHILTGSFDTTARIHGLKAGKTL 120
+I L N A +D H+ ++ + FS + I++ +D ++ L K
Sbjct 131 DRTIKLWNTLAECKYTIVDDQHNDWVSCVRFSPSANKPLIVSCGWDKLVKVWNLSNCKLR 190
Query 121 KEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWD 155
GH + + PD + +G DG +WD
Sbjct 191 TNLVGHTSVLYTVTISPDGSLCASGGKDGVAMLWD 225
> xla:399199 pafah1b1, lis2, mdcr, mds, pafah, pafah1b1-b; platelet-activating
factor acetylhydrolase 1b, regulatory subunit
1 (45kDa) (EC:3.1.1.47); K01062 1-alkyl-2-acetylglycerophosphocholine
esterase [EC:3.1.1.47]
Length=410
Score = 63.2 bits (152), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 41/159 (25%), Positives = 71/159 (44%), Gaps = 13/159 (8%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
++T S D IK+WD +T D F + H + Q + S L+ C+
Sbjct 123 MVTASEDATIKVWDYETGD----FERTLKGHTDSVQDISFDHS---------GKLLASCS 169
Query 63 KSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKE 122
+I L +F G + HD ++S+ + HI++ S D T ++ ++ G +K
Sbjct 170 ADMTIKLWDFQGFECLRTMHGHDHNVSSVAIMPNGDHIVSASRDKTIKMWEVQTGYCVKT 229
Query 123 FRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDC 161
F GH +V D T I + S D +++W T++C
Sbjct 230 FTGHREWVRMVRPNQDGTLIASCSNDQTVRVWVVATKEC 268
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 41/183 (22%), Positives = 81/183 (44%), Gaps = 33/183 (18%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
I++ S D IK+W+ +T C++TFT H + + ++ P G L+ C+
Sbjct 207 IVSASRDKTIKMWEVQTGYCVKTFT----GHREWVR--------MVRPNQDGT-LIASCS 253
Query 63 KSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTH--------------------ILT 102
++ + A K K + H+ + I+++ ++++ +L+
Sbjct 254 NDQTVRVWVVATKECKAELREHEHVVECISWAPESSYSTISDATGSETKRSGKPGPFLLS 313
Query 103 GSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCL 162
GS D T ++ + G L GH +V + P I++ + D I+IWD K + C+
Sbjct 314 GSRDKTIKMWDISIGMCLMTLVGHDNWVRGVQFHPGGKFILSCADDKTIRIWDYKNKRCM 373
Query 163 QTF 165
+T
Sbjct 374 KTL 376
Score = 49.7 bits (117), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 30/143 (20%), Positives = 63/143 (44%), Gaps = 3/143 (2%)
Query 24 QTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCTKSNSITLMNFAGKTIKKIDKA 83
+ + P P + V +I P +M ++ +I + ++ ++ K
Sbjct 92 KEWIPRPPEKYALSGHRSPVTRVIFHPV---FSVMVTASEDATIKVWDYETGDFERTLKG 148
Query 84 HDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRII 143
H ++ I+F + + S D T ++ + + L+ GH V+ +P+ I+
Sbjct 149 HTDSVQDISFDHSGKLLASCSADMTIKLWDFQGFECLRTMHGHDHNVSSVAIMPNGDHIV 208
Query 144 TGSADGKIKIWDSKTQDCLQTFT 166
+ S D IK+W+ +T C++TFT
Sbjct 209 SASRDKTIKMWEVQTGYCVKTFT 231
> dre:406483 gnb3b, gnb3, wu:fk54b04, zgc:73058, zgc:77780; guanine
nucleotide binding protein (G protein), beta polypeptide
3b; K07825 guanine nucleotide binding protein (G protein),
beta 3
Length=338
Score = 63.2 bits (152), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 50/166 (30%), Positives = 77/166 (46%), Gaps = 18/166 (10%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
+++ S DGK+ IWD+ + + + P+ + VM+ AP +L+
Sbjct 70 MVSASQDGKLLIWDAHSGN--KVNAVPLKSSW--------VMTCSYAPS---GNLVASGG 116
Query 63 KSNSITLMNFAG---KTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKT 119
N T+ N KT+K++D AH ++ F D T I+T S DTT + L+ GK
Sbjct 117 LDNMCTVYNLKTPVIKTVKELD-AHTGYLSCCRFISD-TEIVTSSGDTTCALWDLETGKQ 174
Query 120 LKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
F H+ C PDN I+G+ D K+WD + C QTF
Sbjct 175 KTVFLNHIGDCMCLSLSPDNNMFISGACDSLAKLWDIRDGQCKQTF 220
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 40/72 (55%), Gaps = 0/72 (0%)
Query 84 HDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRII 143
H ++ S DN ++G+ D+ A++ ++ G+ + F+GH + +N +LP T I
Sbjct 181 HIGDCMCLSLSPDNNMFISGACDSLAKLWDIRDGQCKQTFQGHTSDINAISFLPSATAFI 240
Query 144 TGSADGKIKIWD 155
TGS D K++D
Sbjct 241 TGSDDCTCKMYD 252
Score = 43.1 bits (100), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 38/160 (23%), Positives = 59/160 (36%), Gaps = 25/160 (15%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAP-------RNVGA 55
I+G+ D K+WD + C QTF + + LPS + I ++ A
Sbjct 197 FISGACDSLAKLWDIRDGQCKQTFQGHT-SDINAISFLPSATAFITGSDDCTCKMYDIRA 255
Query 56 DLMYVCTKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLK 115
D +C + D A ++ + S+ S I G D I
Sbjct 256 DQEVICYQ-----------------DSALNSGVTSVALSVSGRLIFAGYDDFNCNIWDSL 298
Query 116 AGKTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWD 155
G+ + GH V+C D + TGS D +KIW+
Sbjct 299 KGEKVGVLSGHDNRVSCTGVPGDGMCVCTGSWDSFLKIWN 338
Score = 35.8 bits (81), Expect = 0.060, Method: Compositional matrix adjust.
Identities = 40/159 (25%), Positives = 69/159 (43%), Gaps = 17/159 (10%)
Query 1 TRIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYV 60
T I+T S D +WD +T F H+ CL L+P N M++
Sbjct 153 TEIVTSSGDTTCALWDLETGKQKTVFLN----HIGDCMCLS------LSPDNN----MFI 198
Query 61 CTKSNSIT-LMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKT 119
+S+ L + K+ + H + I +I+F T +TGS D T +++ ++A +
Sbjct 199 SGACDSLAKLWDIRDGQCKQTFQGHTSDINAISFLPSATAFITGSDDCTCKMYDIRADQE 258
Query 120 LKEFR-GHLTFVNCALYLPDNTRII-TGSADGKIKIWDS 156
+ ++ L ++ L + R+I G D IWDS
Sbjct 259 VICYQDSALNSGVTSVALSVSGRLIFAGYDDFNCNIWDS 297
Score = 35.4 bits (80), Expect = 0.080, Method: Compositional matrix adjust.
Identities = 15/41 (36%), Positives = 24/41 (58%), Gaps = 0/41 (0%)
Query 118 KTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKT 158
KT K +GHL + + DN +++ S DGK+ IWD+ +
Sbjct 46 KTRKTLKGHLAKIYAMHWCTDNRLMVSASQDGKLLIWDAHS 86
> bbo:BBOV_II003080 18.m06257; WD-repeat protein; K14855 ribosome
assembly protein 4
Length=548
Score = 63.2 bits (152), Expect = 4e-10, Method: Composition-based stats.
Identities = 27/74 (36%), Positives = 44/74 (59%), Gaps = 0/74 (0%)
Query 82 KAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTR 141
+ H ++ + FS D + TGS D++ RI L+ G +K +GH +V C L+ PD TR
Sbjct 125 EGHSESVLCMDFSADGKLLATGSGDSSVRIWDLQTGTPIKTLKGHTNWVMCVLWSPDCTR 184
Query 142 IITGSADGKIKIWD 155
+ +G DG++ IW+
Sbjct 185 LASGGMDGRVIIWE 198
Score = 43.5 bits (101), Expect = 3e-04, Method: Composition-based stats.
Identities = 24/76 (31%), Positives = 38/76 (50%), Gaps = 0/76 (0%)
Query 83 AHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRI 142
H I + FS D + SFD T RI G+ L+ RGH+ V + + +
Sbjct 391 GHQQLINHVAFSADGRLFASASFDRTVRIWCGITGRYLRTLRGHIGRVYRIAWSCCGSLL 450
Query 143 ITGSADGKIKIWDSKT 158
I+ S+D +K+WD++T
Sbjct 451 ISCSSDTTLKLWDAET 466
Score = 35.0 bits (79), Expect = 0.11, Method: Composition-based stats.
Identities = 38/170 (22%), Positives = 70/170 (41%), Gaps = 34/170 (20%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTP--PVPAHMQTAQCL---PSVMS----------II 47
+ TGS D ++IWD LQT TP + H C+ P II
Sbjct 143 LATGSGDSSVRIWD------LQTGTPIKTLKGHTNWVMCVLWSPDCTRLASGGMDGRVII 196
Query 48 LAPRNVGADLMYVCTKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDT 107
P ++ + + + + +T + A + + ID A + +GS D+
Sbjct 197 WEPDSLSSHHITLSGHTKGVTTL--AWQPLHHIDLQVRAY----------PLLASGSMDS 244
Query 108 TARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTR-IITGSADGKIKIWDS 156
+ +I +K + L+ GH ++ L+ + + + + S D IK+WD+
Sbjct 245 SIKIWDVKVAQCLRTLSGHTRGISQVLWSGEKSNWLFSASRDTLIKVWDT 294
Score = 34.3 bits (77), Expect = 0.20, Method: Composition-based stats.
Identities = 44/195 (22%), Positives = 65/195 (33%), Gaps = 32/195 (16%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPR----------- 51
+ +GS D IKIWD K CL+T + Q + A R
Sbjct 237 LASGSMDSSIKIWDVKVAQCLRTLSGHTRGISQVLWSGEKSNWLFSASRDTLIKVWDTDK 296
Query 52 --------------NVGADLMYVCTKSNSITLMNFA-GKT----IKKIDKAHDAAIASIT 92
N Y KS T MNF G+T ++++ +
Sbjct 297 GGLVKDLKGHGHWINTLTSNTYRTIKSGPFTAMNFERGRTHFENLREMIEESRVVYNKFI 356
Query 93 FSKDNTHILTGSFDTTARI--HGLKAGKTLKEFRGHLTFVNCALYLPDNTRIITGSADGK 150
+L+GS D T I ++ K L GH +N + D + S D
Sbjct 357 KESGQERLLSGSDDNTMFIWLPHQQSRKPLHRLTGHQQLINHVAFSADGRLFASASFDRT 416
Query 151 IKIWDSKTQDCLQTF 165
++IW T L+T
Sbjct 417 VRIWCGITGRYLRTL 431
Score = 30.8 bits (68), Expect = 2.1, Method: Composition-based stats.
Identities = 14/43 (32%), Positives = 21/43 (48%), Gaps = 0/43 (0%)
Query 123 FRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
GH V C + D + TGS D ++IWD +T ++T
Sbjct 124 LEGHSESVLCMDFSADGKLLATGSGDSSVRIWDLQTGTPIKTL 166
Score = 30.0 bits (66), Expect = 3.5, Method: Composition-based stats.
Identities = 28/134 (20%), Positives = 58/134 (43%), Gaps = 8/134 (5%)
Query 1 TRIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLP--SVMSIILAPRNVGADLM 58
TR+ +G DG++ IW+ D L + + H + L + I L R L+
Sbjct 183 TRLASGGMDGRVIIWEP---DSLSSHHITLSGHTKGVTTLAWQPLHHIDLQVR--AYPLL 237
Query 59 YVCTKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTH-ILTGSFDTTARIHGLKAG 117
+ +SI + + + H I+ + +S + ++ + + S DT ++ G
Sbjct 238 ASGSMDSSIKIWDVKVAQCLRTLSGHTRGISQVLWSGEKSNWLFSASRDTLIKVWDTDKG 297
Query 118 KTLKEFRGHLTFVN 131
+K+ +GH ++N
Sbjct 298 GLVKDLKGHGHWIN 311
> ath:AT3G49660 transducin family protein / WD-40 repeat family
protein; K14963 COMPASS component SWD3
Length=317
Score = 62.4 bits (150), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 46/166 (27%), Positives = 79/166 (47%), Gaps = 19/166 (11%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQ--TAQCLPSVMSIILAPRNVGADLMYV 60
I++GS D ++IWD T CL+ +PAH TA S+I++ G +
Sbjct 128 IVSGSFDETVRIWDVTTGKCLKV----LPAHSDPVTAVDFNRDGSLIVSSSYDG-----L 178
Query 61 CTKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTL 120
C +S T G +K + + ++ + FS + IL G+ D T R+ + + K L
Sbjct 179 CRIWDSGT-----GHCVKTLIDDENPPVSFVRFSPNGKFILVGTLDNTLRLWNISSAKFL 233
Query 121 KEFRGHLTFVNC---ALYLPDNTRIITGSADGKIKIWDSKTQDCLQ 163
K + GH+ C A + + RI++GS D + +W+ ++ LQ
Sbjct 234 KTYTGHVNAQYCISSAFSVTNGKRIVSGSEDNCVHMWELNSKKLLQ 279
Score = 57.0 bits (136), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 25/83 (30%), Positives = 43/83 (51%), Gaps = 0/83 (0%)
Query 83 AHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRI 142
H+ I+ + FS D I++ S D T ++ ++ G +K GH + C + P + I
Sbjct 69 GHENGISDVAFSSDARFIVSASDDKTLKLWDVETGSLIKTLIGHTNYAFCVNFNPQSNMI 128
Query 143 ITGSADGKIKIWDSKTQDCLQTF 165
++GS D ++IWD T CL+
Sbjct 129 VSGSFDETVRIWDVTTGKCLKVL 151
Score = 52.4 bits (124), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 27/75 (36%), Positives = 40/75 (53%), Gaps = 0/75 (0%)
Query 91 ITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRIITGSADGK 150
+ F+ + I++GSFD T RI + GK LK H V + D + I++ S DG
Sbjct 119 VNFNPQSNMIVSGSFDETVRIWDVTTGKCLKVLPAHSDPVTAVDFNRDGSLIVSSSYDGL 178
Query 151 IKIWDSKTQDCLQTF 165
+IWDS T C++T
Sbjct 179 CRIWDSGTGHCVKTL 193
Score = 51.6 bits (122), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 46/161 (28%), Positives = 70/161 (43%), Gaps = 17/161 (10%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
I++ S DG +IWDS T C++T P V + +P + V T
Sbjct 170 IVSSSYDGLCRIWDSGTGHCVKTLIDDEN---------PPVSFVRFSPN---GKFILVGT 217
Query 63 KSNSITLMNFAGKTIKKIDKAHDAAIASIT--FSKDN-THILTGSFDTTARIHGLKAGKT 119
N++ L N + K H A I+ FS N I++GS D + L + K
Sbjct 218 LDNTLRLWNISSAKFLKTYTGHVNAQYCISSAFSVTNGKRIVSGSEDNCVHMWELNSKKL 277
Query 120 LKEFRGHL-TFVNCALYLPDNTRIITGSADGKIKIWDSKTQ 159
L++ GH T +N A + P I +GS D ++IW K +
Sbjct 278 LQKLEGHTETVMNVACH-PTENLIASGSLDKTVRIWTQKKE 317
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 34/168 (20%), Positives = 75/168 (44%), Gaps = 20/168 (11%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
I++ S D +K+WD +T ++T H A C+ N ++++ +
Sbjct 86 IVSASDDKTLKLWDVETGSLIKTLI----GHTNYAFCVN---------FNPQSNMIVSGS 132
Query 63 KSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKE 122
++ + + K+ AH + ++ F++D + I++ S+D RI G +K
Sbjct 133 FDETVRIWDVTTGKCLKVLPAHSDPVTAVDFNRDGSLIVSSSYDGLCRIWDSGTGHCVKT 192
Query 123 F----RGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTFT 166
++FV + P+ I+ G+ D +++W+ + L+T+T
Sbjct 193 LIDDENPPVSFVR---FSPNGKFILVGTLDNTLRLWNISSAKFLKTYT 237
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 23/88 (26%), Positives = 45/88 (51%), Gaps = 5/88 (5%)
Query 83 AHDAAIASITFSKDNTHILTGSFDTTARIHGLKA-----GKTLKEFRGHLTFVNCALYLP 137
+H+ A++S+ FS D + + S D T R + + + ++EF GH ++ +
Sbjct 22 SHNRAVSSVKFSSDGRLLASASADKTIRTYTINTINDPIAEPVQEFTGHENGISDVAFSS 81
Query 138 DNTRIITGSADGKIKIWDSKTQDCLQTF 165
D I++ S D +K+WD +T ++T
Sbjct 82 DARFIVSASDDKTLKLWDVETGSLIKTL 109
> ath:AT4G02730 transducin family protein / WD-40 repeat family
protein
Length=333
Score = 62.4 bits (150), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 34/114 (29%), Positives = 60/114 (52%), Gaps = 3/114 (2%)
Query 55 ADLMYVCTKSNSITLMNFAGKT---IKKIDKAHDAAIASITFSKDNTHILTGSFDTTARI 111
+D Y C+ S+ TL + ++ K+ + H + + F+ + I++GSFD T RI
Sbjct 95 SDSHYTCSASDDCTLRIWDARSPYECLKVLRGHTNFVFCVNFNPPSNLIVSGSFDETIRI 154
Query 112 HGLKAGKTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
+K GK ++ + H ++ + D + I++ S DG KIWD+K CL+T
Sbjct 155 WEVKTGKCVRMIKAHSMPISSVHFNRDGSLIVSASHDGSCKIWDAKEGTCLKTL 208
Score = 55.5 bits (132), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 43/165 (26%), Positives = 73/165 (44%), Gaps = 17/165 (10%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
I++GS D I+IW+ KT C++ M A +P I N L+ +
Sbjct 143 IVSGSFDETIRIWEVKTGKCVR---------MIKAHSMP----ISSVHFNRDGSLIVSAS 189
Query 63 KSNSITLMNF-AGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
S + + G +K + A++ FS + IL + D+T ++ GK LK
Sbjct 190 HDGSCKIWDAKEGTCLKTLIDDKSPAVSFAKFSPNGKFILVATLDSTLKLSNYATGKFLK 249
Query 122 EFRGHLTFVNC---ALYLPDNTRIITGSADGKIKIWDSKTQDCLQ 163
+ GH V C A + + I++GS D + +WD + ++ LQ
Sbjct 250 VYTGHTNKVFCITSAFSVTNGKYIVSGSEDNCVYLWDLQARNILQ 294
Score = 46.2 bits (108), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 43/157 (27%), Positives = 66/157 (42%), Gaps = 17/157 (10%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
I++ S DG KIWD+K CL+T + A+ P+ I++A T
Sbjct 185 IVSASHDGSCKIWDAKEGTCLKTLIDDKSPAVSFAKFSPNGKFILVA------------T 232
Query 63 KSNSITLMNFAGKTIKKIDKAHDAAIASIT--FSKDN-THILTGSFDTTARIHGLKAGKT 119
+++ L N+A K+ H + IT FS N +I++GS D + L+A
Sbjct 233 LDSTLKLSNYATGKFLKVYTGHTNKVFCITSAFSVTNGKYIVSGSEDNCVYLWDLQARNI 292
Query 120 LKEFRGHLTFVNCALYLPDNTRIITGSA--DGKIKIW 154
L+ GH V P I + D I+IW
Sbjct 293 LQRLEGHTDAVISVSCHPVQNEISSSGNHLDKTIRIW 329
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 32/125 (25%), Positives = 47/125 (37%), Gaps = 43/125 (34%)
Query 82 KAHDAAIASITFSKDNTHILTGSFDTTA------------RIHGLKAG------------ 117
+ H AAI+ + FS D + + S D T R G +G
Sbjct 40 EGHTAAISCVKFSNDGNLLASASVDKTMILWSATNYSLIHRYEGHSSGISDLAWSSDSHY 99
Query 118 -------------------KTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKT 158
+ LK RGH FV C + P + I++GS D I+IW+ KT
Sbjct 100 TCSASDDCTLRIWDARSPYECLKVLRGHTNFVFCVNFNPPSNLIVSGSFDETIRIWEVKT 159
Query 159 QDCLQ 163
C++
Sbjct 160 GKCVR 164
> dre:406291 katnb1, wu:fj32f02, wu:fj65h01, zgc:56071; katanin
p80 (WD repeat containing) subunit B 1
Length=694
Score = 62.0 bits (149), Expect = 8e-10, Method: Composition-based stats.
Identities = 45/154 (29%), Positives = 71/154 (46%), Gaps = 13/154 (8%)
Query 2 RIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVC 61
R++ GS G +++WD + L+T H S+ S+ P +G L
Sbjct 77 RVVAGSLSGSLRLWDLEAAKILRTLM----GHKA------SISSLDFHP--MGEYLASGS 124
Query 62 TKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLK 121
SN I L + K K H A+ + FS D + + S D+T ++ L AGK +
Sbjct 125 VDSN-IKLWDVRRKGCVFRYKGHTQAVRCLAFSPDGKWLASASDDSTVKLWDLIAGKMIT 183
Query 122 EFRGHLTFVNCALYLPDNTRIITGSADGKIKIWD 155
EF H + VN + P+ + +GSAD +K+WD
Sbjct 184 EFTSHTSAVNVVQFHPNEYLLASGSADRTVKLWD 217
Score = 57.4 bits (137), Expect = 2e-08, Method: Composition-based stats.
Identities = 35/164 (21%), Positives = 68/164 (41%), Gaps = 13/164 (7%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
+ TG D ++ IW +C+ + T H C+ N + + +
Sbjct 36 LATGGEDCRVNIWAVSKPNCIMSLT----GHTSAVGCIQF---------NSSEERVVAGS 82
Query 63 KSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKE 122
S S+ L + I + H A+I+S+ F ++ +GS D+ ++ ++ +
Sbjct 83 LSGSLRLWDLEAAKILRTLMGHKASISSLDFHPMGEYLASGSVDSNIKLWDVRRKGCVFR 142
Query 123 FRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTFT 166
++GH V C + PD + + S D +K+WD + FT
Sbjct 143 YKGHTQAVRCLAFSPDGKWLASASDDSTVKLWDLIAGKMITEFT 186
Score = 44.3 bits (103), Expect = 2e-04, Method: Composition-based stats.
Identities = 35/149 (23%), Positives = 67/149 (44%), Gaps = 19/149 (12%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
+ +GS D IK+WD + + C+ + H Q +CL +P D ++ +
Sbjct 120 LASGSVDSNIKLWDVRRKGCVFRYK----GHTQAVRCLA------FSP-----DGKWLAS 164
Query 63 KSNSITLMNF---AGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKT 119
S+ T+ + AGK I + +H +A+ + F + + +GS D T ++ L+
Sbjct 165 ASDDSTVKLWDLIAGKMITEF-TSHTSAVNVVQFHPNEYLLASGSADRTVKLWDLEKFNM 223
Query 120 LKEFRGHLTFVNCALYLPDNTRIITGSAD 148
+ G V L+ PD + + +GS +
Sbjct 224 IGSSEGETGVVRSVLFNPDGSCLYSGSEN 252
> dre:394246 pafah1b1a; platelet-activating factor acetylhydrolase,
isoform Ib, alpha subunit a; K01062 1-alkyl-2-acetylglycerophosphocholine
esterase [EC:3.1.1.47]
Length=410
Score = 62.0 bits (149), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 40/159 (25%), Positives = 70/159 (44%), Gaps = 13/159 (8%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
I++ S D IK+WD +T D F + H + Q + + L+ C+
Sbjct 123 IVSASEDATIKVWDHETGD----FERTLKGHTDSVQDISF---------DHTGKLLASCS 169
Query 63 KSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKE 122
+I L +F G + HD ++S+ + HI++ S D T ++ + G +K
Sbjct 170 ADMTIKLWDFQGFECIRTMHGHDHNVSSVAIMPNGDHIVSASRDKTIKMWEVATGYCVKT 229
Query 123 FRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDC 161
F GH +V D T I + S D +++W T++C
Sbjct 230 FTGHREWVRMVRPNQDGTLIASSSNDQTVRVWVVATKEC 268
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 41/184 (22%), Positives = 78/184 (42%), Gaps = 33/184 (17%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
I++ S D IK+W+ T C++TFT H + + ++ P G L+ +
Sbjct 207 IVSASRDKTIKMWEVATGYCVKTFT----GHREWVR--------MVRPNQDGT-LIASSS 253
Query 63 KSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTH--------------------ILT 102
++ + A K K + H+ + I+++ ++ H +L+
Sbjct 254 NDQTVRVWVVATKECKAELREHEHVVECISWAPESAHPTILEATGSETKKSGKPGPFLLS 313
Query 103 GSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCL 162
GS D T ++ + G L GH +V L P I++ + D ++IWD K + C
Sbjct 314 GSRDKTIKMWDVSIGMCLMTLVGHDNWVRGVLVHPGGKYIVSCADDKTLRIWDYKNKRCT 373
Query 163 QTFT 166
+T +
Sbjct 374 KTLS 377
Score = 46.2 bits (108), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 41/179 (22%), Positives = 69/179 (38%), Gaps = 33/179 (18%)
Query 7 SADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCTKSNS 66
SAD IK+WD + +C++T H +V S+ + P D + ++ +
Sbjct 169 SADMTIKLWDFQGFECIRTMH----GHDH------NVSSVAIMP---NGDHIVSASRDKT 215
Query 67 ITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGH 126
I + A K H + + ++D T I + S D T R+ + + E R H
Sbjct 216 IKMWEVATGYCVKTFTGHREWVRMVRPNQDGTLIASSSNDQTVRVWVVATKECKAELREH 275
Query 127 LTFVNCALYLPDNTR--------------------IITGSADGKIKIWDSKTQDCLQTF 165
V C + P++ +++GS D IK+WD CL T
Sbjct 276 EHVVECISWAPESAHPTILEATGSETKKSGKPGPFLLSGSRDKTIKMWDVSIGMCLMTL 334
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 22/85 (25%), Positives = 42/85 (49%), Gaps = 0/85 (0%)
Query 82 KAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTR 141
K H ++ I+F + + S D T ++ + + ++ GH V+ +P+
Sbjct 147 KGHTDSVQDISFDHTGKLLASCSADMTIKLWDFQGFECIRTMHGHDHNVSSVAIMPNGDH 206
Query 142 IITGSADGKIKIWDSKTQDCLQTFT 166
I++ S D IK+W+ T C++TFT
Sbjct 207 IVSASRDKTIKMWEVATGYCVKTFT 231
Score = 38.1 bits (87), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 37/174 (21%), Positives = 63/174 (36%), Gaps = 27/174 (15%)
Query 1 TRIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYV 60
T I + S D +++W T++C + H +C I AP + ++
Sbjct 247 TLIASSSNDQTVRVWVVATKEC----KAELREHEHVVEC------ISWAPESAHPTILEA 296
Query 61 -------CTKSNSITLMNFAGKTIKKIDKA----------HDAAIASITFSKDNTHILTG 103
K L KTIK D + HD + + +I++
Sbjct 297 TGSETKKSGKPGPFLLSGSRDKTIKMWDVSIGMCLMTLVGHDNWVRGVLVHPGGKYIVSC 356
Query 104 SFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSK 157
+ D T RI K + K H FV + ++TGS D +K+W+ +
Sbjct 357 ADDKTLRIWDYKNKRCTKTLSAHEHFVTSLDFHKTAPYVVTGSVDQTVKVWECR 410
Score = 32.0 bits (71), Expect = 0.97, Method: Compositional matrix adjust.
Identities = 19/83 (22%), Positives = 38/83 (45%), Gaps = 0/83 (0%)
Query 83 AHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRI 142
H + + + F + I++ S D T ++ + G + +GH V + +
Sbjct 106 GHRSPVTRVIFHPVFSVIVSASEDATIKVWDHETGDFERTLKGHTDSVQDISFDHTGKLL 165
Query 143 ITGSADGKIKIWDSKTQDCLQTF 165
+ SAD IK+WD + +C++T
Sbjct 166 ASCSADMTIKLWDFQGFECIRTM 188
> hsa:5048 PAFAH1B1, LIS1, LIS2, MDCR, MDS, PAFAH; platelet-activating
factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
(EC:3.1.1.47); K01062 1-alkyl-2-acetylglycerophosphocholine
esterase [EC:3.1.1.47]
Length=410
Score = 62.0 bits (149), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 40/159 (25%), Positives = 71/159 (44%), Gaps = 13/159 (8%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
+++ S D IK+WD +T D F + H + Q + S L+ C+
Sbjct 123 MVSASEDATIKVWDYETGD----FERTLKGHTDSVQDISFDHS---------GKLLASCS 169
Query 63 KSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKE 122
+I L +F G + HD ++S+ + HI++ S D T ++ ++ G +K
Sbjct 170 ADMTIKLWDFQGFECIRTMHGHDHNVSSVAIMPNGDHIVSASRDKTIKMWEVQTGYCVKT 229
Query 123 FRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDC 161
F GH +V D T I + S D +++W T++C
Sbjct 230 FTGHREWVRMVRPNQDGTLIASCSNDQTVRVWVVATKEC 268
Score = 55.1 bits (131), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 39/183 (21%), Positives = 81/183 (44%), Gaps = 33/183 (18%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
I++ S D IK+W+ +T C++TFT H + + ++ P G L+ C+
Sbjct 207 IVSASRDKTIKMWEVQTGYCVKTFT----GHREWVR--------MVRPNQDGT-LIASCS 253
Query 63 KSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTH--------------------ILT 102
++ + A K K + H+ + I+++ ++++ +L+
Sbjct 254 NDQTVRVWVVATKECKAELREHEHVVECISWAPESSYSSISEATGSETKKSGKPGPFLLS 313
Query 103 GSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCL 162
GS D T ++ + G L GH +V L+ I++ + D +++WD K + C+
Sbjct 314 GSRDKTIKMWDVSTGMCLMTLVGHDNWVRGVLFHSGGKFILSCADDKTLRVWDYKNKRCM 373
Query 163 QTF 165
+T
Sbjct 374 KTL 376
Score = 48.1 bits (113), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 29/143 (20%), Positives = 63/143 (44%), Gaps = 3/143 (2%)
Query 24 QTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCTKSNSITLMNFAGKTIKKIDKA 83
+ + P P + V +I P +M ++ +I + ++ ++ K
Sbjct 92 KEWIPRPPEKYALSGHRSPVTRVIFHPV---FSVMVSASEDATIKVWDYETGDFERTLKG 148
Query 84 HDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRII 143
H ++ I+F + + S D T ++ + + ++ GH V+ +P+ I+
Sbjct 149 HTDSVQDISFDHSGKLLASCSADMTIKLWDFQGFECIRTMHGHDHNVSSVAIMPNGDHIV 208
Query 144 TGSADGKIKIWDSKTQDCLQTFT 166
+ S D IK+W+ +T C++TFT
Sbjct 209 SASRDKTIKMWEVQTGYCVKTFT 231
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 38/174 (21%), Positives = 64/174 (36%), Gaps = 27/174 (15%)
Query 1 TRIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYV 60
T I + S D +++W T++C + H +C I AP + + +
Sbjct 247 TLIASCSNDQTVRVWVVATKEC----KAELREHEHVVEC------ISWAPESSYSSISEA 296
Query 61 -------CTKSNSITLMNFAGKTIKKIDKA----------HDAAIASITFSKDNTHILTG 103
K L KTIK D + HD + + F IL+
Sbjct 297 TGSETKKSGKPGPFLLSGSRDKTIKMWDVSTGMCLMTLVGHDNWVRGVLFHSGGKFILSC 356
Query 104 SFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSK 157
+ D T R+ K + +K H FV + ++TGS D +K+W+ +
Sbjct 357 ADDKTLRVWDYKNKRCMKTLNAHEHFVTSLDFHKTAPYVVTGSVDQTVKVWECR 410
> mmu:18472 Pafah1b1, LIS-1, Lis1, MGC25297, MMS10-U, Mdsh, Ms10u,
Pafaha; platelet-activating factor acetylhydrolase, isoform
1b, subunit 1 (EC:3.1.1.47); K01062 1-alkyl-2-acetylglycerophosphocholine
esterase [EC:3.1.1.47]
Length=410
Score = 62.0 bits (149), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 40/159 (25%), Positives = 71/159 (44%), Gaps = 13/159 (8%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
+++ S D IK+WD +T D F + H + Q + S L+ C+
Sbjct 123 MVSASEDATIKVWDYETGD----FERTLKGHTDSVQDISFDHS---------GKLLASCS 169
Query 63 KSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKE 122
+I L +F G + HD ++S+ + HI++ S D T ++ ++ G +K
Sbjct 170 ADMTIKLWDFQGFECIRTMHGHDHNVSSVAIMPNGDHIVSASRDKTIKMWEVQTGYCVKT 229
Query 123 FRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDC 161
F GH +V D T I + S D +++W T++C
Sbjct 230 FTGHREWVRMVRPNQDGTLIASCSNDQTVRVWVVATKEC 268
Score = 55.1 bits (131), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 39/183 (21%), Positives = 81/183 (44%), Gaps = 33/183 (18%)
Query 3 IITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCT 62
I++ S D IK+W+ +T C++TFT H + + ++ P G L+ C+
Sbjct 207 IVSASRDKTIKMWEVQTGYCVKTFT----GHREWVR--------MVRPNQDGT-LIASCS 253
Query 63 KSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTH--------------------ILT 102
++ + A K K + H+ + I+++ ++++ +L+
Sbjct 254 NDQTVRVWVVATKECKAELREHEHVVECISWAPESSYSSISEATGSETKKSGKPGPFLLS 313
Query 103 GSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCL 162
GS D T ++ + G L GH +V L+ I++ + D +++WD K + C+
Sbjct 314 GSRDKTIKMWDVSTGMCLMTLVGHDNWVRGVLFHSGGKFILSCADDKTLRVWDYKNKRCM 373
Query 163 QTF 165
+T
Sbjct 374 KTL 376
Score = 48.1 bits (113), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 29/143 (20%), Positives = 63/143 (44%), Gaps = 3/143 (2%)
Query 24 QTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYVCTKSNSITLMNFAGKTIKKIDKA 83
+ + P P + V +I P +M ++ +I + ++ ++ K
Sbjct 92 KEWIPRPPEKYALSGHRSPVTRVIFHPV---FSVMVSASEDATIKVWDYETGDFERTLKG 148
Query 84 HDAAIASITFSKDNTHILTGSFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRII 143
H ++ I+F + + S D T ++ + + ++ GH V+ +P+ I+
Sbjct 149 HTDSVQDISFDHSGKLLASCSADMTIKLWDFQGFECIRTMHGHDHNVSSVAIMPNGDHIV 208
Query 144 TGSADGKIKIWDSKTQDCLQTFT 166
+ S D IK+W+ +T C++TFT
Sbjct 209 SASRDKTIKMWEVQTGYCVKTFT 231
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 38/174 (21%), Positives = 64/174 (36%), Gaps = 27/174 (15%)
Query 1 TRIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCLPSVMSIILAPRNVGADLMYV 60
T I + S D +++W T++C + H +C I AP + + +
Sbjct 247 TLIASCSNDQTVRVWVVATKEC----KAELREHEHVVEC------ISWAPESSYSSISEA 296
Query 61 -------CTKSNSITLMNFAGKTIKKIDKA----------HDAAIASITFSKDNTHILTG 103
K L KTIK D + HD + + F IL+
Sbjct 297 TGSETKKSGKPGPFLLSGSRDKTIKMWDVSTGMCLMTLVGHDNWVRGVLFHSGGKFILSC 356
Query 104 SFDTTARIHGLKAGKTLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSK 157
+ D T R+ K + +K H FV + ++TGS D +K+W+ +
Sbjct 357 ADDKTLRVWDYKNKRCMKTLNAHEHFVTSLDFHKTAPYVVTGSVDQTVKVWECR 410
> dre:556760 wdr69, si:dkey-223n17.5, zgc:153626, zgc:153780;
WD repeat domain 69
Length=418
Score = 62.0 bits (149), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 46/167 (27%), Positives = 70/167 (41%), Gaps = 19/167 (11%)
Query 2 RIITGSADGKIKIWDSKTQDCLQTFTPPVPAHMQTAQCL---PSVMSIILAPRNVGADLM 58
++ TGS D K+W ++T C TF H CL P + + A L
Sbjct 149 KVATGSFDKTCKLWSAETGKCFYTF----RGHTAEIVCLAFNPQSTLVATGSMDTTAKLW 204
Query 59 YVCTKSNSITLMNFAGKTIKKIDKAHDAAIASITFSKDNTHILTGSFDTTARIHGLKAGK 118
V + TL H A I S+ F+ ++TGSFD TA + + +G+
Sbjct 205 DVESGEEVSTL------------AGHFAEIISLCFNTTGDRLVTGSFDHTAILWDVPSGR 252
Query 119 TLKEFRGHLTFVNCALYLPDNTRIITGSADGKIKIWDSKTQDCLQTF 165
+ GH ++C + D + I T S D K+WD++ CL T
Sbjct 253 KVHVLSGHRGEISCVQFNWDCSLIATASLDKSCKVWDAEGGQCLATL 299
Lambda K H
0.321 0.133 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4027755848
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40