bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0150_orf2
Length=142
Score E
Sequences producing significant alignments: (Bits) Value
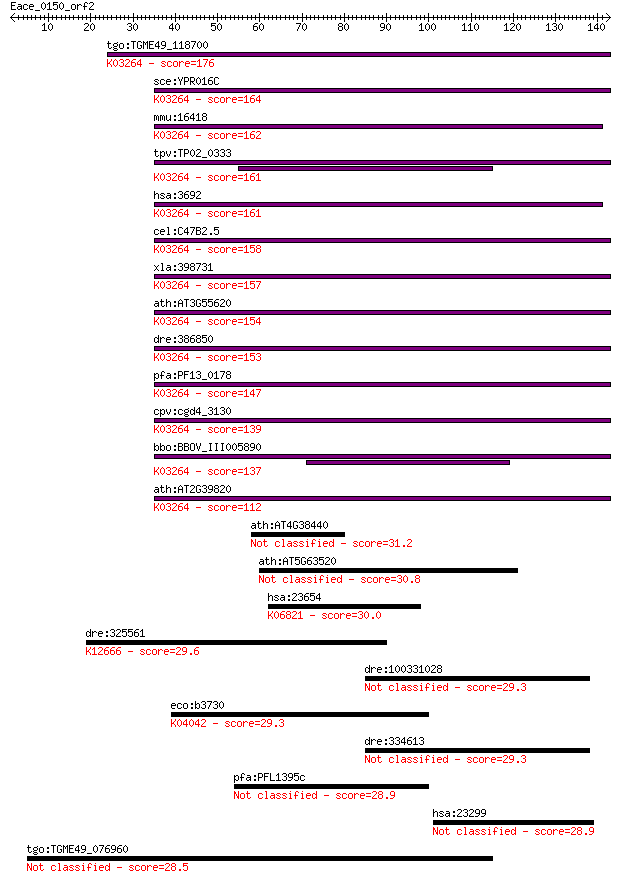
tgo:TGME49_118700 eukaryotic translation initiation factor 6, ... 176 2e-44
sce:YPR016C TIF6, CDC95; Constituent of 66S pre-ribosomal part... 164 7e-41
mmu:16418 Eif6, 1110004P11, AA408895, CAB, Itgb4bp, eIF-6, imc... 162 4e-40
tpv:TP02_0333 translation initiation factor 6; K03264 translat... 161 5e-40
hsa:3692 EIF6, CAB, EIF3A, ITGB4BP, b(2)gcn, eIF-6, p27(BBP), ... 161 8e-40
cel:C47B2.5 eif-6; Eukaryotic Initiation Factor family member ... 158 6e-39
xla:398731 eif6, eIF-6, itgb4bp, p27(BBP), p27BBP; eukaryotic ... 157 1e-38
ath:AT3G55620 emb1624 (embryo defective 1624); ribosome bindin... 154 1e-37
dre:386850 eif6, Itgb4bp, MGC56562, eIF-6, itgb4bp4, wu:fc28a0... 153 2e-37
pfa:PF13_0178 translation initiation factor 6, putative; K0326... 147 1e-35
cpv:cgd4_3130 eIF6, translation initiation factor 6 ; K03264 t... 139 3e-33
bbo:BBOV_III005890 17.m07522; translation initiation factor eI... 137 1e-32
ath:AT2G39820 eukaryotic translation initiation factor 6, puta... 112 3e-25
ath:AT4G38440 hypothetical protein 31.2 1.1
ath:AT5G63520 hypothetical protein 30.8 1.6
hsa:23654 PLXNB2, KIAA0315, MM1, Nbla00445, PLEXB2, dJ402G11.3... 30.0 2.5
dre:325561 rpn1, wu:fa12h07, wu:fc68b07, wu:fc88a12; ribophori... 29.6 3.5
dre:100331028 fibronectin 1b-like 29.3 3.7
eco:b3730 glmU, ECK3723, JW3708, tms, yieA; fused N-acetyl glu... 29.3 3.8
dre:334613 fn1b, cb1057, fn1l, fn3, wu:fa14f11, wu:fb03c02; fi... 29.3 3.9
pfa:PFL1395c conserved Plasmodium protein 28.9 4.9
hsa:23299 BICD2, KIAA0699, bA526D8.1; bicaudal D homolog 2 (Dr... 28.9 5.4
tgo:TGME49_076960 hypothetical protein 28.5 7.1
> tgo:TGME49_118700 eukaryotic translation initiation factor 6,
putative ; K03264 translation initiation factor 6
Length=264
Score = 176 bits (447), Expect = 2e-44, Method: Compositional matrix adjust.
Identities = 85/119 (71%), Positives = 98/119 (82%), Gaps = 0/119 (0%)
Query 24 CSTVSSACSAAMAIRAHCEASCEVGVFGRLTNSYCLVSCGGSENFYSLLEAELGPHIPVV 83
C+ + S S+ MA RA E+S EVGVF +LTNSYCLV+ GGSE+FYS LEAEL PHIPVV
Sbjct 5 CAALWSLGSSRMATRAQFESSNEVGVFAKLTNSYCLVALGGSEHFYSTLEAELAPHIPVV 64
Query 84 HASIAGTAVVGSVSVGNKHGLLVPSTTTDLELQHLRNSLPDSVKIRRVEERLSALGNCV 142
HA++ GT V+G V VGN+ GL+VPS TTD ELQHLRNSLPDSV+IRRVEERLSALGN V
Sbjct 65 HATVGGTRVIGRVCVGNRRGLIVPSITTDQELQHLRNSLPDSVEIRRVEERLSALGNNV 123
> sce:YPR016C TIF6, CDC95; Constituent of 66S pre-ribosomal particles,
has similarity to human translation initiation factor
6 (eIF6); may be involved in the biogenesis and or stability
of 60S ribosomal subunits; K03264 translation initiation
factor 6
Length=245
Score = 164 bits (416), Expect = 7e-41, Method: Compositional matrix adjust.
Identities = 74/108 (68%), Positives = 88/108 (81%), Gaps = 0/108 (0%)
Query 35 MAIRAHCEASCEVGVFGRLTNSYCLVSCGGSENFYSLLEAELGPHIPVVHASIAGTAVVG 94
MA R E S E+GVF +LTN+YCLV+ GGSENFYS EAELG IP+VH +IAGT ++G
Sbjct 1 MATRTQFENSNEIGVFSKLTNTYCLVAVGGSENFYSAFEAELGDAIPIVHTTIAGTRIIG 60
Query 95 SVSVGNKHGLLVPSTTTDLELQHLRNSLPDSVKIRRVEERLSALGNCV 142
++ GN+ GLLVP+ TTD ELQHLRNSLPDSVKI+RVEERLSALGN +
Sbjct 61 RMTAGNRRGLLVPTQTTDQELQHLRNSLPDSVKIQRVEERLSALGNVI 108
> mmu:16418 Eif6, 1110004P11, AA408895, CAB, Itgb4bp, eIF-6, imc-415,
p27(BBP), p27BBP; eukaryotic translation initiation
factor 6 (EC:3.6.5.3); K03264 translation initiation factor
6
Length=245
Score = 162 bits (410), Expect = 4e-40, Method: Compositional matrix adjust.
Identities = 76/106 (71%), Positives = 89/106 (83%), Gaps = 0/106 (0%)
Query 35 MAIRAHCEASCEVGVFGRLTNSYCLVSCGGSENFYSLLEAELGPHIPVVHASIAGTAVVG 94
MA+RA E +CEVG F +LTN+YCLV+ GGSENFYS+ E EL IPVVHASIAG ++G
Sbjct 1 MAVRASFENNCEVGCFAKLTNAYCLVAIGGSENFYSVFEGELSDAIPVVHASIAGCRIIG 60
Query 95 SVSVGNKHGLLVPSTTTDLELQHLRNSLPDSVKIRRVEERLSALGN 140
+ VGN+HGLLVP+ TTD ELQH+RNSLPDSV+IRRVEERLSALGN
Sbjct 61 RMCVGNRHGLLVPNNTTDQELQHIRNSLPDSVQIRRVEERLSALGN 106
> tpv:TP02_0333 translation initiation factor 6; K03264 translation
initiation factor 6
Length=249
Score = 161 bits (408), Expect = 5e-40, Method: Compositional matrix adjust.
Identities = 79/108 (73%), Positives = 86/108 (79%), Gaps = 0/108 (0%)
Query 35 MAIRAHCEASCEVGVFGRLTNSYCLVSCGGSENFYSLLEAELGPHIPVVHASIAGTAVVG 94
MAIRA E S EVGVF LTNSY LVS G S NF SL EAEL PHIPVVH +I GT V+G
Sbjct 1 MAIRAQYENSNEVGVFSTLTNSYALVSLGSSTNFSSLFEAELTPHIPVVHTTIGGTRVIG 60
Query 95 SVSVGNKHGLLVPSTTTDLELQHLRNSLPDSVKIRRVEERLSALGNCV 142
VSVGNK GLLV S TD EL+HLRNSLPDSV+IRR++ERLSALGNC+
Sbjct 61 RVSVGNKKGLLVSSICTDKELRHLRNSLPDSVEIRRIDERLSALGNCI 108
Score = 28.1 bits (61), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 23/63 (36%), Positives = 35/63 (55%), Gaps = 5/63 (7%)
Query 55 NSYC-LVSCGGSENFYSLLEAELGPHIPVVHASIAGTAVVGSVS-VGNKHGLL-VPSTTT 111
N Y L+ + ++E LG I V ASIAG ++GS + NK GL+ V +TT+
Sbjct 111 NDYVGLIHVDMDKETEEIVEDVLG--IEVFRASIAGDVLIGSYTRFQNKGGLVHVKTTTS 168
Query 112 DLE 114
++E
Sbjct 169 EME 171
> hsa:3692 EIF6, CAB, EIF3A, ITGB4BP, b(2)gcn, eIF-6, p27(BBP),
p27BBP; eukaryotic translation initiation factor 6 (EC:3.6.5.3);
K03264 translation initiation factor 6
Length=245
Score = 161 bits (407), Expect = 8e-40, Method: Compositional matrix adjust.
Identities = 74/106 (69%), Positives = 89/106 (83%), Gaps = 0/106 (0%)
Query 35 MAIRAHCEASCEVGVFGRLTNSYCLVSCGGSENFYSLLEAELGPHIPVVHASIAGTAVVG 94
MA+RA E +CE+G F +LTN+YCLV+ GGSENFYS+ E EL IPVVHASIAG ++G
Sbjct 1 MAVRASFENNCEIGCFAKLTNTYCLVAIGGSENFYSVFEGELSDTIPVVHASIAGCRIIG 60
Query 95 SVSVGNKHGLLVPSTTTDLELQHLRNSLPDSVKIRRVEERLSALGN 140
+ VGN+HGLLVP+ TTD ELQH+RNSLPD+V+IRRVEERLSALGN
Sbjct 61 RMCVGNRHGLLVPNNTTDQELQHIRNSLPDTVQIRRVEERLSALGN 106
> cel:C47B2.5 eif-6; Eukaryotic Initiation Factor family member
(eif-6); K03264 translation initiation factor 6
Length=246
Score = 158 bits (399), Expect = 6e-39, Method: Compositional matrix adjust.
Identities = 75/108 (69%), Positives = 87/108 (80%), Gaps = 0/108 (0%)
Query 35 MAIRAHCEASCEVGVFGRLTNSYCLVSCGGSENFYSLLEAELGPHIPVVHASIAGTAVVG 94
MA+R E S +VGVF LTNSYCLV GG++NFYS+LEAEL IPVVH SIA T +VG
Sbjct 1 MALRVDYEGSNDVGVFCTLTNSYCLVGVGGTQNFYSILEAELSDLIPVVHTSIASTRIVG 60
Query 95 SVSVGNKHGLLVPSTTTDLELQHLRNSLPDSVKIRRVEERLSALGNCV 142
++VGN+HGLLVP+ TTD ELQHLRNSLPD V IRRV+ERLSALGN +
Sbjct 61 RLTVGNRHGLLVPNATTDQELQHLRNSLPDEVAIRRVDERLSALGNVI 108
> xla:398731 eif6, eIF-6, itgb4bp, p27(BBP), p27BBP; eukaryotic
translation initiation factor 6 (EC:3.6.5.3); K03264 translation
initiation factor 6
Length=245
Score = 157 bits (397), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 72/108 (66%), Positives = 89/108 (82%), Gaps = 0/108 (0%)
Query 35 MAIRAHCEASCEVGVFGRLTNSYCLVSCGGSENFYSLLEAELGPHIPVVHASIAGTAVVG 94
MA+RA E + E+G F +LTN+YCLV+ GGSENFYS+ E EL IPVVHASIAG ++G
Sbjct 1 MAVRASFENNNEIGCFAKLTNTYCLVAIGGSENFYSVFEGELSETIPVVHASIAGCRIIG 60
Query 95 SVSVGNKHGLLVPSTTTDLELQHLRNSLPDSVKIRRVEERLSALGNCV 142
+ VGN+HGL+VP+ TTD ELQH+RNSLPDSV+I+RVEERLSALGN +
Sbjct 61 RMCVGNRHGLMVPNNTTDQELQHMRNSLPDSVRIQRVEERLSALGNVI 108
> ath:AT3G55620 emb1624 (embryo defective 1624); ribosome binding
/ translation initiation factor; K03264 translation initiation
factor 6
Length=245
Score = 154 bits (388), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 70/108 (64%), Positives = 85/108 (78%), Gaps = 0/108 (0%)
Query 35 MAIRAHCEASCEVGVFGRLTNSYCLVSCGGSENFYSLLEAELGPHIPVVHASIAGTAVVG 94
MA R E +CEVGVF +LTN+YCLV+ GGSENFYS E+EL IP+V SI GT ++G
Sbjct 1 MATRLQFENNCEVGVFSKLTNAYCLVAIGGSENFYSAFESELADVIPIVKTSIGGTRIIG 60
Query 95 SVSVGNKHGLLVPSTTTDLELQHLRNSLPDSVKIRRVEERLSALGNCV 142
+ GNK+GLLVP TTTD ELQHLRNSLPD V ++R++ERLSALGNC+
Sbjct 61 RLCAGNKNGLLVPHTTTDQELQHLRNSLPDQVVVQRIDERLSALGNCI 108
> dre:386850 eif6, Itgb4bp, MGC56562, eIF-6, itgb4bp4, wu:fc28a04,
wu:ft88f05, zgc:56562; eukaryotic translation initiation
factor 6 (EC:3.6.5.3); K03264 translation initiation factor
6
Length=245
Score = 153 bits (386), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 70/108 (64%), Positives = 87/108 (80%), Gaps = 0/108 (0%)
Query 35 MAIRAHCEASCEVGVFGRLTNSYCLVSCGGSENFYSLLEAELGPHIPVVHASIAGTAVVG 94
MA+RA E + E+G F +LTN+YCLV+ GGSENFYS E EL +PV+HASIAG ++G
Sbjct 1 MAVRASFEKNNEIGCFAKLTNTYCLVAIGGSENFYSAFEGELSETMPVIHASIAGCRIIG 60
Query 95 SVSVGNKHGLLVPSTTTDLELQHLRNSLPDSVKIRRVEERLSALGNCV 142
+ VGN+HGLLVP+ TTD ELQH+RN LPDSV+I+RVEERLSALGN +
Sbjct 61 RMCVGNRHGLLVPNNTTDQELQHIRNCLPDSVRIQRVEERLSALGNVI 108
> pfa:PF13_0178 translation initiation factor 6, putative; K03264
translation initiation factor 6
Length=247
Score = 147 bits (371), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 71/108 (65%), Positives = 84/108 (77%), Gaps = 0/108 (0%)
Query 35 MAIRAHCEASCEVGVFGRLTNSYCLVSCGGSENFYSLLEAELGPHIPVVHASIAGTAVVG 94
MAIR E S EVGVF RLTNSY LV+ GGSENF S+ E+EL HIP+V+ +I GT V+G
Sbjct 1 MAIRVQFENSNEVGVFSRLTNSYALVALGGSENFSSVFESELSQHIPLVYTTIGGTRVIG 60
Query 95 SVSVGNKHGLLVPSTTTDLELQHLRNSLPDSVKIRRVEERLSALGNCV 142
V VGN+ GLLV S TD EL HLRN LP++VKI+R+EERLSALGNC+
Sbjct 61 RVCVGNRKGLLVSSICTDQELLHLRNCLPENVKIKRIEERLSALGNCI 108
> cpv:cgd4_3130 eIF6, translation initiation factor 6 ; K03264
translation initiation factor 6
Length=252
Score = 139 bits (349), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 64/108 (59%), Positives = 83/108 (76%), Gaps = 0/108 (0%)
Query 35 MAIRAHCEASCEVGVFGRLTNSYCLVSCGGSENFYSLLEAELGPHIPVVHASIAGTAVVG 94
MA+R E+S E+GVF LTN YCL++ G S F S+ EAEL HIP+++ I GT +VG
Sbjct 1 MALRVAYESSSEIGVFANLTNRYCLLAHGSSAAFTSVFEAELMDHIPIINTLIGGTRLVG 60
Query 95 SVSVGNKHGLLVPSTTTDLELQHLRNSLPDSVKIRRVEERLSALGNCV 142
+VGN++GLLV + TD ELQHLRNSLPD+VK++R+EERLSALGNC+
Sbjct 61 RCTVGNRNGLLVSNMATDQELQHLRNSLPDNVKVQRIEERLSALGNCI 108
> bbo:BBOV_III005890 17.m07522; translation initiation factor
eIF-6 family protein; K03264 translation initiation factor 6
Length=262
Score = 137 bits (344), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 73/121 (60%), Positives = 83/121 (68%), Gaps = 13/121 (10%)
Query 35 MAIRAHCEASCEVGVFGRLTNSYCLVSCGGSENFYSLLEAELGPHIPVVHASIAGTAVVG 94
MAIR E S EVGVF LTNSY LVS G S NF S+ EAEL P IPVV +I GT VVG
Sbjct 1 MAIRTQYENSNEVGVFATLTNSYALVSLGSSCNFASVFEAELMPQIPVVQTTIGGTRVVG 60
Query 95 SVSV-------------GNKHGLLVPSTTTDLELQHLRNSLPDSVKIRRVEERLSALGNC 141
SV+V GN+ GLLV S TD EL+HLRNSLPDSV+IRR+++RLSALGN
Sbjct 61 SVTVGMLYSNYSVILFTGNRKGLLVSSICTDTELRHLRNSLPDSVEIRRIDDRLSALGNV 120
Query 142 V 142
+
Sbjct 121 I 121
Score = 29.3 bits (64), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 22/49 (44%), Positives = 29/49 (59%), Gaps = 4/49 (8%)
Query 71 LLEAELGPHIPVVHASIAGTAVVGSV-SVGNKHGLLVPSTTTDLELQHL 118
++E LG I V ASIAG ++GS NK GL+ TTTD E++ L
Sbjct 141 IVEDVLG--IEVFRASIAGNVLIGSYCRFQNKGGLVHVKTTTD-EMEEL 186
> ath:AT2G39820 eukaryotic translation initiation factor 6, putative
/ eIF-6, putative; K03264 translation initiation factor
6
Length=247
Score = 112 bits (281), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 57/110 (51%), Positives = 78/110 (70%), Gaps = 2/110 (1%)
Query 35 MAIRAHCEAS-CEVGVFGRLTNSYCLVSC-GGSENFYSLLEAELGPHIPVVHASIAGTAV 92
MA R + + CE+GVF +LTN+YCLVS S NF++ E++L IP+V SI G+
Sbjct 1 MATRLQYDNNNCEIGVFSKLTNAYCLVSATSASANFFTGYESKLKGVIPIVTTSIGGSGT 60
Query 93 VGSVSVGNKHGLLVPSTTTDLELQHLRNSLPDSVKIRRVEERLSALGNCV 142
+GS+ VGNK+GLL+ T TD ELQHLR+SLPD V ++R+EE + ALGN +
Sbjct 61 IGSLCVGNKNGLLLSHTITDQELQHLRDSLPDEVVVQRIEEPICALGNAI 110
> ath:AT4G38440 hypothetical protein
Length=1465
Score = 31.2 bits (69), Expect = 1.1, Method: Composition-based stats.
Identities = 12/22 (54%), Positives = 18/22 (81%), Gaps = 1/22 (4%)
Query 58 CLVSCGGSENFYSLLEAELGPH 79
CL+SC +ENF+++LE +GPH
Sbjct 457 CLLSCSLNENFFNILE-NMGPH 477
> ath:AT5G63520 hypothetical protein
Length=519
Score = 30.8 bits (68), Expect = 1.6, Method: Composition-based stats.
Identities = 15/61 (24%), Positives = 33/61 (54%), Gaps = 2/61 (3%)
Query 60 VSCGGSENFYSLLEAELGPHIPVVHASIAGTAVVGSVSVGNKHGLLVPSTTTDLELQHLR 119
++CG E +L+ +G +P++ + + G ++G + +K G + +T+D EL +
Sbjct 101 ITCGNMEETLTLITERVGSRVPIIVSVVTG--ILGKEACNDKAGEVRLHSTSDDELFDVA 158
Query 120 N 120
N
Sbjct 159 N 159
> hsa:23654 PLXNB2, KIAA0315, MM1, Nbla00445, PLEXB2, dJ402G11.3;
plexin B2; K06821 plexin B
Length=1838
Score = 30.0 bits (66), Expect = 2.5, Method: Composition-based stats.
Identities = 14/36 (38%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 62 CGGSENFYSLLEAELGPHIPVVHASIAGTAVVGSVS 97
C N+YS LE +L P +HA+ GT + SV+
Sbjct 250 CREDPNYYSYLEMDLQCRDPDIHAAAFGTCLAASVA 285
> dre:325561 rpn1, wu:fa12h07, wu:fc68b07, wu:fc88a12; ribophorin
I (EC:2.4.1.119); K12666 oligosaccharyltransferase complex
subunit alpha (ribophorin I)
Length=598
Score = 29.6 bits (65), Expect = 3.5, Method: Composition-based stats.
Identities = 22/73 (30%), Positives = 34/73 (46%), Gaps = 6/73 (8%)
Query 19 CCFCFCSTVSSACSAAMAIRAHCEASCEVGVFGRLTNSYCLVSCGGSE--NFYSLLEAEL 76
CCF FC +V + ++ + S + ++T L + G S +F LE EL
Sbjct 13 CCF-FCGSVCADGLVNEDVKRTLDLSSHLA---KITAEIQLANRGSSRANSFIIGLEEEL 68
Query 77 GPHIPVVHASIAG 89
PH+ V AS+ G
Sbjct 69 APHLAFVGASVKG 81
> dre:100331028 fibronectin 1b-like
Length=2500
Score = 29.3 bits (64), Expect = 3.7, Method: Composition-based stats.
Identities = 20/54 (37%), Positives = 29/54 (53%), Gaps = 1/54 (1%)
Query 85 ASIAGTAVVGSVSV-GNKHGLLVPSTTTDLELQHLRNSLPDSVKIRRVEERLSA 137
A I G VV + SV G+ L++P T T + L LR L ++ I VE+ L +
Sbjct 836 APITGYRVVYTPSVEGSSTELILPETQTSVTLGDLRPGLSYNISIYSVEDNLES 889
> eco:b3730 glmU, ECK3723, JW3708, tms, yieA; fused N-acetyl glucosamine-1-phosphate
uridyltransferase/glucosamine-1-phosphate
acetyl transferase (EC:2.7.7.23); K04042 bifunctional UDP-N-acetylglucosamine
pyrophosphorylase / Glucosamine-1-phosphate
N-acetyltransferase [EC:2.7.7.23 2.3.1.157]
Length=456
Score = 29.3 bits (64), Expect = 3.8, Method: Composition-based stats.
Identities = 19/62 (30%), Positives = 30/62 (48%), Gaps = 1/62 (1%)
Query 39 AHCEASCEVGVFGRLTNSYCLVSCGGSENFYSLLEAELGPHIPVVHASIAGTAVVG-SVS 97
A+ A+C +G F RL L+ NF + +A LG H + G A +G +V+
Sbjct 318 ANLAAACTIGPFARLRPGAELLEGAHVGNFVEMKKARLGKGSKAGHLTYLGDAEIGDNVN 377
Query 98 VG 99
+G
Sbjct 378 IG 379
> dre:334613 fn1b, cb1057, fn1l, fn3, wu:fa14f11, wu:fb03c02;
fibronectin 1b
Length=2408
Score = 29.3 bits (64), Expect = 3.9, Method: Composition-based stats.
Identities = 20/54 (37%), Positives = 29/54 (53%), Gaps = 1/54 (1%)
Query 85 ASIAGTAVVGSVSV-GNKHGLLVPSTTTDLELQHLRNSLPDSVKIRRVEERLSA 137
A I G VV + SV G+ L++P T T + L LR L ++ I VE+ L +
Sbjct 836 APITGYRVVYTPSVEGSSTELILPGTQTSVTLGDLRPGLSYNISIYSVEDNLES 889
> pfa:PFL1395c conserved Plasmodium protein
Length=3209
Score = 28.9 bits (63), Expect = 4.9, Method: Composition-based stats.
Identities = 16/46 (34%), Positives = 23/46 (50%), Gaps = 0/46 (0%)
Query 54 TNSYCLVSCGGSENFYSLLEAELGPHIPVVHASIAGTAVVGSVSVG 99
+N+YC V CG N YS E+ + +I + S A + V V G
Sbjct 1487 SNTYCNVECGMESNVYSNAESVVHSNIESITYSNAESNVQSDVDKG 1532
> hsa:23299 BICD2, KIAA0699, bA526D8.1; bicaudal D homolog 2 (Drosophila)
Length=855
Score = 28.9 bits (63), Expect = 5.4, Method: Composition-based stats.
Identities = 17/39 (43%), Positives = 22/39 (56%), Gaps = 1/39 (2%)
Query 101 KHGLLVPSTTTDLELQHLRNSLPDSV-KIRRVEERLSAL 138
K GLL T +L+H R SL + K+ R+ E LSAL
Sbjct 359 KAGLLATLQDTQKQLEHTRGSLSEQQEKVTRLTENLSAL 397
> tgo:TGME49_076960 hypothetical protein
Length=1779
Score = 28.5 bits (62), Expect = 7.1, Method: Composition-based stats.
Identities = 26/117 (22%), Positives = 39/117 (33%), Gaps = 7/117 (5%)
Query 5 VATCSTCAYTACCTCCFCFCSTVSSACSAAMAIRAHCEASCEVGVFGRLTNSYCLVSCGG 64
+ T + C+ C C C + AC + A HCEA L ++S
Sbjct 811 IDTLTRCSNAFRCLYCVCSFAASEDACGSQTASEDHCEAFAGTSDLSALREEAAVLSLSE 870
Query 65 SENFYSLLEAELGPHI-------PVVHASIAGTAVVGSVSVGNKHGLLVPSTTTDLE 114
+ + + L P I P + A A + S N P+T D E
Sbjct 871 TCDASAAALLSLFPDIMAGEAFSPSIRAFAASDELPKSARDANSKAPGTPTTGRDTE 927
Lambda K H
0.325 0.134 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2683748972
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40