bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0156_orf1
Length=144
Score E
Sequences producing significant alignments: (Bits) Value
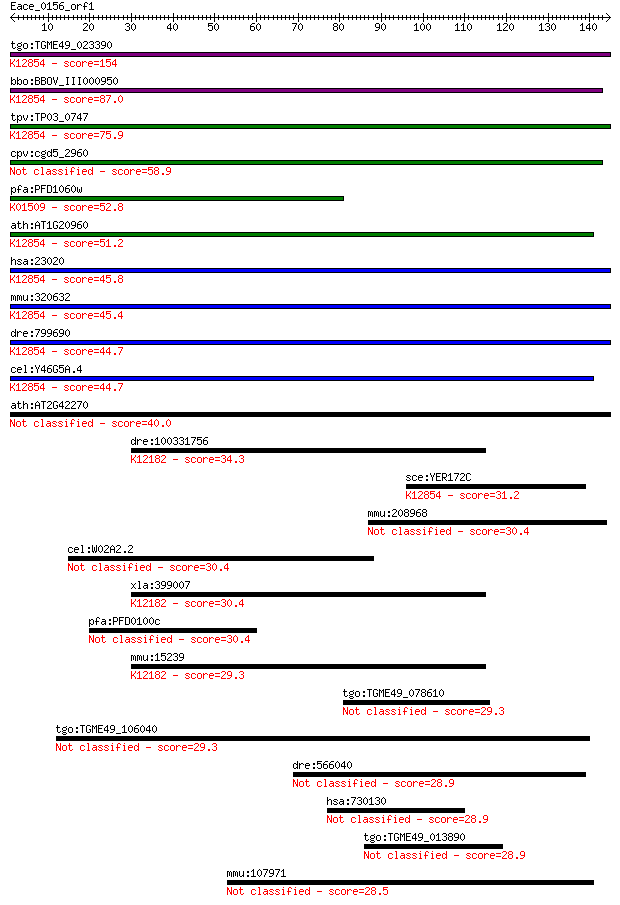
tgo:TGME49_023390 sec63 domain-containing DEAD/DEAH box helica... 154 6e-38
bbo:BBOV_III000950 17.m07111; sec63 domain containing protein ... 87.0 2e-17
tpv:TP03_0747 ATP-dependent RNA helicase; K12854 pre-mRNA-spli... 75.9 5e-14
cpv:cgd5_2960 U5snrp Brr2 SFII RNA helicase (sec63 and the sec... 58.9 6e-09
pfa:PFD1060w U5 small nuclear ribonucleoprotein-specific prote... 52.8 3e-07
ath:AT1G20960 emb1507 (embryo defective 1507); ATP binding / A... 51.2 1e-06
hsa:23020 SNRNP200, ASCC3L1, BRR2, FLJ11521, HELIC2, RP33, U5-... 45.8 4e-05
mmu:320632 Snrnp200, A330064G03Rik, Ascc3l1, BC011390, HELIC2,... 45.4 7e-05
dre:799690 fb63a09; wu:fb63a09 (EC:3.6.4.13); K12854 pre-mRNA-... 44.7 1e-04
cel:Y46G5A.4 hypothetical protein; K12854 pre-mRNA-splicing he... 44.7 1e-04
ath:AT2G42270 U5 small nuclear ribonucleoprotein helicase, put... 40.0 0.003
dre:100331756 hepatocyte growth factor-regulated tyrosine kina... 34.3 0.13
sce:YER172C BRR2, PRP44, RSS1, SLT22, SNU246; Brr2p (EC:3.6.1.... 31.2 1.2
mmu:208968 Zfp280c, BC028839, MGC38010, Suhw3, Zfp633, Znf280c... 30.4 1.9
cel:W02A2.2 far-6; Fatty Acid/Retinol binding protein family m... 30.4 1.9
xla:399007 hgs, MGC68804; hepatocyte growth factor-regulated t... 30.4 2.0
pfa:PFD0100c PFD0105c; 3D7Surf4.1 30.4 2.1
mmu:15239 Hgs, Hgr, Hrs, ZFYVE8; HGF-regulated tyrosine kinase... 29.3 4.1
tgo:TGME49_078610 hypothetical protein 29.3 4.2
tgo:TGME49_106040 zinc finger (CHY type) protein, putative 29.3 4.8
dre:566040 hypothetical LOC566040 28.9 5.3
hsa:730130 TMEM229A, MGC189723; transmembrane protein 229A 28.9 5.4
tgo:TGME49_013890 hypothetical protein 28.9 5.5
mmu:107971 Frs3, 4930417B13Rik, AI449674, Frs2beta, MGC25496, ... 28.5 6.9
> tgo:TGME49_023390 sec63 domain-containing DEAD/DEAH box helicase,
putative ; K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2198
Score = 154 bits (390), Expect = 6e-38, Method: Compositional matrix adjust.
Identities = 90/165 (54%), Positives = 112/165 (67%), Gaps = 21/165 (12%)
Query 1 RWKLFYCIRLGQAQTTEEKDGIREEMKNTQEGQEVLELLDALTSRRNKEKEVTLNVRKEA 60
RWK+F+ +RLGQAQ+ EEK+ I EEM+++ EGQEVLELLDAL+SRRNKEKE+ +NVRKEA
Sbjct 327 RWKIFFAVRLGQAQSPEEKNAIFEEMRSSPEGQEVLELLDALSSRRNKEKEIAINVRKEA 386
Query 61 ATLAAKAQARDASLRAAEADDDSTALGGGAHGTP---------------------GGPGG 99
A+LAAKAQAR ASLRAAE +D G G H + GG
Sbjct 387 ASLAAKAQARAASLRAAEFAEDDAGGGAGLHASHASSAFPKKTGNARDAETKNEGGGAEA 446
Query 100 PSSQAAQRKPTGSVDLQNIAFQQGSHLMANAKVKLPDGTQRIETK 144
P + +KPT +VDL +AF QG H M+N++VKLPDG +RIETK
Sbjct 447 PQASRGAKKPTAAVDLAAVAFHQGGHFMSNSRVKLPDGARRIETK 491
> bbo:BBOV_III000950 17.m07111; sec63 domain containing protein
(EC:3.6.1.-); K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2133
Score = 87.0 bits (214), Expect = 2e-17, Method: Composition-based stats.
Identities = 52/142 (36%), Positives = 78/142 (54%), Gaps = 15/142 (10%)
Query 1 RWKLFYCIRLGQAQTTEEKDGIREEMKNTQEGQEVLELLDALTSRRNKEKEVTLNVRKEA 60
R K+ YC RLGQAQT EK+ I EEM+ + EG VL+ L+ + RRN+++++ LNV +EA
Sbjct 316 RHKIMYCTRLGQAQTDAEKESIYEEMRQSPEGTAVLQELEEVHLRRNRDQKMALNVTREA 375
Query 61 ATLAAKAQARDASLRAAEADDDSTALGGGAHGTPGGPGGPSSQAAQRKPTGSVDLQNIAF 120
+ LA ++ A D +AL P ++ P +DL ++AF
Sbjct 376 SNLARQS-----------AKPDDSALQDLEDLVPEN----TTSGDVSTPIEVLDLDSLAF 420
Query 121 QQGSHLMANAKVKLPDGTQRIE 142
+ G+ M N KV LP ++RIE
Sbjct 421 KDGAQHMTNTKVVLPPESERIE 442
> tpv:TP03_0747 ATP-dependent RNA helicase; K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2249
Score = 75.9 bits (185), Expect = 5e-14, Method: Composition-based stats.
Identities = 47/147 (31%), Positives = 73/147 (49%), Gaps = 15/147 (10%)
Query 1 RWKLFYCIRLGQAQTTEEKDGIREEMKNTQEGQEVLELLDALTSRRNKEKEVTLNVRKEA 60
R+K+ YC RLGQAQ+ +EK+ I +EM T +GQ VL+ L + ++ K++ +T N+ KE
Sbjct 316 RYKIIYCTRLGQAQSQDEKNKIFDEMSKTAQGQLVLQELSLINLKKTKQQLLTHNLEKEL 375
Query 61 ATLAAKAQARDASLR---AAEADDDSTALGGGAHGTPGGPGGPSSQAAQRKPTGSVDLQN 117
+ +D + E D+ Q A +DL+N
Sbjct 376 INMKT-GHHKDLETKNEDELEVPDEFVETKEDV-----------VQTATTTGLQVLDLEN 423
Query 118 IAFQQGSHLMANAKVKLPDGTQRIETK 144
+ + G+ M N KV LP+GT+RIE K
Sbjct 424 LVIKDGAQHMTNVKVVLPEGTERIEHK 450
> cpv:cgd5_2960 U5snrp Brr2 SFII RNA helicase (sec63 and the second
part of the RNA
Length=1204
Score = 58.9 bits (141), Expect = 6e-09, Method: Composition-based stats.
Identities = 40/143 (27%), Positives = 65/143 (45%), Gaps = 27/143 (18%)
Query 1 RWKLFYCIRLGQAQTTEEKDGIREEMKNTQEGQEVLELLDALTSRRNKEKEVTLNVRKEA 60
RW +++C LGQA + E++ I + M+N Q+G EVL+L + +NK+ + ++ K
Sbjct 367 RWSIYFCTILGQAPSDSEREKIIDRMRNHQKGSEVLDLFSKPSLWKNKDSDFFKSINKYI 426
Query 61 ATLAAKAQARDASLRAAEADDDSTALGGGAHGTPGGPGGPSSQAAQRKPTGS-VDLQNIA 119
D D+D + RKP G +DL+ I
Sbjct 427 EETTGVMTGLD--------DEDKNLI------------------TSRKPHGKLLDLERIY 460
Query 120 FQQGSHLMANAKVKLPDGTQRIE 142
+Q +L + KV LP G++RIE
Sbjct 461 QEQSLNLNLSTKVVLPQGSERIE 483
> pfa:PFD1060w U5 small nuclear ribonucleoprotein-specific protein,
putative; K01509 adenosinetriphosphatase [EC:3.6.1.3]
Length=2874
Score = 52.8 bits (125), Expect = 3e-07, Method: Composition-based stats.
Identities = 37/81 (45%), Positives = 54/81 (66%), Gaps = 1/81 (1%)
Query 1 RWKLFYCIRLGQAQTTEEKDGIREEMKNTQEGQEVL-ELLDALTSRRNKEKEVTLNVRKE 59
RWK++YC LGQAQT EEK+ I++EMK T+EGQ++L EL + R+NK+ E + +RKE
Sbjct 429 RWKIYYCTLLGQAQTEEEKEEIKKEMKKTEEGQDILDELCNFKNVRKNKQSEFSKIIRKE 488
Query 60 AATLAAKAQARDASLRAAEAD 80
A L K + D + + +D
Sbjct 489 ADNLFGKKKINDYNKKGNMSD 509
> ath:AT1G20960 emb1507 (embryo defective 1507); ATP binding /
ATP-dependent helicase/ helicase/ nucleic acid binding / nucleoside-triphosphatase/
nucleotide binding; K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2171
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 47/144 (32%), Positives = 69/144 (47%), Gaps = 24/144 (16%)
Query 1 RWKLFYCIRLGQAQTTEEKDGIREEMKNTQEGQEVLELLDALTSRRNKEKEVTLNVRK-- 58
R K+ +C RL +A+ EE++ I EEM+ G E+ +++ L + R KE N++K
Sbjct 338 RLKVVWCTRLARAEDQEERNRIEEEMRGL--GPELTAIVEQLHATRATAKEREENLQKSI 395
Query 59 --EAATLAAKAQARDASLRAAEADDDSTALGGGAHGTPGGPGGPSSQAAQRKPTGSVDLQ 116
EA L + R AD DS + G G QR+ +DL+
Sbjct 396 NEEARRLKDETGGDGGRGRRDVADRDSES--GWVKG-------------QRQ---MLDLE 437
Query 117 NIAFQQGSHLMANAKVKLPDGTQR 140
++AF QG LMAN K LP G+ R
Sbjct 438 SLAFDQGGLLMANKKCDLPPGSYR 461
> hsa:23020 SNRNP200, ASCC3L1, BRR2, FLJ11521, HELIC2, RP33, U5-200KD;
small nuclear ribonucleoprotein 200kDa (U5) (EC:3.6.4.13);
K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2136
Score = 45.8 bits (107), Expect = 4e-05, Method: Composition-based stats.
Identities = 39/144 (27%), Positives = 57/144 (39%), Gaps = 28/144 (19%)
Query 1 RWKLFYCIRLGQAQTTEEKDGIREEMKNTQEGQEVLELLDALTSRRNKEKEVTLNVRKEA 60
R + YC L AQ+ EK+ I +M+ E + L L EKE L + +
Sbjct 325 RMMILYCTLLASAQSEAEKERIMGKMEADPELSKFLYQL------HETEKE-DLIREERS 377
Query 61 ATLAAKAQARDASLRAAEADDDSTALGGGAHGTPGGPGGPSSQAAQRKPTGSVDLQNIAF 120
+ D L + D AL P +DL+++ F
Sbjct 378 RRERVRQSRMDTDLETMDLDQGGEALA---------------------PRQVLDLEDLVF 416
Query 121 QQGSHLMANAKVKLPDGTQRIETK 144
QGSH MAN + +LPDG+ R + K
Sbjct 417 TQGSHFMANKRCQLPDGSFRRQRK 440
> mmu:320632 Snrnp200, A330064G03Rik, Ascc3l1, BC011390, HELIC2,
KIAA0788, U5-200-KD, U5-200KD; small nuclear ribonucleoprotein
200 (U5) (EC:3.6.4.13); K12854 pre-mRNA-splicing helicase
BRR2 [EC:3.6.4.13]
Length=2136
Score = 45.4 bits (106), Expect = 7e-05, Method: Composition-based stats.
Identities = 39/144 (27%), Positives = 57/144 (39%), Gaps = 28/144 (19%)
Query 1 RWKLFYCIRLGQAQTTEEKDGIREEMKNTQEGQEVLELLDALTSRRNKEKEVTLNVRKEA 60
R + YC L AQ+ EK+ I +M+ E + L L EKE L + +
Sbjct 325 RMMILYCTLLASAQSEPEKERIVGKMEADPELSKFLYQL------HETEKE-DLIREERS 377
Query 61 ATLAAKAQARDASLRAAEADDDSTALGGGAHGTPGGPGGPSSQAAQRKPTGSVDLQNIAF 120
+ D L + D AL P +DL+++ F
Sbjct 378 RRERVRQSRMDTDLETMDLDQGGEALA---------------------PRQVLDLEDLVF 416
Query 121 QQGSHLMANAKVKLPDGTQRIETK 144
QGSH MAN + +LPDG+ R + K
Sbjct 417 TQGSHFMANKRCQLPDGSFRRQRK 440
> dre:799690 fb63a09; wu:fb63a09 (EC:3.6.4.13); K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2134
Score = 44.7 bits (104), Expect = 1e-04, Method: Composition-based stats.
Identities = 40/144 (27%), Positives = 63/144 (43%), Gaps = 30/144 (20%)
Query 1 RWKLFYCIRLGQAQTTEEKDGIREEMKNTQEGQEVLELLDALTSRRNKEKEVTLNVRKEA 60
R + YC L AQ+ EK+ I +M+ Q+ +VL L + EKE + +
Sbjct 324 RRMILYCTMLASAQSEAEKEKIINKMEADQDLSKVLYQL------QETEKEDIIREERSR 377
Query 61 ATLAAKAQARDASLRAAEADDDSTALGGGAHGTPGGPGGPSSQAAQRKPTGSVDLQNIAF 120
K++ D L + + D G S + Q +DL+++ F
Sbjct 378 RERMRKSRVDD--LESMDIDH-----------------GESVSSRQL-----LDLEDLTF 413
Query 121 QQGSHLMANAKVKLPDGTQRIETK 144
QGSH MAN + +LPDG+ R + K
Sbjct 414 TQGSHFMANKRCQLPDGSFRKQRK 437
> cel:Y46G5A.4 hypothetical protein; K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2145
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 40/142 (28%), Positives = 63/142 (44%), Gaps = 35/142 (24%)
Query 1 RWKLFYCIRLGQAQTTEEKDGIREEMKNTQEGQEVLELLDALTSRRNKEKEVTLNVRKEA 60
R + YC L QA +E+ I ++M++ E +L LL E
Sbjct 322 RLMILYCTLLRQA-NEKERLQIEDDMRSRPELHPILALLQE---------------TDEG 365
Query 61 ATLAAKAQARDA--SLRAAEADDDSTALGGGAHGTPGGPGGPSSQAAQRKPTGSVDLQNI 118
+ + + RDA S +AA A +++ + G G RK +DL ++
Sbjct 366 SVVQVEKSKRDAEKSKKAATAANEAISAGQWQAG--------------RK---MLDLNDL 408
Query 119 AFQQGSHLMANAKVKLPDGTQR 140
F QGSHLM+N + +LPDG+ R
Sbjct 409 TFSQGSHLMSNKRCELPDGSYR 430
> ath:AT2G42270 U5 small nuclear ribonucleoprotein helicase, putative
Length=2172
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 41/148 (27%), Positives = 66/148 (44%), Gaps = 23/148 (15%)
Query 1 RWKLFYCIRLGQAQTTEEKDGIREEMKNTQEGQEVLELLDALTSRR----NKEKEVTLNV 56
R K+ +C RL + + EE++ I EEM G E+ ++ L ++R +E++ ++
Sbjct 338 RLKVVWCTRLARGRDQEERNQIEEEMLGL--GSELAAIVKELHAKRATAKEREEKREKDI 395
Query 57 RKEAATLAAKAQARDASLRAAEADDDSTALGGGAHGTPGGPGGPSSQAAQRKPTGSVDLQ 116
++EA L D R DD G G QR+ +DL+
Sbjct 396 KEEAQHLMDDDSGVDGD-RGMRDVDDLDLENGWLKG-------------QRQ---VMDLE 438
Query 117 NIAFQQGSHLMANAKVKLPDGTQRIETK 144
++AF QG N K +LPD + RI K
Sbjct 439 SLAFNQGGFTRENNKCELPDRSFRIRGK 466
> dre:100331756 hepatocyte growth factor-regulated tyrosine kinase
substrate-like; K12182 hepatocyte growth factor-regulated
tyrosine kinase substrate
Length=757
Score = 34.3 bits (77), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 26/87 (29%), Positives = 40/87 (45%), Gaps = 5/87 (5%)
Query 30 QEGQEVLELLDALTSRR--NKEKEVTLNVRKEAATLAAKAQARDASLRAAEADDDSTALG 87
Q+ QE LE+ L +R +EKE + + ++ T+ +AQ SL A+ +
Sbjct 506 QKKQEYLEMQRQLAIQRLQEQEKERQMRLEQQKHTIQMRAQMPAFSLPYAQMQSLPPNVA 565
Query 88 GGAHGTPGGPGGPSSQAAQRKPTGSVD 114
GG P GP S P+GSV+
Sbjct 566 GGVVYP---PAGPPSYPGTFSPSGSVE 589
> sce:YER172C BRR2, PRP44, RSS1, SLT22, SNU246; Brr2p (EC:3.6.1.-);
K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2163
Score = 31.2 bits (69), Expect = 1.2, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 24/49 (48%), Gaps = 6/49 (12%)
Query 96 GPGGPSSQAAQRKPTGS------VDLQNIAFQQGSHLMANAKVKLPDGT 138
G P S A+R + +DL+ I F + S LM KV LP+G+
Sbjct 404 GDDQPQSSEAKRTKFSNPAIPPVIDLEKIKFDESSKLMTVTKVSLPEGS 452
> mmu:208968 Zfp280c, BC028839, MGC38010, Suhw3, Zfp633, Znf280c,
mKIAA1584; zinc finger protein 280C
Length=742
Score = 30.4 bits (67), Expect = 1.9, Method: Composition-based stats.
Identities = 16/57 (28%), Positives = 25/57 (43%), Gaps = 0/57 (0%)
Query 87 GGGAHGTPGGPGGPSSQAAQRKPTGSVDLQNIAFQQGSHLMANAKVKLPDGTQRIET 143
G G + P + RK S+DL+N+ QGSH+ + K+ D + T
Sbjct 587 GTGTTKSKAKPSYKQKRQRTRKNKFSIDLKNLRCHQGSHMCIECRSKIKDFSSHFST 643
> cel:W02A2.2 far-6; Fatty Acid/Retinol binding protein family
member (far-6)
Length=184
Score = 30.4 bits (67), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 21/73 (28%), Positives = 35/73 (47%), Gaps = 13/73 (17%)
Query 15 TTEEKDGIREEMKNTQEGQEVLELLDALTSRRNKEKEVTLNVRKEAATLAAKAQARDASL 74
TTEEK I+E +K+ G + +E E++ ++++ + +L AK + D L
Sbjct 50 TTEEKAAIKEFIKSVMGGNKSVE-------------ELSADIKERSPSLYAKVEKLDVLL 96
Query 75 RAAEADDDSTALG 87
R A D AL
Sbjct 97 RTKLAKLDPAALA 109
> xla:399007 hgs, MGC68804; hepatocyte growth factor-regulated
tyrosine kinase substrate; K12182 hepatocyte growth factor-regulated
tyrosine kinase substrate
Length=751
Score = 30.4 bits (67), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 27/87 (31%), Positives = 40/87 (45%), Gaps = 7/87 (8%)
Query 30 QEGQEVLELLDALTSRR--NKEKEVTLNVRKEAATLAAKAQARDASLRAAEADDDSTALG 87
Q+ QE LE+ L +R +EKE L + ++ T+ +AQ SL A+ S
Sbjct 496 QKKQEYLEMQRQLAIQRLQEQEKERQLRLEQQKQTIQMRAQMPAFSLPYAQLQ--SLPAT 553
Query 88 GGAHGTPGGPGGPSSQAAQRKPTGSVD 114
GG P GP + P+GSV+
Sbjct 554 GGVMYP---PSGPPAYPGSFSPSGSVE 577
> pfa:PFD0100c PFD0105c; 3D7Surf4.1
Length=2156
Score = 30.4 bits (67), Expect = 2.1, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 26/44 (59%), Gaps = 4/44 (9%)
Query 20 DGIREEMKNTQEGQEVLELLDALTS----RRNKEKEVTLNVRKE 59
D I+E+ NT+E L+L+ A+ + ++NKEKE+ + E
Sbjct 1101 DDIKEKQTNTEETNNTLKLIRAMENKYILKKNKEKEIINQFKNE 1144
> mmu:15239 Hgs, Hgr, Hrs, ZFYVE8; HGF-regulated tyrosine kinase
substrate; K12182 hepatocyte growth factor-regulated tyrosine
kinase substrate
Length=776
Score = 29.3 bits (64), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 28/87 (32%), Positives = 41/87 (47%), Gaps = 7/87 (8%)
Query 30 QEGQEVLELLDALTSRR--NKEKEVTLNVRKEAATLAAKAQARDASLRAAEADDDSTALG 87
Q+ QE LE+ L +R +EKE + + ++ T+ +AQ L A+ TA
Sbjct 518 QKKQEYLEVQRQLAIQRLQEQEKERQMRLEQQKQTVQMRAQMPAFPLPYAQLQAMPTA-- 575
Query 88 GGAHGTPGGPGGPSSQAAQRKPTGSVD 114
GG P GP+S A P GSV+
Sbjct 576 GGVLYQ---PSGPTSFPATFSPAGSVE 599
> tgo:TGME49_078610 hypothetical protein
Length=216
Score = 29.3 bits (64), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 15/35 (42%), Positives = 20/35 (57%), Gaps = 2/35 (5%)
Query 81 DDSTALGGGAHGTPGGPGGPSSQAAQRKPTGSVDL 115
D S+ G + TPGG GPSS + R+P+ V L
Sbjct 87 DTSSTFGSKSPSTPGG--GPSSCFSSRRPSADVSL 119
> tgo:TGME49_106040 zinc finger (CHY type) protein, putative
Length=1024
Score = 29.3 bits (64), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 38/132 (28%), Positives = 56/132 (42%), Gaps = 14/132 (10%)
Query 12 QAQTTEEKDG--IREEMKNTQEGQEVLELLDALTSRRNKEKEVTLNVRKEAATLAAKA-- 67
+A+ EK G EE K +EG L + K K + VR A A+
Sbjct 596 KAENEPEKKGDWTLEEQKRLEEG---LVWYRRVADPVTKWKLIGSFVRTRTAKECAQRFR 652
Query 68 QARDASLRAAEADDDSTALGGGAHGTPGGPGGPSSQAAQRKPTGSVDLQNIAFQQGSHLM 127
R+A LR AEA + A G +P P P++ AA+ G++ A +G
Sbjct 653 DCREAILRRAEA---AVASNSGGTASPANPSDPATAAAE----GTLQEATFAGDEGGESN 705
Query 128 ANAKVKLPDGTQ 139
+++ V PDG Q
Sbjct 706 SSSAVSAPDGDQ 717
> dre:566040 hypothetical LOC566040
Length=577
Score = 28.9 bits (63), Expect = 5.3, Method: Composition-based stats.
Identities = 22/73 (30%), Positives = 28/73 (38%), Gaps = 12/73 (16%)
Query 69 ARDASLRAAEADDDSTALGGG---AHGTPGGPGGPSSQAAQRKPTGSVDLQNIAFQQGSH 125
A +A+ EAD +S G G PGG G A+ KP + L H
Sbjct 121 AIEATQHGPEADPNSGLAGAGDSSKETNPGGRGSSQQSASVNKPADDMKL---------H 171
Query 126 LMANAKVKLPDGT 138
+ N V DGT
Sbjct 172 FLKNTAVTCNDGT 184
> hsa:730130 TMEM229A, MGC189723; transmembrane protein 229A
Length=380
Score = 28.9 bits (63), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 15/33 (45%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 77 AEADDDSTALGGGAHGTPGGPGGPSSQAAQRKP 109
++ D + A GGA PG PGGP S+AA P
Sbjct 4 SDVDSEGPARRGGAARRPGAPGGPGSEAAAGCP 36
> tgo:TGME49_013890 hypothetical protein
Length=1022
Score = 28.9 bits (63), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 17/33 (51%), Positives = 18/33 (54%), Gaps = 3/33 (9%)
Query 86 LGGGAHGTPGGPGGPSSQAAQRKPTGSVDLQNI 118
LG HG P GP GPS Q A PTGS L +
Sbjct 699 LGPSLHGAPNGPRGPSPQLA---PTGSPQLAQM 728
> mmu:107971 Frs3, 4930417B13Rik, AI449674, Frs2beta, MGC25496,
Snt2; fibroblast growth factor receptor substrate 3
Length=492
Score = 28.5 bits (62), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 25/100 (25%), Positives = 41/100 (41%), Gaps = 13/100 (13%)
Query 53 TLNVRKEAATLAAKAQARDASLRAAEADDDSTALGGGAHGTPGGPG-GPSSQAAQRKPTG 111
++NV +E + + + L + G ++G PG PG GP +A R+P+
Sbjct 109 SINVTEEPVIITRSSHPPELDLPRGPPQPAGYTVSGFSNGFPGCPGEGPRFSSAPRRPST 168
Query 112 SVDLQN-----------IAFQQGSHLMANAKVKLPDGTQR 140
S L++ IA ++ SH N DG R
Sbjct 169 S-SLRHPSPGEESTHTLIASEEQSHTYVNTPTGDEDGRSR 207
Lambda K H
0.309 0.126 0.346
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2814663556
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40