bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0182_orf1
Length=117
Score E
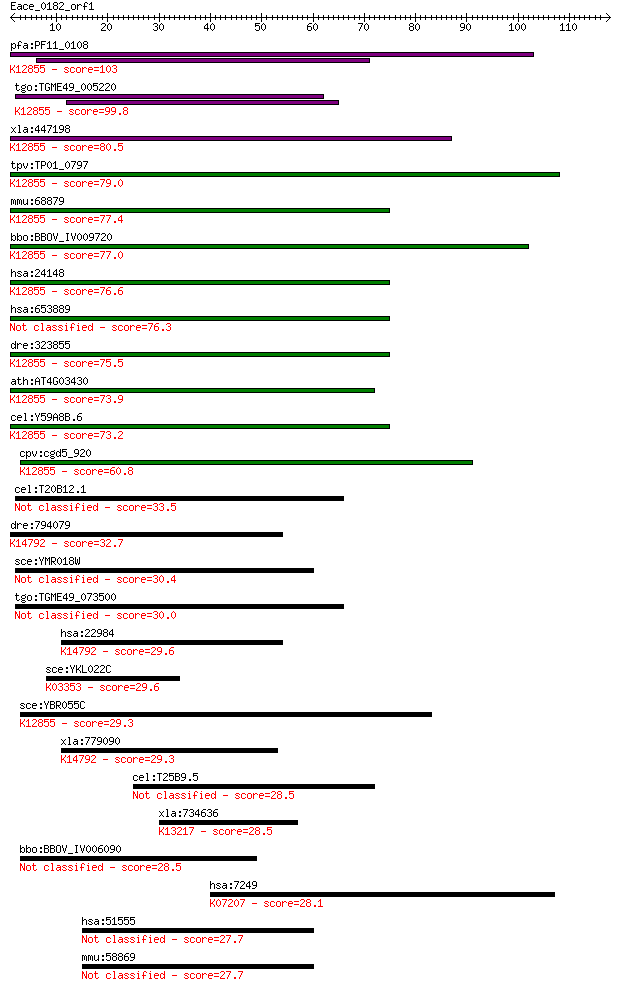
Sequences producing significant alignments: (Bits) Value
pfa:PF11_0108 U5 snRNP-associated protein, putative; K12855 pr... 103 1e-22
tgo:TGME49_005220 U5 snRNP-associated 102 kDa protein, putativ... 99.8 2e-21
xla:447198 prpf6, MGC80263; PRP6 pre-mRNA processing factor 6 ... 80.5 1e-15
tpv:TP01_0797 hypothetical protein; K12855 pre-mRNA-processing... 79.0 4e-15
mmu:68879 Prpf6, 1190003A07Rik, 2610031L17Rik, ANT-1, MGC11655... 77.4 1e-14
bbo:BBOV_IV009720 23.m06014; u5 snRNP-associated subunit, puta... 77.0 1e-14
hsa:24148 PRPF6, ANT-1, C20orf14, Prp6, TOM, U5-102K, hPrp6; P... 76.6 2e-14
hsa:653889 pre-mRNA-processing factor 6-like 76.3 2e-14
dre:323855 c20orf14, fc12b02, prpf6, wu:fa05f07, wu:fc12b02, z... 75.5 4e-14
ath:AT4G03430 EMB2770 (EMBRYO DEFECTIVE 2770); RNA splicing fa... 73.9 1e-13
cel:Y59A8B.6 hypothetical protein; K12855 pre-mRNA-processing ... 73.2 2e-13
cpv:cgd5_920 Pre-mRNA splicing factor Pro1/Prp6. HAT repeat pr... 60.8 9e-10
cel:T20B12.1 hypothetical protein 33.5 0.19
dre:794079 pdcd11, MGC162501, cb680, im:7148359, sb:cb680, wu:... 32.7 0.28
sce:YMR018W Putative protein of unknown function with similari... 30.4 1.6
tgo:TGME49_073500 signal transduction protein, putative 30.0 1.9
hsa:22984 PDCD11, ALG-4, ALG4, KIAA0185, NFBP, RRP5; programme... 29.6 2.2
sce:YKL022C CDC16; Cdc16p; K03353 anaphase-promoting complex s... 29.6 2.4
sce:YBR055C PRP6, RNA6, TSM7269; Prp6p; K12855 pre-mRNA-proces... 29.3 2.9
xla:779090 pdcd11, alg-4, alg4, nfbp, rrp5; programmed cell de... 29.3 3.6
cel:T25B9.5 hypothetical protein 28.5 5.0
xla:734636 prpf39.1, MGC115228, prpf39; PRP39 pre-mRNA process... 28.5 5.0
bbo:BBOV_IV006090 23.m05922; tetratricopeptide repeat containi... 28.5 5.1
hsa:7249 TSC2, FLJ43106, LAM, TSC4; tuberous sclerosis 2; K072... 28.1 7.8
hsa:51555 PEX5L, PEX5R, PXR2, PXR2B; peroxisomal biogenesis fa... 27.7 9.2
mmu:58869 Pex5l, 1700016J08Rik, PXR2, Pex2, TRIP8b; peroxisoma... 27.7 9.7
> pfa:PF11_0108 U5 snRNP-associated protein, putative; K12855
pre-mRNA-processing factor 6
Length=1329
Score = 103 bits (258), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 48/102 (47%), Positives = 63/102 (61%), Gaps = 0/102 (0%)
Query 1 RLFWKDQKIAKARKWMNRSVTLDPSLGDAWASFLAFEIEHGGEKECIDVINRCALAQPNR 60
+LFW + KI KARKW R + L+P GD WA+FLAFEI+ E D+IN+C A+PNR
Sbjct 1134 KLFWVNFKIQKARKWFYRVINLNPHFGDGWATFLAFEIDQQNEINQKDIINKCIKAEPNR 1193
Query 61 GLAWNKTTKQVRCWSFSPAQKLNAVLEYMYPNALKDGISQVV 102
G WNK TK+V W QKL ++ +YP+ L IS +
Sbjct 1194 GYLWNKITKRVENWRLKYPQKLYKYIKDIYPHVLNKKISDPI 1235
Score = 28.5 bits (62), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 14/65 (21%), Positives = 30/65 (46%), Gaps = 0/65 (0%)
Query 6 DQKIAKARKWMNRSVTLDPSLGDAWASFLAFEIEHGGEKECIDVINRCALAQPNRGLAWN 65
++ I AR N ++ + + W + E+ HG ++ +V++R + P+ + W
Sbjct 669 NKSIHTARTLYNEALKIFKTKKSLWLALANLELTHGKREDVDEVLHRAVQSCPHSSVLWL 728
Query 66 KTTKQ 70
KQ
Sbjct 729 MLAKQ 733
> tgo:TGME49_005220 U5 snRNP-associated 102 kDa protein, putative
; K12855 pre-mRNA-processing factor 6
Length=985
Score = 99.8 bits (247), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 45/61 (73%), Positives = 55/61 (90%), Gaps = 1/61 (1%)
Query 2 LFWKDQKIAKARKWMNRSVTLDPSLGDAWASFLAFEIEH-GGEKECIDVINRCALAQPNR 60
LFWK+ KI+KARKW+NRSVTLD S GDAWA+FLAFE+E+ GGEKEC ++IN+ +LAQPNR
Sbjct 919 LFWKEGKISKARKWLNRSVTLDASFGDAWAAFLAFELENGGGEKECRNIINKASLAQPNR 978
Query 61 G 61
G
Sbjct 979 G 979
Score = 28.5 bits (62), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 14/53 (26%), Positives = 25/53 (47%), Gaps = 0/53 (0%)
Query 12 ARKWMNRSVTLDPSLGDAWASFLAFEIEHGGEKECIDVINRCALAQPNRGLAW 64
AR + ++ +P D W + + E+E G ++ V ++ PN GL W
Sbjct 831 ARAILEKAKLRNPKNPDLWHAAIRIEVEAGNKQMAQHVASKAVQECPNSGLVW 883
> xla:447198 prpf6, MGC80263; PRP6 pre-mRNA processing factor
6 homolog; K12855 pre-mRNA-processing factor 6
Length=948
Score = 80.5 bits (197), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 35/86 (40%), Positives = 55/86 (63%), Gaps = 4/86 (4%)
Query 1 RLFWKDQKIAKARKWMNRSVTLDPSLGDAWASFLAFEIEHGGEKECIDVINRCALAQPNR 60
+LFW ++KI KAR+W +R+V +D LGDAWA+F FE++HG E++ ++ RC A+P
Sbjct 856 KLFWSERKITKAREWFHRTVKIDSDLGDAWATFYKFELQHGTEEQQEEIRKRCENAEPRH 915
Query 61 GLAWNKTTKQVRCWSFSPAQKLNAVL 86
G W +K ++ W QK+ +L
Sbjct 916 GELWCAVSKDIKNWQ----QKIGEIL 937
> tpv:TP01_0797 hypothetical protein; K12855 pre-mRNA-processing
factor 6
Length=1032
Score = 79.0 bits (193), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 40/117 (34%), Positives = 63/117 (53%), Gaps = 10/117 (8%)
Query 1 RLFWKDQKIAKARKWMNRSVTLDPSLGDAWA----------SFLAFEIEHGGEKECIDVI 50
++FW + I KAR+W +TLD S G +W +F+AFE++ G E+ I
Sbjct 907 KIFWNCKMIDKARRWFQTCITLDESNGISWGIFVKFMNVLGTFIAFELDCGTEESMKQAI 966
Query 51 NRCALAQPNRGLAWNKTTKQVRCWSFSPAQKLNAVLEYMYPNALKDGISQVVLDALH 107
N+ A+PNRG W + TK+V W+ QKL ++ YP L D + + V++ L+
Sbjct 967 NKFIEAEPNRGYEWCRVTKKVENWNIPLPQKLYKFIQEYYPEVLTDKVPKDVMEVLN 1023
> mmu:68879 Prpf6, 1190003A07Rik, 2610031L17Rik, ANT-1, MGC11655,
MGC36967, MGC38351, U5-102K; PRP6 pre-mRNA splicing factor
6 homolog (yeast); K12855 pre-mRNA-processing factor 6
Length=941
Score = 77.4 bits (189), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 33/74 (44%), Positives = 48/74 (64%), Gaps = 0/74 (0%)
Query 1 RLFWKDQKIAKARKWMNRSVTLDPSLGDAWASFLAFEIEHGGEKECIDVINRCALAQPNR 60
+LFW ++KI KAR+W +R+V +D LGDAWA F FE++HG E++ +V RC A+P
Sbjct 849 KLFWSERKITKAREWFHRTVKIDSDLGDAWAFFYKFELQHGTEEQQEEVRKRCENAEPRH 908
Query 61 GLAWNKTTKQVRCW 74
G W +K + W
Sbjct 909 GELWCAVSKDITNW 922
> bbo:BBOV_IV009720 23.m06014; u5 snRNP-associated subunit, putaitve;
K12855 pre-mRNA-processing factor 6
Length=1040
Score = 77.0 bits (188), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 35/101 (34%), Positives = 56/101 (55%), Gaps = 0/101 (0%)
Query 1 RLFWKDQKIAKARKWMNRSVTLDPSLGDAWASFLAFEIEHGGEKECIDVINRCALAQPNR 60
+LFW+D K+ K RKW R++ ++ S G W +FLAFE++ G DVIN C A+P+
Sbjct 920 KLFWRDGKVLKTRKWFKRALAIEESNGVIWGTFLAFELDSGDNDAIKDVINGCTKAEPST 979
Query 61 GLAWNKTTKQVRCWSFSPAQKLNAVLEYMYPNALKDGISQV 101
G W + K+V W + K+ +E Y + L + ++
Sbjct 980 GYDWCRVVKRVVNWRLTWPHKMYKFVEEHYNDILTKALEKL 1020
> hsa:24148 PRPF6, ANT-1, C20orf14, Prp6, TOM, U5-102K, hPrp6;
PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae);
K12855 pre-mRNA-processing factor 6
Length=941
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 33/74 (44%), Positives = 47/74 (63%), Gaps = 0/74 (0%)
Query 1 RLFWKDQKIAKARKWMNRSVTLDPSLGDAWASFLAFEIEHGGEKECIDVINRCALAQPNR 60
+LFW +KI KAR+W +R+V +D LGDAWA F FE++HG E++ +V RC A+P
Sbjct 849 KLFWSQRKITKAREWFHRTVKIDSDLGDAWAFFYKFELQHGTEEQQEEVRKRCESAEPRH 908
Query 61 GLAWNKTTKQVRCW 74
G W +K + W
Sbjct 909 GELWCAVSKDIANW 922
> hsa:653889 pre-mRNA-processing factor 6-like
Length=406
Score = 76.3 bits (186), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 33/74 (44%), Positives = 47/74 (63%), Gaps = 0/74 (0%)
Query 1 RLFWKDQKIAKARKWMNRSVTLDPSLGDAWASFLAFEIEHGGEKECIDVINRCALAQPNR 60
+LFW +KI KAR+W +R+V +D LGDAWA F FE++HG E++ +V RC A+P
Sbjct 314 KLFWSQRKITKAREWFHRTVKIDSDLGDAWAFFYKFELQHGTEEQQEEVRKRCESAEPRH 373
Query 61 GLAWNKTTKQVRCW 74
G W +K + W
Sbjct 374 GELWCAVSKDIANW 387
> dre:323855 c20orf14, fc12b02, prpf6, wu:fa05f07, wu:fc12b02,
zgc:65913; c20orf14 homolog (H. sapiens); K12855 pre-mRNA-processing
factor 6
Length=944
Score = 75.5 bits (184), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 32/74 (43%), Positives = 47/74 (63%), Gaps = 0/74 (0%)
Query 1 RLFWKDQKIAKARKWMNRSVTLDPSLGDAWASFLAFEIEHGGEKECIDVINRCALAQPNR 60
+LFW ++KI KAR+W R+V ++P LGDAW F FE++HG E++ +V RC A+P
Sbjct 852 KLFWSERKITKAREWFLRTVKIEPDLGDAWGFFYKFELQHGTEEQQHEVKKRCENAEPRH 911
Query 61 GLAWNKTTKQVRCW 74
G W +K + W
Sbjct 912 GELWCAESKHILNW 925
> ath:AT4G03430 EMB2770 (EMBRYO DEFECTIVE 2770); RNA splicing
factor, transesterification mechanism; K12855 pre-mRNA-processing
factor 6
Length=1029
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 29/71 (40%), Positives = 46/71 (64%), Gaps = 0/71 (0%)
Query 1 RLFWKDQKIAKARKWMNRSVTLDPSLGDAWASFLAFEIEHGGEKECIDVINRCALAQPNR 60
+LFW+D+K+ KAR W R+VT+ P +GD WA F FE++HG +++ +V+ +C +P
Sbjct 934 KLFWQDKKVEKARAWFERAVTVGPDIGDFWALFYKFELQHGSDEDRKEVVAKCVACEPKH 993
Query 61 GLAWNKTTKQV 71
G W +K V
Sbjct 994 GEKWQAISKAV 1004
> cel:Y59A8B.6 hypothetical protein; K12855 pre-mRNA-processing
factor 6
Length=968
Score = 73.2 bits (178), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 34/74 (45%), Positives = 46/74 (62%), Gaps = 0/74 (0%)
Query 1 RLFWKDQKIAKARKWMNRSVTLDPSLGDAWASFLAFEIEHGGEKECIDVINRCALAQPNR 60
RLFW ++KI KAR+W R+V LDP GDA+A+FLAFE HG E++ V +C ++P
Sbjct 876 RLFWSERKIKKAREWFVRAVNLDPDNGDAFANFLAFEQIHGKEEDRKSVFKKCVTSEPRY 935
Query 61 GLAWNKTTKQVRCW 74
G W +K W
Sbjct 936 GDLWQSVSKDPINW 949
> cpv:cgd5_920 Pre-mRNA splicing factor Pro1/Prp6. HAT repeat
protein ; K12855 pre-mRNA-processing factor 6
Length=923
Score = 60.8 bits (146), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 28/88 (31%), Positives = 44/88 (50%), Gaps = 0/88 (0%)
Query 3 FWKDQKIAKARKWMNRSVTLDPSLGDAWASFLAFEIEHGGEKECIDVINRCALAQPNRGL 62
FWK+ K+RKW ++ +D +GD W ++AFE+ +G + D +N A PN+G
Sbjct 817 FWKENDFHKSRKWFKSALEIDNKIGDTWIHYIAFELLNGDFQSQRDALNDFINATPNKGF 876
Query 63 AWNKTTKQVRCWSFSPAQKLNAVLEYMY 90
WN + W + L LE +Y
Sbjct 877 EWNNIRRTHFFWDQKSNEILIICLELIY 904
> cel:T20B12.1 hypothetical protein
Length=771
Score = 33.5 bits (75), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 33/66 (50%), Gaps = 4/66 (6%)
Query 2 LFWKDQKIAKARKWMNRSVTLDPSLGDAW--ASFLAFEIEHGGEKECIDVINRCALAQPN 59
L D+K +A K + RS+ L P W A + A+++E+ KE +RC QP+
Sbjct 460 LLLMDKKFEEAYKHLRRSLELQPIQLGTWFNAGYCAWKLENF--KESTQCYHRCVSLQPD 517
Query 60 RGLAWN 65
AWN
Sbjct 518 HFEAWN 523
> dre:794079 pdcd11, MGC162501, cb680, im:7148359, sb:cb680, wu:fc68c10;
programmed cell death 11; K14792 rRNA biogenesis
protein RRP5
Length=1816
Score = 32.7 bits (73), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 14/53 (26%), Positives = 28/53 (52%), Gaps = 0/53 (0%)
Query 1 RLFWKDQKIAKARKWMNRSVTLDPSLGDAWASFLAFEIEHGGEKECIDVINRC 53
RL ++ KA+ ++ +T P D W+ F+ ++HG +KE ++ +R
Sbjct 1714 RLEFQFGNSEKAKSMFDKVLTTYPKRTDLWSVFIDLMVKHGSQKEVRELFDRV 1766
> sce:YMR018W Putative protein of unknown function with similarity
to human PEX5Rp (peroxin protein 5 related protein); transcription
increases during colony development similar to genes
involved in peroxisome biogenesis; YMR018W is not an essential
gene
Length=514
Score = 30.4 bits (67), Expect = 1.6, Method: Composition-based stats.
Identities = 15/58 (25%), Positives = 27/58 (46%), Gaps = 0/58 (0%)
Query 2 LFWKDQKIAKARKWMNRSVTLDPSLGDAWASFLAFEIEHGGEKECIDVINRCALAQPN 59
L++ DQKI +++K + + P+ G W + A I+ N+C +PN
Sbjct 369 LYYSDQKIKQSQKCLEFLLLEKPNNGTIWNRYGAILANTKSYHSAINAYNKCKQLRPN 426
> tgo:TGME49_073500 signal transduction protein, putative
Length=978
Score = 30.0 bits (66), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 13/64 (20%), Positives = 32/64 (50%), Gaps = 0/64 (0%)
Query 2 LFWKDQKIAKARKWMNRSVTLDPSLGDAWASFLAFEIEHGGEKECIDVINRCALAQPNRG 61
L+ KI +A + R++ ++P+ +A+ + + G ++ + ++C L PN
Sbjct 467 LYTCTGKIGEALHFAKRAIEVNPNYAEAYNNLGVLYRDQGDIEDSVKAYDKCLLLDPNSP 526
Query 62 LAWN 65
A++
Sbjct 527 NAFH 530
> hsa:22984 PDCD11, ALG-4, ALG4, KIAA0185, NFBP, RRP5; programmed
cell death 11; K14792 rRNA biogenesis protein RRP5
Length=1871
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 10/43 (23%), Positives = 22/43 (51%), Gaps = 0/43 (0%)
Query 11 KARKWMNRSVTLDPSLGDAWASFLAFEIEHGGEKECIDVINRC 53
+A+ +++ P D W+ ++ I+HG +K+ D+ R
Sbjct 1779 RAKAIFENTLSTYPKRTDVWSVYIDMTIKHGSQKDVRDIFERV 1821
> sce:YKL022C CDC16; Cdc16p; K03353 anaphase-promoting complex
subunit 6
Length=840
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 11/26 (42%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 8 KIAKARKWMNRSVTLDPSLGDAWASF 33
+I++A+K+ ++S LDPS AW F
Sbjct 546 RISEAQKYYSKSSILDPSFAAAWLGF 571
> sce:YBR055C PRP6, RNA6, TSM7269; Prp6p; K12855 pre-mRNA-processing
factor 6
Length=899
Score = 29.3 bits (64), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 18/80 (22%), Positives = 33/80 (41%), Gaps = 2/80 (2%)
Query 3 FWKDQKIAKARKWMNRSVTLDPSLGDAWASFLAFEIEHGGEKECIDVINRCALAQPNRGL 62
F+ + + + KW+ R++ GD W G K+ +D+ N +P G
Sbjct 815 FYAEAQYETSLKWLERALKKCSRYGDTWVWLFRTYARLG--KDTVDLYNMFDQCEPTYGP 872
Query 63 AWNKTTKQVRCWSFSPAQKL 82
W +K V+ +P + L
Sbjct 873 EWIAASKNVKMQYCTPREIL 892
> xla:779090 pdcd11, alg-4, alg4, nfbp, rrp5; programmed cell
death 11; K14792 rRNA biogenesis protein RRP5
Length=1812
Score = 29.3 bits (64), Expect = 3.6, Method: Composition-based stats.
Identities = 10/42 (23%), Positives = 22/42 (52%), Gaps = 0/42 (0%)
Query 11 KARKWMNRSVTLDPSLGDAWASFLAFEIEHGGEKECIDVINR 52
+A+ +++ P D W+ ++ ++HG +KE D+ R
Sbjct 1720 RAKALFESTLSSYPKRTDLWSVYIDMMVKHGSQKEVRDIFER 1761
> cel:T25B9.5 hypothetical protein
Length=541
Score = 28.5 bits (62), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 26/51 (50%), Gaps = 4/51 (7%)
Query 25 SLGDAWASFLAFE----IEHGGEKECIDVINRCALAQPNRGLAWNKTTKQV 71
SL DA A L E +E+G KE + +I+ C PN L +N +Q+
Sbjct 381 SLKDAKAWILTEERPHKMENGDPKELMVLIDACCERNPNERLNFNTVKRQI 431
> xla:734636 prpf39.1, MGC115228, prpf39; PRP39 pre-mRNA processing
factor 39 homolog, gene 1; K13217 pre-mRNA-processing
factor 39
Length=641
Score = 28.5 bits (62), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 9/27 (33%), Positives = 16/27 (59%), Gaps = 0/27 (0%)
Query 30 WASFLAFEIEHGGEKECIDVINRCALA 56
W +L FE+E+G + + + RC +A
Sbjct 327 WKEYLEFELENGSNERIVILFERCVIA 353
> bbo:BBOV_IV006090 23.m05922; tetratricopeptide repeat containing
domain protein
Length=976
Score = 28.5 bits (62), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 15/49 (30%), Positives = 25/49 (51%), Gaps = 3/49 (6%)
Query 3 FWKDQKIAKARKWMNRSVTLDPSLGDAWASFLAFEIEHGGEKE---CID 48
+ K + +A R V+++PS DAWA+ + + G KE CI+
Sbjct 713 YLKKGSLERAISVFARVVSMNPSCHDAWANMCSAHLNIGNMKEATICIE 761
> hsa:7249 TSC2, FLJ43106, LAM, TSC4; tuberous sclerosis 2; K07207
tuberous sclerosis 2
Length=1807
Score = 28.1 bits (61), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 19/72 (26%), Positives = 31/72 (43%), Gaps = 8/72 (11%)
Query 40 HGGEKECIDVINRCALAQPNRGLAWNKTTKQVRCWSFSPA-----QKLNAVLEYMYPNAL 94
HG ++ +++ RCA +P L R S PA Q L A++E + +
Sbjct 401 HGSQERYFELVERCADQRPESSLL---NLISYRAQSIHPAKDGWIQNLQALMERFFRSES 457
Query 95 KDGISQVVLDAL 106
+ + VLD L
Sbjct 458 RGAVRIKVLDVL 469
> hsa:51555 PEX5L, PEX5R, PXR2, PXR2B; peroxisomal biogenesis
factor 5-like
Length=626
Score = 27.7 bits (60), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 13/45 (28%), Positives = 21/45 (46%), Gaps = 0/45 (0%)
Query 15 WMNRSVTLDPSLGDAWASFLAFEIEHGGEKECIDVINRCALAQPN 59
+M ++ DP +AW + E+ E+ I + RC QPN
Sbjct 348 FMEAAILQDPGDAEAWQFLGITQAENENEQAAIVALQRCLELQPN 392
> mmu:58869 Pex5l, 1700016J08Rik, PXR2, Pex2, TRIP8b; peroxisomal
biogenesis factor 5-like
Length=567
Score = 27.7 bits (60), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 13/45 (28%), Positives = 21/45 (46%), Gaps = 0/45 (0%)
Query 15 WMNRSVTLDPSLGDAWASFLAFEIEHGGEKECIDVINRCALAQPN 59
+M ++ DP +AW + E+ E+ I + RC QPN
Sbjct 289 FMEAAILQDPGDAEAWQFLGITQAENENEQAAIVALQRCLELQPN 333
Lambda K H
0.319 0.132 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2032807080
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40