bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
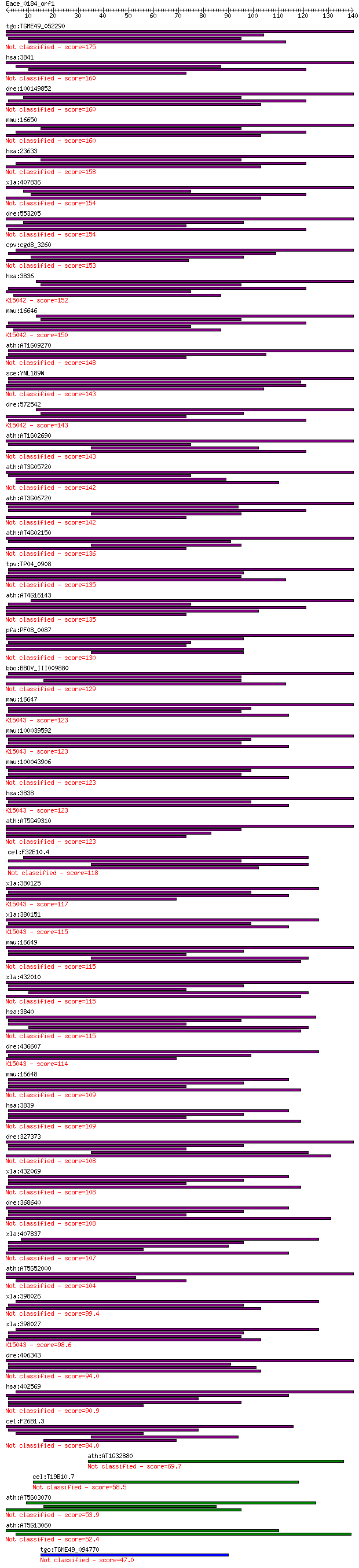
Query= Eace_0184_orf1
Length=139
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_052290 importin alpha, putative 175 3e-44
hsa:3841 KPNA5, IPOA6, SRP6; karyopherin alpha 5 (importin alp... 160 9e-40
dre:100149852 karyopherin alpha 6-like 160 1e-39
mmu:16650 Kpna6, IPOA7, Kpna5, NPI-2; karyopherin (importin) a... 160 2e-39
hsa:23633 KPNA6, FLJ11249, IPOA7, KPNA7, MGC17918; karyopherin... 158 5e-39
xla:407836 kpna6; karyopherin alpha 6 (importin alpha 7) 154 6e-38
dre:553205 kpna5, im:7151662, zgc:110662; karyopherin alpha 5 ... 154 7e-38
cpv:cgd8_3260 importin alpha subunit 153 2e-37
hsa:3836 KPNA1, IPOA5, NPI-1, RCH2, SRP1; karyopherin alpha 1 ... 152 4e-37
mmu:16646 Kpna1, AW494490, IPOA5, NPI1, Rch2, mSRP1; karyopher... 150 1e-36
ath:AT1G09270 IMPA-4; IMPA-4 (IMPORTIN ALPHA ISOFORM 4); bindi... 148 6e-36
sce:YNL189W SRP1, KAP60, SCM1; Karyopherin alpha homolog, form... 143 1e-34
dre:572542 karyopherin alpha 6-like; K15042 importin subunit a... 143 1e-34
ath:AT1G02690 IMPA-6; IMPA-6 (IMPORTIN ALPHA ISOFORM 6); bindi... 143 2e-34
ath:AT3G05720 IMPA-7; IMPA-7 (IMPORTIN ALPHA ISOFORM 7); bindi... 142 4e-34
ath:AT3G06720 IMPA-1; IMPA-1 (IMPORTIN ALPHA ISOFORM 1); bindi... 142 4e-34
ath:AT4G02150 MOS6; MOS6 (MODIFIER OF SNC1, 6); binding / prot... 136 2e-32
tpv:TP04_0908 importin alpha 135 4e-32
ath:AT4G16143 IMPA-2; IMPA-2 (IMPORTIN ALPHA ISOFORM 2); bindi... 135 4e-32
pfa:PF08_0087 importin alpha, putative 130 9e-31
bbo:BBOV_III009880 17.m07856; armadillo/beta-catenin-like repe... 129 2e-30
mmu:16647 Kpna2, 2410044B12Rik, IPOA1, MGC91246, Rch1; karyoph... 123 2e-28
mmu:100039592 Gm10259; predicted pseudogene 10259; K15043 impo... 123 2e-28
mmu:100043906 Gm10184; predicted pseudogene 10184 123 2e-28
hsa:3838 KPNA2, IPOA1, QIP2, RCH1, SRP1alpha; karyopherin alph... 123 2e-28
ath:AT5G49310 IMPA-5; IMPA-5 (IMPORTIN ALPHA ISOFORM 5); bindi... 123 2e-28
cel:F32E10.4 ima-3; IMportin Alpha family member (ima-3) 118 7e-27
xla:380125 kpna2, MGC53789, ipoa1, pen, qip2, rch1, srp1alpha;... 117 1e-26
xla:380151 kpna2, MGC52989; karyopherin alpha 2 (RAG cohort 1,... 115 3e-26
mmu:16649 Kpna4, 1110058D08Rik, IPOA3; karyopherin (importin) ... 115 5e-26
xla:432010 kpna4, MGC78839, ipoa3, qip1, srp3; karyopherin alp... 115 5e-26
hsa:3840 KPNA4, IPOA3, MGC12217, MGC26703, QIP1, SRP3; karyoph... 115 6e-26
dre:436607 kpna2, MGC113902, hm:zeh0389, hm:zeh0389r, zgc:1139... 114 7e-26
mmu:16648 Kpna3, IPOA4; karyopherin (importin) alpha 3 109 3e-24
hsa:3839 KPNA3, IPOA4, SRP1, SRP1gamma, SRP4, hSRP1; karyopher... 109 3e-24
dre:327373 kpna4, impa3, wu:fa56d07, wu:fa66g10, wu:fe14c04, w... 108 4e-24
xla:432069 kpna3, MGC78841, ipoa4, srp1, srp1gamma, srp4; kary... 108 5e-24
dre:368640 kpna3, si:dz79f24.2; karyopherin (importin) alpha 3 108 6e-24
xla:407837 imp_alpha_4; importin alpha 4 protein 107 9e-24
ath:AT5G52000 IMPA-8; IMPA-8 (IMPORTIN ALPHA ISOFORM 8); bindi... 104 7e-23
xla:398026 kpna7, kpna1; karyopherin alpha 7 (importin alpha 8) 99.4 3e-21
xla:398027 kpna2, MGC154872, MGC86340; importin alpha 1b; K150... 98.6 5e-21
dre:406343 fi21b05, wu:fi21b05, wu:fj93d06; zgc:55877 94.0 1e-19
hsa:402569 KPNA7; karyopherin alpha 7 (importin alpha 8) 90.9 1e-18
cel:F26B1.3 ima-2; IMportin Alpha family member (ima-2) 84.0 1e-16
ath:AT1G32880 importin alpha-1 subunit, putative 69.7 3e-12
cel:T19B10.7 ima-1; IMportin Alpha family member (ima-1) 58.5 5e-09
ath:AT5G03070 IMPA-9; IMPA-9 (IMPORTIN ALPHA ISOFORM 9); binding 53.9 1e-07
ath:AT5G13060 ABAP1; ABAP1 (ARMADILLO BTB ARABIDOPSIS PROTEIN ... 52.4 5e-07
tgo:TGME49_094770 hypothetical protein 47.0 2e-05
> tgo:TGME49_052290 importin alpha, putative
Length=545
Score = 175 bits (444), Expect = 3e-44, Method: Compositional matrix adjust.
Identities = 87/139 (62%), Positives = 106/139 (76%), Gaps = 0/139 (0%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
A+P LL LLSS +K ++KEACWTISNITAGN++QIQ+V+++G + L+ LL TADFD +K
Sbjct 340 AVPALLMLLSSPKKAIRKEACWTISNITAGNRDQIQQVIDAGLIHPLIELLSTADFDVRK 399
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
EAAWA+SNA SGGS+ QV LV GC PLC LL DSKIV V+LEAL ILR GK K
Sbjct 400 EAAWAISNAASGGSNAQVEALVECGCIKPLCSLLAVQDSKIVSVALEALENILRVGKVKK 459
Query 121 ETEGLSINPYATLIEQADG 139
E + L+ NP+ LIEQADG
Sbjct 460 EQQQLAENPFCELIEQADG 478
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 38/105 (36%), Positives = 57/105 (54%), Gaps = 4/105 (3%)
Query 1 ALPNLLSLLS-SNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTK 59
A+P + L S++ +Q EA W ++NI +G QEQ Q V+E GAVP + LL + D +
Sbjct 128 AVPLFVQFLKRSDQPRMQFEAAWALTNIASGTQEQTQVVIEHGAVPIFVELLSSPTEDVR 187
Query 60 KEAAWAVSNAVSGGSDMQVAELV-RLGCAPPLCQLLGCMDSKIVM 103
++A WA+ N G Q +LV + G PL L ++K M
Sbjct 188 EQAVWALGNI--AGDSPQCRDLVLQAGVLSPLLAQLNDSEAKFTM 230
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 30/95 (31%), Positives = 53/95 (55%), Gaps = 3/95 (3%)
Query 2 LPNLLSLLSSNRKTVQKEACWTISN-ITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
LP ++++LSS + EA ++ ++ IQEV+E+GAVP + L+ +D +
Sbjct 86 LPQMMAMLSSGDPQQEFEATEQFRRALSIESRPPIQEVIEAGAVPLFVQFLKRSDQPRMQ 145
Query 61 -EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLL 94
EAAWA++N ++ G+ Q ++ G P +LL
Sbjct 146 FEAAWALTN-IASGTQEQTQVVIEHGAVPIFVELL 179
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 29/103 (28%), Positives = 53/103 (51%), Gaps = 5/103 (4%)
Query 10 SSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAAWAVSNA 69
S + T+Q+ A WT+SN+ G + E ++ A+ L L+ + D + +A WA+S
Sbjct 224 SEAKFTMQRNATWTLSNLCRGKPQPPFEWVQP-ALTTLAKLIYSTDTEVLTDACWALS-Y 281
Query 70 VSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAI 112
+S G + ++ ++ G + L +LLG K +V AL +
Sbjct 282 ISDGPNERIEAVIEAGVSRRLVELLG---HKSTLVQTPALRTV 321
> hsa:3841 KPNA5, IPOA6, SRP6; karyopherin alpha 5 (importin alpha
6)
Length=539
Score = 160 bits (406), Expect = 9e-40, Method: Compositional matrix adjust.
Identities = 77/139 (55%), Positives = 102/139 (73%), Gaps = 0/139 (0%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
ALP LL LLSS +++++KEACWT+SNITAGN+ QIQ V+++ P L+ +L+ A+F T+K
Sbjct 340 ALPCLLHLLSSPKESIRKEACWTVSNITAGNRAQIQAVIDANIFPVLIEILQKAEFRTRK 399
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
EAAWA++NA SGG+ Q+ LV LGC PLC LL MDSKIV V+L L ILR G++
Sbjct 400 EAAWAITNATSGGTPEQIRYLVALGCIKPLCDLLTVMDSKIVQVALNGLENILRLGEQES 459
Query 121 ETEGLSINPYATLIEQADG 139
+ G+ INPY LIE+A G
Sbjct 460 KQNGIGINPYCALIEEAYG 478
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 27/83 (32%), Positives = 44/83 (53%), Gaps = 4/83 (4%)
Query 5 LLSLLSSNRK-TVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAA 63
+ L N T+Q EA W ++NI +G + V+E+GAVP + LL + D +++A
Sbjct 132 FVKFLERNENCTLQFEAAWALTNIASGTFLHTKVVIETGAVPIFIKLLNSEHEDVQEQAV 191
Query 64 WAVSNAVSGGSDMQVAELVRLGC 86
WA+ N G + + + V L C
Sbjct 192 WALGNI--AGDNAECRDFV-LNC 211
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 33/111 (29%), Positives = 54/111 (48%), Gaps = 2/111 (1%)
Query 10 SSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAAWAVSNA 69
+SNR T + A W +SN+ G S + L LL ++D D + WA+S
Sbjct 223 NSNRLTTTRNAVWALSNLCRGKNPPPNFSKVSPCLNVLSRLLFSSDPDVLADVCWALS-Y 281
Query 70 VSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
+S G + ++ ++ G L +LL D K+V +L A+ I+ TG I+
Sbjct 282 LSDGPNDKIQAVIDSGVCRRLVELLMHNDYKVVSPALRAVGNIV-TGDDIQ 331
Score = 35.4 bits (80), Expect = 0.049, Method: Compositional matrix adjust.
Identities = 27/73 (36%), Positives = 39/73 (53%), Gaps = 1/73 (1%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETAD-FDTK 59
A+P + LL+S + VQ++A W + NI N E VL +P LL LL ++ T
Sbjct 171 AVPIFIKLLNSEHEDVQEQAVWALGNIAGDNAECRDFVLNCEILPPLLELLTNSNRLTTT 230
Query 60 KEAAWAVSNAVSG 72
+ A WA+SN G
Sbjct 231 RNAVWALSNLCRG 243
> dre:100149852 karyopherin alpha 6-like
Length=537
Score = 160 bits (405), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 77/139 (55%), Positives = 103/139 (74%), Gaps = 0/139 (0%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
ALP LL LLSS +++++KEACWTISNITAGN+ QIQ V+++ P L+ +L+ A+F T+K
Sbjct 338 ALPCLLHLLSSAKESIRKEACWTISNITAGNRAQIQAVIDANIFPVLIEILQKAEFRTRK 397
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
EAAWA++NA SGG+ Q+ LV LGC PLC LL MDSKIV+V+L L ILR G++
Sbjct 398 EAAWAITNATSGGTPEQIRYLVNLGCIKPLCDLLTVMDSKIVLVALNGLENILRLGEQEA 457
Query 121 ETEGLSINPYATLIEQADG 139
+ G +NPY +LIE+A G
Sbjct 458 KQNGSGLNPYCSLIEEAYG 476
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 31/87 (35%), Positives = 50/87 (57%), Gaps = 1/87 (1%)
Query 8 LLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAAWAVS 67
L S T+Q EA W ++NI +G +Q + V+E+GAVP + LL + D +++A WA+
Sbjct 134 LQKSTNCTLQFEAAWALTNIASGTSQQTKFVIEAGAVPVFIELLNSDFEDVQEQAVWALG 193
Query 68 NAVSGGSDMQVAELVRLGCAPPLCQLL 94
N ++G S + ++ PPL LL
Sbjct 194 N-IAGDSAVCRDYVLNCNILPPLLVLL 219
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 40/120 (33%), Positives = 61/120 (50%), Gaps = 3/120 (2%)
Query 2 LPNLLSLLS-SNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
LP LL LL+ S R T+ + A W +SN+ G S +P L LL ++D D
Sbjct 212 LPPLLVLLTKSTRLTMTRNAVWALSNLCRGKSPPPDFEKVSPCLPVLSRLLFSSDSDLLA 271
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
+A WA+S +S G + ++ ++ G L +LL D K+ SL A+ I+ TG I+
Sbjct 272 DACWALS-YLSDGPNEKIQAVIDSGVCRRLVELLMHSDYKVASPSLRAVGNIV-TGDDIQ 329
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 34/103 (33%), Positives = 51/103 (49%), Gaps = 2/103 (1%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLL-ETADFDTK 59
A+P + LL+S+ + VQ++A W + NI + VL +P LL LL ++
Sbjct 169 AVPVFIELLNSDFEDVQEQAVWALGNIAGDSAVCRDYVLNCNILPPLLVLLTKSTRLTMT 228
Query 60 KEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIV 102
+ A WA+SN G S E V C P L +LL DS ++
Sbjct 229 RNAVWALSNLCRGKSPPPDFEKVS-PCLPVLSRLLFSSDSDLL 270
> mmu:16650 Kpna6, IPOA7, Kpna5, NPI-2; karyopherin (importin)
alpha 6
Length=536
Score = 160 bits (404), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 77/139 (55%), Positives = 102/139 (73%), Gaps = 0/139 (0%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
ALP LL LLSS++++++KEACWTISNITAGN+ QIQ V+++ P L+ +L+ A+F T+K
Sbjct 337 ALPCLLHLLSSSKESIRKEACWTISNITAGNRAQIQAVIDANIFPVLIEILQKAEFRTRK 396
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
EAAWA++NA SGG+ Q+ LV LGC PLC LL MDSKIV V+L L ILR G++
Sbjct 397 EAAWAITNATSGGTPEQIRYLVSLGCIKPLCDLLTVMDSKIVQVALNGLENILRLGEQES 456
Query 121 ETEGLSINPYATLIEQADG 139
+ G +NPY LIE+A G
Sbjct 457 KRSGSGVNPYCGLIEEAYG 475
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 28/80 (35%), Positives = 47/80 (58%), Gaps = 1/80 (1%)
Query 15 TVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAAWAVSNAVSGGS 74
T+Q EA W ++NI +G +Q + V+E+GAVP + LL + D +++A WA+ N ++G S
Sbjct 140 TLQFEAAWALTNIASGTSQQTKIVIEAGAVPIFIELLNSDFEDVQEQAVWALGN-IAGDS 198
Query 75 DMQVAELVRLGCAPPLCQLL 94
+ ++ PL LL
Sbjct 199 SLCRDYVLNCSILNPLLTLL 218
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 37/117 (31%), Positives = 61/117 (52%), Gaps = 3/117 (2%)
Query 5 LLSLLS-SNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAA 63
LL+LL+ S R T+ + A W +SN+ G + S +P L LL ++D D +A
Sbjct 214 LLTLLTKSTRLTMTRNAVWALSNLCRGKNPPPEFAKVSPCLPVLSRLLFSSDSDLLADAC 273
Query 64 WAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
WA+S +S G + ++ ++ G L +LL D K+ +L A+ I+ TG I+
Sbjct 274 WALS-YLSDGPNEKIQAVIDSGVCRRLVELLMHNDYKVASPALRAVGNIV-TGDDIQ 328
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 30/105 (28%), Positives = 55/105 (52%), Gaps = 6/105 (5%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAV--PALLHLLETADFDT 58
A+P + LL+S+ + VQ++A W + NI AG+ ++ + + ++ P L L ++
Sbjct 168 AVPIFIELLNSDFEDVQEQAVWALGNI-AGDSSLCRDYVLNCSILNPLLTLLTKSTRLTM 226
Query 59 KKEAAWAVSNAVSGGSDMQVAELVRLG-CAPPLCQLLGCMDSKIV 102
+ A WA+SN G + E ++ C P L +LL DS ++
Sbjct 227 TRNAVWALSNLCRGKN--PPPEFAKVSPCLPVLSRLLFSSDSDLL 269
> hsa:23633 KPNA6, FLJ11249, IPOA7, KPNA7, MGC17918; karyopherin
alpha 6 (importin alpha 7)
Length=536
Score = 158 bits (400), Expect = 5e-39, Method: Compositional matrix adjust.
Identities = 77/139 (55%), Positives = 101/139 (72%), Gaps = 0/139 (0%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
ALP LL LLSS +++++KEACWTISNITAGN+ QIQ V+++ P L+ +L+ A+F T+K
Sbjct 337 ALPCLLHLLSSPKESIRKEACWTISNITAGNRAQIQAVIDANIFPVLIEILQKAEFRTRK 396
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
EAAWA++NA SGG+ Q+ LV LGC PLC LL MDSKIV V+L L ILR G++
Sbjct 397 EAAWAITNATSGGTPEQIRYLVSLGCIKPLCDLLTVMDSKIVQVALNGLENILRLGEQEG 456
Query 121 ETEGLSINPYATLIEQADG 139
+ G +NPY LIE+A G
Sbjct 457 KRSGSGVNPYCGLIEEAYG 475
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 28/80 (35%), Positives = 47/80 (58%), Gaps = 1/80 (1%)
Query 15 TVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAAWAVSNAVSGGS 74
T+Q EA W ++NI +G +Q + V+E+GAVP + LL + D +++A WA+ N ++G S
Sbjct 140 TLQFEAAWALTNIASGTSQQTKIVIEAGAVPIFIELLNSDFEDVQEQAVWALGN-IAGDS 198
Query 75 DMQVAELVRLGCAPPLCQLL 94
+ ++ PL LL
Sbjct 199 SVCRDYVLNCSILNPLLTLL 218
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 37/117 (31%), Positives = 61/117 (52%), Gaps = 3/117 (2%)
Query 5 LLSLLS-SNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAA 63
LL+LL+ S R T+ + A W +SN+ G + S +P L LL ++D D +A
Sbjct 214 LLTLLTKSTRLTMTRNAVWALSNLCRGKNPPPEFAKVSPCLPVLSRLLFSSDSDLLADAC 273
Query 64 WAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
WA+S +S G + ++ ++ G L +LL D K+ +L A+ I+ TG I+
Sbjct 274 WALS-YLSDGPNEKIQAVIDSGVCRRLVELLMHNDYKVASPALRAVGNIV-TGDDIQ 328
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 31/104 (29%), Positives = 51/104 (49%), Gaps = 4/104 (3%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLL-ETADFDTK 59
A+P + LL+S+ + VQ++A W + NI + VL + LL LL ++
Sbjct 168 AVPIFIELLNSDFEDVQEQAVWALGNIAGDSSVCRDYVLNCSILNPLLTLLTKSTRLTMT 227
Query 60 KEAAWAVSNAVSGGSDMQVAELVRLG-CAPPLCQLLGCMDSKIV 102
+ A WA+SN G + E ++ C P L +LL DS ++
Sbjct 228 RNAVWALSNLCRGKN--PPPEFAKVSPCLPVLSRLLFSSDSDLL 269
> xla:407836 kpna6; karyopherin alpha 6 (importin alpha 7)
Length=537
Score = 154 bits (390), Expect = 6e-38, Method: Compositional matrix adjust.
Identities = 77/139 (55%), Positives = 100/139 (71%), Gaps = 0/139 (0%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
ALP LL LLSS +++++KEACWTISNITAGN+ QIQ V ++ P L+ +L+ A+F T+K
Sbjct 338 ALPCLLHLLSSPKESIRKEACWTISNITAGNRGQIQVVADANIFPVLIEILQKAEFRTRK 397
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
EAAWA++NA SGG+ Q+ LV +GC PLC LL MDSKIV V+L L ILR G++
Sbjct 398 EAAWAITNATSGGTPEQIRYLVNIGCIKPLCDLLTVMDSKIVQVALNGLENILRLGEQEA 457
Query 121 ETEGLSINPYATLIEQADG 139
+ G INPY LIE+A G
Sbjct 458 KHGGNGINPYCGLIEEAYG 476
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 26/67 (38%), Positives = 43/67 (64%), Gaps = 1/67 (1%)
Query 8 LLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAAWAVS 67
L S+ T+Q EA W ++NI +G +Q + V+E+GAVP + LL + D +++A WA+
Sbjct 134 LKKSDNYTLQFEAAWALTNIASGTSQQTKIVIEAGAVPIFIQLLNSDYEDVQEQAVWALG 193
Query 68 NAVSGGS 74
N ++G S
Sbjct 194 N-IAGDS 199
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 33/110 (30%), Positives = 55/110 (50%), Gaps = 2/110 (1%)
Query 11 SNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAAWAVSNAV 70
S R T+ + A W +SN+ G + S +P L LL ++D D +A WA+S +
Sbjct 222 STRLTMTRNAVWALSNLCRGKNPPPEFDKVSQCLPVLSRLLFSSDSDLLADACWALS-YL 280
Query 71 SGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
S G + ++ ++ G L +LL D K+ +L A+ I+ TG I+
Sbjct 281 SDGPNEKIQAVIDSGVCRRLVELLMHNDYKVASPALRAVGNIV-TGDDIQ 329
Score = 35.8 bits (81), Expect = 0.047, Method: Compositional matrix adjust.
Identities = 31/104 (29%), Positives = 51/104 (49%), Gaps = 4/104 (3%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETAD--FDT 58
A+P + LL+S+ + VQ++A W + NI AG+ ++ + S + L L T
Sbjct 169 AVPIFIQLLNSDYEDVQEQAVWALGNI-AGDSSVCRDYVLSCDILLPLLNLLTKSTRLTM 227
Query 59 KKEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIV 102
+ A WA+SN G + + V C P L +LL DS ++
Sbjct 228 TRNAVWALSNLCRGKNPPPEFDKVS-QCLPVLSRLLFSSDSDLL 270
> dre:553205 kpna5, im:7151662, zgc:110662; karyopherin alpha
5 (importin alpha 6)
Length=539
Score = 154 bits (389), Expect = 7e-38, Method: Compositional matrix adjust.
Identities = 75/139 (53%), Positives = 99/139 (71%), Gaps = 0/139 (0%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
ALP LL LLSS +++++KEACWT+SNITAGN+ QIQ V++S P L+ +L+ A+F T+K
Sbjct 340 ALPCLLHLLSSPKESIKKEACWTVSNITAGNRAQIQAVIDSNVFPVLIEILQKAEFRTRK 399
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
EAAWA++NA SGG+ Q+ LV L P+C LL MDSKIV V+L L ILR G++
Sbjct 400 EAAWAITNATSGGTPEQIRYLVSLNTIKPMCDLLTVMDSKIVQVALNGLENILRLGEQES 459
Query 121 ETEGLSINPYATLIEQADG 139
+ G INPY LIE+A G
Sbjct 460 KQNGTGINPYCALIEEAYG 478
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 30/88 (34%), Positives = 50/88 (56%), Gaps = 1/88 (1%)
Query 8 LLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAAWAVS 67
L S+ T+Q EA W ++NI +G + + V+E+GAVP + LL + D +++A WA+
Sbjct 136 LRRSDNCTLQFEAAWALTNIASGTFQHTKVVIETGAVPIFIELLNSEYEDVQEQAVWALG 195
Query 68 NAVSGGSDMQVAELVRLGCAPPLCQLLG 95
N ++G + ++ G P L QLL
Sbjct 196 N-IAGDNAECRDYVLNCGILPSLQQLLA 222
Score = 49.3 bits (116), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 27/73 (36%), Positives = 40/73 (54%), Gaps = 1/73 (1%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETAD-FDTK 59
A+P + LL+S + VQ++A W + NI N E VL G +P+L LL ++ T
Sbjct 171 AVPIFIELLNSEYEDVQEQAVWALGNIAGDNAECRDYVLNCGILPSLQQLLAKSNRLTTT 230
Query 60 KEAAWAVSNAVSG 72
+ A WA+SN G
Sbjct 231 RNAVWALSNLCRG 243
Score = 48.5 bits (114), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 39/120 (32%), Positives = 61/120 (50%), Gaps = 3/120 (2%)
Query 2 LPNLLSLLS-SNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
LP+L LL+ SNR T + A W +SN+ G S + L LL ++D D
Sbjct 214 LPSLQQLLAKSNRLTTTRNAVWALSNLCRGKNPPPDFAKVSPCLSVLSRLLFSSDPDVLA 273
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
+A WA+S +S G + ++ ++ G L +LL D K+V +L A+ I+ TG I+
Sbjct 274 DACWALS-YLSDGPNDKIQTVIDSGVCRRLVELLMHSDYKVVSPALRAVGNIV-TGDDIQ 331
> cpv:cgd8_3260 importin alpha subunit
Length=552
Score = 153 bits (386), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 74/135 (54%), Positives = 99/135 (73%), Gaps = 0/135 (0%)
Query 5 LLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAAW 64
LL LLSS +K ++KEACWT+SNITAGN+EQIQE++++G + L++LL TA+FD KKEAAW
Sbjct 356 LLQLLSSPKKVIRKEACWTVSNITAGNKEQIQEIIDNGLIVPLVNLLNTAEFDVKKEAAW 415
Query 65 AVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIKETEG 124
A+SNA +GG+ Q+ LV G PLC LL D+K++ V+LEA+ IL+TG + G
Sbjct 416 AISNATTGGTVEQIEYLVSQGVIKPLCDLLSIEDAKVINVALEAIENILKTGAIRQHERG 475
Query 125 LSINPYATLIEQADG 139
L NPY L+EQA G
Sbjct 476 LQENPYCALVEQAYG 490
Score = 62.8 bits (151), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 35/112 (31%), Positives = 65/112 (58%), Gaps = 6/112 (5%)
Query 2 LPNLLSLLSS-NRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
+P L+ LS + +Q EA WT++NI++G EQ EV+ G++P + LL + + K+
Sbjct 118 IPRLVHFLSDYDHPNLQFEAAWTLTNISSGTTEQTCEVVRHGSIPKCVELLNSPKLEVKE 177
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQL----LGCMDSKIVMVSLEA 108
+A W + N ++G S ++R G PP+ QL +G +D+ M+++++
Sbjct 178 QAIWTLGN-IAGDSANCRDLVLRAGALPPILQLIALEIGPLDNNEQMMNVKS 228
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 26/85 (30%), Positives = 47/85 (55%), Gaps = 2/85 (2%)
Query 11 SNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAAWAVSNAV 70
S + ++ + A WT++N+ G E++ S ++P L LL +D + +A WA+S +
Sbjct 237 SGKTSILRTATWTVNNLCRGRPPPPFEMV-SHSLPVLCRLLYFSDLEVMTDACWALS-YI 294
Query 71 SGGSDMQVAELVRLGCAPPLCQLLG 95
S G + +V ++R P L +LLG
Sbjct 295 SDGPNDRVEAVLRSEACPRLVELLG 319
Score = 38.1 bits (87), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 25/76 (32%), Positives = 43/76 (56%), Gaps = 4/76 (5%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQE-VLESGAVPALLHL--LETADFD 57
++P + LL+S + V+++A WT+ NI AG+ ++ VL +GA+P +L L LE D
Sbjct 160 SIPKCVELLNSPKLEVKEQAIWTLGNI-AGDSANCRDLVLRAGALPPILQLIALEIGPLD 218
Query 58 TKKEAAWAVSNAVSGG 73
++ S + GG
Sbjct 219 NNEQMMNVKSPSTVGG 234
> hsa:3836 KPNA1, IPOA5, NPI-1, RCH2, SRP1; karyopherin alpha
1 (importin alpha 5); K15042 importin subunit alpha-1
Length=538
Score = 152 bits (383), Expect = 4e-37, Method: Compositional matrix adjust.
Identities = 72/127 (56%), Positives = 94/127 (74%), Gaps = 0/127 (0%)
Query 13 RKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAAWAVSNAVSG 72
+++++KEACWTISNITAGN+ QIQ V+++ PAL+ +L+TA+F T+KEAAWA++NA SG
Sbjct 351 KESIKKEACWTISNITAGNRAQIQTVIDANIFPALISILQTAEFRTRKEAAWAITNATSG 410
Query 73 GSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIKETEGLSINPYAT 132
GS Q+ LV LGC PLC LL MDSKIV V+L L ILR G++ + G INPY
Sbjct 411 GSAEQIKYLVELGCIKPLCDLLTVMDSKIVQVALNGLENILRLGEQEAKRNGTGINPYCA 470
Query 133 LIEQADG 139
LIE+A G
Sbjct 471 LIEEAYG 477
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 29/80 (36%), Positives = 48/80 (60%), Gaps = 1/80 (1%)
Query 15 TVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAAWAVSNAVSGGS 74
T+Q E+ W ++NI +GN Q + V+++GAVP + LL + D +++A WA+ N ++G S
Sbjct 142 TLQFESAWVLTNIASGNSLQTRIVIQAGAVPIFIELLSSEFEDVQEQAVWALGN-IAGDS 200
Query 75 DMQVAELVRLGCAPPLCQLL 94
M ++ PPL QL
Sbjct 201 TMCRDYVLDCNILPPLLQLF 220
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 39/120 (32%), Positives = 60/120 (50%), Gaps = 3/120 (2%)
Query 2 LPNLLSLLSS-NRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
LP LL L S NR T+ + A W +SN+ G + S + L LL +D D
Sbjct 213 LPPLLQLFSKQNRLTMTRNAVWALSNLCRGKSPPPEFAKVSPCLNVLSWLLFVSDTDVLA 272
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
+A WA+S +S G + ++ ++ G L +LL D K+V +L A+ I+ TG I+
Sbjct 273 DACWALS-YLSDGPNDKIQAVIDAGVCRRLVELLMHNDYKVVSPALRAVGNIV-TGDDIQ 330
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 26/75 (34%), Positives = 38/75 (50%), Gaps = 1/75 (1%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDT-K 59
A+P + LLSS + VQ++A W + NI + VL+ +P LL L + T
Sbjct 170 AVPIFIELLSSEFEDVQEQAVWALGNIAGDSTMCRDYVLDCNILPPLLQLFSKQNRLTMT 229
Query 60 KEAAWAVSNAVSGGS 74
+ A WA+SN G S
Sbjct 230 RNAVWALSNLCRGKS 244
Score = 41.6 bits (96), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 30/84 (35%), Positives = 45/84 (53%), Gaps = 4/84 (4%)
Query 4 NLLS-LLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEA 62
N+LS LL + V +ACW +S ++ G ++IQ V+++G L+ LL D+ A
Sbjct 257 NVLSWLLFVSDTDVLADACWALSYLSDGPNDKIQAVIDAGVCRRLVELLMHNDYKVVSPA 316
Query 63 AWAVSNAVSGGSDMQVAELVRLGC 86
AV N V+ G D+Q V L C
Sbjct 317 LRAVGNIVT-GDDIQTQ--VILNC 337
> mmu:16646 Kpna1, AW494490, IPOA5, NPI1, Rch2, mSRP1; karyopherin
(importin) alpha 1; K15042 importin subunit alpha-1
Length=538
Score = 150 bits (379), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 71/127 (55%), Positives = 94/127 (74%), Gaps = 0/127 (0%)
Query 13 RKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAAWAVSNAVSG 72
+++++KEACWTISNITAGN+ QIQ V+++ PAL+ +L+TA+F T+KEAAWA++NA SG
Sbjct 351 KESIKKEACWTISNITAGNRAQIQTVIDANMFPALISILQTAEFRTRKEAAWAITNATSG 410
Query 73 GSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIKETEGLSINPYAT 132
GS Q+ LV LGC PLC LL MD+KIV V+L L ILR G++ + G INPY
Sbjct 411 GSAEQIKYLVELGCIKPLCDLLTVMDAKIVQVALNGLENILRLGEQEAKRNGSGINPYCA 470
Query 133 LIEQADG 139
LIE+A G
Sbjct 471 LIEEAYG 477
Score = 59.3 bits (142), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 29/80 (36%), Positives = 48/80 (60%), Gaps = 1/80 (1%)
Query 15 TVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAAWAVSNAVSGGS 74
T+Q E+ W ++NI +GN Q + V+++GAVP + LL + D +++A WA+ N ++G S
Sbjct 142 TLQFESAWVLTNIASGNSLQTRNVIQAGAVPIFIELLSSEFEDVQEQAVWALGN-IAGDS 200
Query 75 DMQVAELVRLGCAPPLCQLL 94
M ++ PPL QL
Sbjct 201 TMCRDYVLNCNILPPLLQLF 220
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 39/120 (32%), Positives = 60/120 (50%), Gaps = 3/120 (2%)
Query 2 LPNLLSLLSS-NRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
LP LL L S NR T+ + A W +SN+ G + S + L LL +D D
Sbjct 213 LPPLLQLFSKQNRLTMTRNAVWALSNLCRGKSPPPEFAKVSPCLNVLSWLLFVSDTDVLA 272
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
+A WA+S +S G + ++ ++ G L +LL D K+V +L A+ I+ TG I+
Sbjct 273 DACWALS-YLSDGPNDKIQAVIDAGVCRRLVELLMHNDYKVVSPALRAVGNIV-TGDDIQ 330
Score = 41.6 bits (96), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 26/75 (34%), Positives = 37/75 (49%), Gaps = 1/75 (1%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDT-K 59
A+P + LLSS + VQ++A W + NI + VL +P LL L + T
Sbjct 170 AVPIFIELLSSEFEDVQEQAVWALGNIAGDSTMCRDYVLNCNILPPLLQLFSKQNRLTMT 229
Query 60 KEAAWAVSNAVSGGS 74
+ A WA+SN G S
Sbjct 230 RNAVWALSNLCRGKS 244
Score = 41.6 bits (96), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 30/84 (35%), Positives = 45/84 (53%), Gaps = 4/84 (4%)
Query 4 NLLS-LLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEA 62
N+LS LL + V +ACW +S ++ G ++IQ V+++G L+ LL D+ A
Sbjct 257 NVLSWLLFVSDTDVLADACWALSYLSDGPNDKIQAVIDAGVCRRLVELLMHNDYKVVSPA 316
Query 63 AWAVSNAVSGGSDMQVAELVRLGC 86
AV N V+ G D+Q V L C
Sbjct 317 LRAVGNIVT-GDDIQTQ--VILNC 337
> ath:AT1G09270 IMPA-4; IMPA-4 (IMPORTIN ALPHA ISOFORM 4); binding
/ protein transporter
Length=456
Score = 148 bits (373), Expect = 6e-36, Method: Compositional matrix adjust.
Identities = 78/141 (55%), Positives = 103/141 (73%), Gaps = 4/141 (2%)
Query 2 LPNLLSLLSSN-RKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
LP+L +LL+ N +K+++KEACWTISNITAGN+ QI+ V+ +G + L+HLL+ A+FD KK
Sbjct 253 LPHLYNLLTQNHKKSIKKEACWTISNITAGNKLQIEAVVGAGIILPLVHLLQNAEFDIKK 312
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
EAAWA+SNA SGGS Q+ LV GC PLC LL C D +IV V LE L IL+ G+ K
Sbjct 313 EAAWAISNATSGGSHEQIQYLVTQGCIKPLCDLLICPDPRIVTVCLEGLENILKVGEADK 372
Query 121 ETEGLS--INPYATLIEQADG 139
E GL+ +N YA +IE++DG
Sbjct 373 EM-GLNSGVNLYAQIIEESDG 392
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 32/104 (30%), Positives = 55/104 (52%), Gaps = 3/104 (2%)
Query 2 LPNLLSLLS-SNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
+P + L + +Q EA W ++N+ +G + + V+E GAVP + LL +A D ++
Sbjct 42 IPRFVEFLGRHDHPQLQFEAAWALTNVASGTSDHTRVVIEQGAVPIFVKLLTSASDDVRE 101
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMV 104
+A WA+ N V+G S ++ G PL L +SK+ M+
Sbjct 102 QAVWALGN-VAGDSPNCRNLVLNYGALEPLLAQLN-ENSKLSML 143
Score = 37.0 bits (84), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 22/73 (30%), Positives = 35/73 (47%), Gaps = 1/73 (1%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAV-PALLHLLETADFDTK 59
A+P + LL+S V+++A W + N+ + VL GA+ P L L E +
Sbjct 84 AVPIFVKLLTSASDDVREQAVWALGNVAGDSPNCRNLVLNYGALEPLLAQLNENSKLSML 143
Query 60 KEAAWAVSNAVSG 72
+ A W +SN G
Sbjct 144 RNATWTLSNFCRG 156
> sce:YNL189W SRP1, KAP60, SCM1; Karyopherin alpha homolog, forms
a dimer with karyopherin beta Kap95p to mediate import of
nuclear proteins, binds the nuclear localization signal of
the substrate during import; may also play a role in regulation
of protein degradation
Length=542
Score = 143 bits (361), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 74/142 (52%), Positives = 99/142 (69%), Gaps = 6/142 (4%)
Query 2 LPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKE 61
LP L LLSS ++ ++KEACWTISNITAGN EQIQ V+++ +P L+ LLE A++ TKKE
Sbjct 343 LPALRLLLSSPKENIKKEACWTISNITAGNTEQIQAVIDANLIPPLVKLLEVAEYKTKKE 402
Query 62 AAWAVSNAVSGGSDMQVAELVRL----GCAPPLCQLLGCMDSKIVMVSLEALHAILRTGK 117
A WA+SNA SGG +Q +++R GC PLC LL D++I+ V+L+AL IL+ G+
Sbjct 403 ACWAISNASSGG--LQRPDIIRYLVSQGCIKPLCDLLEIADNRIIEVTLDALENILKMGE 460
Query 118 KIKETEGLSINPYATLIEQADG 139
KE GL+IN A IE+A G
Sbjct 461 ADKEARGLNINENADFIEKAGG 482
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 33/118 (27%), Positives = 60/118 (50%), Gaps = 3/118 (2%)
Query 2 LPNLLSLLSSNR-KTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
+P L+ + N+ + +Q EA W ++NI +G Q + V+++ AVP + LL T + K+
Sbjct 132 VPRLVEFMRENQPEMLQLEAAWALTNIASGTSAQTKVVVDADAVPLFIQLLYTGSVEVKE 191
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKK 118
+A WA+ N V+G S +++ P+ L ++ + L + R GKK
Sbjct 192 QAIWALGN-VAGDSTDYRDYVLQCNAMEPILGLFNSNKPSLIRTATWTLSNLCR-GKK 247
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 33/120 (27%), Positives = 63/120 (52%), Gaps = 2/120 (1%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
A+ +L L +SN+ ++ + A WT+SN+ G + Q + S A+P L L+ + D +T
Sbjct 216 AMEPILGLFNSNKPSLIRTATWTLSNLCRGKKPQPDWSVVSQALPTLAKLIYSMDTETLV 275
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
+A WA+S +S G + ++ + L +LL + + +L A+ I+ TG ++
Sbjct 276 DACWAIS-YLSDGPQEAIQAVIDVRIPKRLVELLSHESTLVQTPALRAVGNIV-TGNDLQ 333
Score = 35.8 bits (81), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 24/103 (23%), Positives = 47/103 (45%), Gaps = 1/103 (0%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
A+P + LL + V+++A W + N+ + + VL+ A+ +L L + +
Sbjct 174 AVPLFIQLLYTGSVEVKEQAIWALGNVAGDSTDYRDYVLQCNAMEPILGLFNSNKPSLIR 233
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVM 103
A W +SN G +V P L +L+ MD++ ++
Sbjct 234 TATWTLSNLCRGKKPQPDWSVVSQAL-PTLAKLIYSMDTETLV 275
> dre:572542 karyopherin alpha 6-like; K15042 importin subunit
alpha-1
Length=580
Score = 143 bits (361), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 70/127 (55%), Positives = 92/127 (72%), Gaps = 1/127 (0%)
Query 13 RKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAAWAVSNAVSG 72
+++++KEACWTISNITAGN+ QIQ V+++ P L+ +L+ A+F T+KEAAWA++NA SG
Sbjct 394 KESIKKEACWTISNITAGNRAQIQMVIDADLFPPLISILQVAEFRTRKEAAWAITNATSG 453
Query 73 GSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIKETEGLSINPYAT 132
GS Q+ LV LGC PLC LL MDSKIV V+L L ILR G +++ G INPY
Sbjct 454 GSAEQIRHLVELGCIKPLCDLLTLMDSKIVQVALNGLENILRLG-ELEAKRGGGINPYCA 512
Query 133 LIEQADG 139
LIE+A G
Sbjct 513 LIEEAYG 519
Score = 51.6 bits (122), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 27/81 (33%), Positives = 46/81 (56%), Gaps = 1/81 (1%)
Query 15 TVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAAWAVSNAVSGGS 74
T+Q EA W ++NI +G Q + V+++GAVP + +L + D +++A WA+ N ++G S
Sbjct 185 TLQFEAAWVLTNIASGTSHQTRVVIQAGAVPIFIEMLSSDFEDVQEQAVWALGN-IAGDS 243
Query 75 DMQVAELVRLGCAPPLCQLLG 95
++ P L QLL
Sbjct 244 TECRDYVLDCNILPSLVQLLA 264
Score = 44.7 bits (104), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 24/73 (32%), Positives = 40/73 (54%), Gaps = 1/73 (1%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETAD-FDTK 59
A+P + +LSS+ + VQ++A W + NI + E VL+ +P+L+ LL +
Sbjct 213 AVPIFIEMLSSDFEDVQEQAVWALGNIAGDSTECRDYVLDCNILPSLVQLLAKQNRLTMM 272
Query 60 KEAAWAVSNAVSG 72
+ A WA+SN G
Sbjct 273 RNAVWALSNLCRG 285
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 36/120 (30%), Positives = 60/120 (50%), Gaps = 3/120 (2%)
Query 2 LPNLLSLLSS-NRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
LP+L+ LL+ NR T+ + A W +SN+ G S + L LL D D
Sbjct 256 LPSLVQLLAKQNRLTMMRNAVWALSNLCRGKNPPPDFAKVSPCLSVLSWLLFVNDTDILA 315
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
+A WA+S +S G + ++ ++ G L +LL + K+V +L A+ I+ TG ++
Sbjct 316 DACWALS-YLSDGPNEKIQAVIDSGVCRRLVELLLHAEYKVVSPALRAVGNIV-TGDDLQ 373
> ath:AT1G02690 IMPA-6; IMPA-6 (IMPORTIN ALPHA ISOFORM 6); binding
/ protein transporter
Length=538
Score = 143 bits (360), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 74/141 (52%), Positives = 98/141 (69%), Gaps = 2/141 (1%)
Query 1 ALPNLLSLL-SSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTK 59
ALP LL+LL ++ +K+++KEACWTISNITAGN QIQEV ++G + L++LLE +F+ K
Sbjct 331 ALPGLLNLLKNTYKKSIKKEACWTISNITAGNTSQIQEVFQAGIIRPLINLLEIGEFEIK 390
Query 60 KEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKI 119
KEA WA+SNA SGG+ Q+ LV GC PLC LL C D ++V V+LE L IL+ G+
Sbjct 391 KEAVWAISNATSGGNHDQIKFLVSQGCIRPLCDLLPCPDPRVVTVTLEGLENILKVGEAE 450
Query 120 KETEGL-SINPYATLIEQADG 139
K + N YA +IE ADG
Sbjct 451 KNLGNTGNDNLYAQMIEDADG 471
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 27/74 (36%), Positives = 48/74 (64%), Gaps = 2/74 (2%)
Query 2 LPNLLSLLSSNRKT-VQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
+P+++ LS + T +Q EA W ++NI +G E + +++SGAVP + LL +A + ++
Sbjct 121 VPHIVQFLSRDDFTQLQFEAAWALTNIASGTSENTRVIIDSGAVPLFVKLLSSASEEVRE 180
Query 61 EAAWAVSNAVSGGS 74
+A WA+ N V+G S
Sbjct 181 QAVWALGN-VAGDS 193
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 36/68 (52%), Gaps = 2/68 (2%)
Query 35 IQEVLESGAVPALLHLLETADFDT-KKEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQL 93
I EV++SG VP ++ L DF + EAAWA++N SG S+ ++ G P +L
Sbjct 112 INEVVQSGVVPHIVQFLSRDDFTQLQFEAAWALTNIASGTSE-NTRVIIDSGAVPLFVKL 170
Query 94 LGCMDSKI 101
L ++
Sbjct 171 LSSASEEV 178
Score = 38.1 bits (87), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 36/162 (22%), Positives = 66/162 (40%), Gaps = 44/162 (27%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLE-------------------- 40
A+P + LLSS + V+++A W + N+ + + VL
Sbjct 163 AVPLFVKLLSSASEEVREQAVWALGNVAGDSPKCRDHVLSCEAMMSLLAQFHEHSKLSML 222
Query 41 ----------------------SGAVPALLHLLETADFDTKKEAAWAVSNAVSGGSDMQV 78
A+PAL LL + D + +A+WA+S +S G++ ++
Sbjct 223 RNATWTLSNFCRGKPQPAFEQTKAALPALERLLHSTDEEVLTDASWALS-YLSDGTNEKI 281
Query 79 AELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
++ G P L QLL +++ +L + I+ TG I+
Sbjct 282 QTVIDAGVIPRLVQLLAHPSPSVLIPALRTIGNIV-TGDDIQ 322
> ath:AT3G05720 IMPA-7; IMPA-7 (IMPORTIN ALPHA ISOFORM 7); binding
/ protein transporter
Length=528
Score = 142 bits (358), Expect = 4e-34, Method: Compositional matrix adjust.
Identities = 75/148 (50%), Positives = 104/148 (70%), Gaps = 9/148 (6%)
Query 1 ALPNLLSLLS-SNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTK 59
ALP L++LL S KT++KEACWT+SNITAG Q QIQ V ++ PAL++LL+ ++ D K
Sbjct 315 ALPCLVNLLRGSYNKTIRKEACWTVSNITAGCQSQIQAVFDADICPALVNLLQNSEGDVK 374
Query 60 KEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKI 119
KEAAWA+ NA++GGS Q+ LV+ C PLC LL C D+++VMV LEAL IL+ G+
Sbjct 375 KEAAWAICNAIAGGSYKQIMFLVKQECIKPLCDLLTCSDTQLVMVCLEALKKILKVGEVF 434
Query 120 --KETEGL------SINPYATLIEQADG 139
+ EG+ ++NP+A LIE+A+G
Sbjct 435 SSRHAEGIYQCPQTNVNPHAQLIEEAEG 462
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 23/74 (31%), Positives = 41/74 (55%), Gaps = 2/74 (2%)
Query 2 LPNLLSLLS-SNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
+P + L+ + +Q EA W ++NI +G E + V++ GAV L+ LL + ++
Sbjct 105 VPRFVEFLTWDDSPQLQFEAAWALTNIASGTSENTEVVIDHGAVAILVRLLNSPYDVVRE 164
Query 61 EAAWAVSNAVSGGS 74
+ WA+ N +SG S
Sbjct 165 QVVWALGN-ISGDS 177
Score = 35.4 bits (80), Expect = 0.052, Method: Compositional matrix adjust.
Identities = 30/95 (31%), Positives = 48/95 (50%), Gaps = 11/95 (11%)
Query 5 LLSLLSSNRKTVQKEACWTISNITAGN--QEQIQEVLESGAVPALLHLLETADF-DTKKE 61
L+S + S+ + + EA I + G +++EV+++G VP + L D + E
Sbjct 64 LISCIWSDERDLLIEATTQIRTLLCGEMFNVRVEEVIQAGLVPRFVEFLTWDDSPQLQFE 123
Query 62 AAWAVSNAVSGGSDMQ--------VAELVRLGCAP 88
AAWA++N SG S+ VA LVRL +P
Sbjct 124 AAWALTNIASGTSENTEVVIDHGAVAILVRLLNSP 158
Score = 33.9 bits (76), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 29/106 (27%), Positives = 47/106 (44%), Gaps = 3/106 (2%)
Query 5 LLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPA-LLHLLETADFDTKKEAA 63
L+ LL+S V+++ W + NI+ + VL A+P+ LL L A AA
Sbjct 151 LVRLLNSPYDVVREQVVWALGNISGDSPRCRDIVLGHAALPSLLLQLNHGAKLSMLVNAA 210
Query 64 WAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEAL 109
W +SN G ++ P L QL+ D +++ + AL
Sbjct 211 WTLSNLCRGKPQPPFDQVS--AALPALAQLIRLDDKELLAYTCWAL 254
> ath:AT3G06720 IMPA-1; IMPA-1 (IMPORTIN ALPHA ISOFORM 1); binding
/ protein transporter
Length=532
Score = 142 bits (357), Expect = 4e-34, Method: Compositional matrix adjust.
Identities = 75/141 (53%), Positives = 98/141 (69%), Gaps = 2/141 (1%)
Query 1 ALPNLLSLLSSN-RKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTK 59
ALP L +LL+ N +K+++KEACWTISNITAGN++QIQ V+E+ + L+ LL+ A+FD K
Sbjct 327 ALPCLANLLTQNHKKSIKKEACWTISNITAGNKDQIQTVVEANLISPLVSLLQNAEFDIK 386
Query 60 KEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKI 119
KEAAWA+SNA SGGS Q+ LV GC PLC LL C D +I+ V LE L IL+ G+
Sbjct 387 KEAAWAISNATSGGSHDQIKYLVEQGCIKPLCDLLVCPDPRIITVCLEGLENILKVGEAE 446
Query 120 KE-TEGLSINPYATLIEQADG 139
K +N YA LI+ A+G
Sbjct 447 KNLGHTGDMNYYAQLIDDAEG 467
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 30/96 (31%), Positives = 49/96 (51%), Gaps = 7/96 (7%)
Query 2 LPNLLSLLSS-NRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
+P + L + +Q EA W ++NI +G + + V++ AVP + LL + D ++
Sbjct 117 VPRFVEFLKKEDYPAIQFEAAWALTNIASGTSDHTKVVIDHNAVPIFVQLLASPSDDVRE 176
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCA---PPLCQL 93
+A WA+ N G + +LV LGC P L QL
Sbjct 177 QAVWALGNV--AGDSPRCRDLV-LGCGALLPLLNQL 209
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 34/120 (28%), Positives = 62/120 (51%), Gaps = 5/120 (4%)
Query 2 LPNLLSLLSSNRKTVQKEACWTISNITAGN-QEQIQEVLESGAVPALLHLLETADFDTKK 60
LP L L + ++ + A WT+SN G Q +V A+PAL L+ + D +
Sbjct 203 LPLLNQLNEHAKLSMLRNATWTLSNFCRGKPQPHFDQV--KPALPALERLIHSDDEEVLT 260
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
+A WA+S +S G++ ++ +++ G P L +LL +++ +L + I+ TG I+
Sbjct 261 DACWALS-YLSDGTNDKIQTVIQAGVVPKLVELLLHHSPSVLIPALRTVGNIV-TGDDIQ 318
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 22/61 (36%), Positives = 33/61 (54%), Gaps = 2/61 (3%)
Query 35 IQEVLESGAVPALLHLLETADFDT-KKEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQL 93
I+EV+ +G VP + L+ D+ + EAAWA++N SG SD ++ P QL
Sbjct 108 IEEVISAGVVPRFVEFLKKEDYPAIQFEAAWALTNIASGTSD-HTKVVIDHNAVPIFVQL 166
Query 94 L 94
L
Sbjct 167 L 167
Score = 35.8 bits (81), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 25/74 (33%), Positives = 40/74 (54%), Gaps = 3/74 (4%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQE-VLESGA-VPALLHLLETADFDT 58
A+P + LL+S V+++A W + N+ AG+ + ++ VL GA +P L L E A
Sbjct 159 AVPIFVQLLASPSDDVREQAVWALGNV-AGDSPRCRDLVLGCGALLPLLNQLNEHAKLSM 217
Query 59 KKEAAWAVSNAVSG 72
+ A W +SN G
Sbjct 218 LRNATWTLSNFCRG 231
> ath:AT4G02150 MOS6; MOS6 (MODIFIER OF SNC1, 6); binding / protein
transporter
Length=531
Score = 136 bits (343), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 72/141 (51%), Positives = 100/141 (70%), Gaps = 2/141 (1%)
Query 1 ALPNLLSLLSSN-RKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTK 59
ALP LL+LL +N +K+++KEACWTISNITAGN +QIQ V+++G + +L+ +L++A+F+ K
Sbjct 330 ALPCLLNLLKNNYKKSIKKEACWTISNITAGNADQIQAVIDAGIIQSLVWVLQSAEFEVK 389
Query 60 KEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKI 119
KEAAW +SNA SGG+ Q+ +V GC PLC LL C D K+V V LEAL IL G+
Sbjct 390 KEAAWGISNATSGGTHDQIKFMVSQGCIKPLCDLLTCPDLKVVTVCLEALENILVVGEAE 449
Query 120 KET-EGLSINPYATLIEQADG 139
K N YA +I++A+G
Sbjct 450 KNLGHTGEDNLYAQMIDEAEG 470
Score = 55.5 bits (132), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 31/90 (34%), Positives = 50/90 (55%), Gaps = 2/90 (2%)
Query 2 LPNLLSLLSSNR-KTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
+P ++ LS + +Q EA W ++NI +G E ++ESGAVP + LL +A D ++
Sbjct 120 VPRVVKFLSRDDFPKLQFEAAWALTNIASGTSENTNVIIESGAVPIFIQLLSSASEDVRE 179
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPL 90
+A WA+ N V+G S ++ G PL
Sbjct 180 QAVWALGN-VAGDSPKCRDLVLSYGAMTPL 208
Score = 41.6 bits (96), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 24/61 (39%), Positives = 34/61 (55%), Gaps = 2/61 (3%)
Query 35 IQEVLESGAVPALLHLLETADFDT-KKEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQL 93
I EV++SG VP ++ L DF + EAAWA++N SG S+ ++ G P QL
Sbjct 111 INEVVQSGVVPRVVKFLSRDDFPKLQFEAAWALTNIASGTSE-NTNVIIESGAVPIFIQL 169
Query 94 L 94
L
Sbjct 170 L 170
Score = 37.0 bits (84), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 24/74 (32%), Positives = 39/74 (52%), Gaps = 3/74 (4%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQE-VLESGAV-PALLHLLETADFDT 58
A+P + LLSS + V+++A W + N+ AG+ + ++ VL GA+ P L E
Sbjct 162 AVPIFIQLLSSASEDVREQAVWALGNV-AGDSPKCRDLVLSYGAMTPLLSQFNENTKLSM 220
Query 59 KKEAAWAVSNAVSG 72
+ A W +SN G
Sbjct 221 LRNATWTLSNFCRG 234
> tpv:TP04_0908 importin alpha
Length=538
Score = 135 bits (340), Expect = 4e-32, Method: Compositional matrix adjust.
Identities = 64/138 (46%), Positives = 88/138 (63%), Gaps = 0/138 (0%)
Query 2 LPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKE 61
+P L LL S +KT++KEACWT+SNI+AG + QI+ L+S V L+ L+ DFD ++E
Sbjct 343 IPVLYKLLFSEKKTIKKEACWTLSNISAGTRSQIESFLQSDVVEKLVELMSCNDFDIQRE 402
Query 62 AAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIKE 121
A+WA+ NA SGG Q L GC P+C +L D+K++ V+L AL IL G+ I E
Sbjct 403 ASWAICNAASGGDLKQAENLASRGCIKPICSILTSTDTKLISVALRALENILTVGQHIME 462
Query 122 TEGLSINPYATLIEQADG 139
LS NPY +IE+ DG
Sbjct 463 INNLSSNPYTAVIEECDG 480
Score = 58.2 bits (139), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 35/96 (36%), Positives = 53/96 (55%), Gaps = 4/96 (4%)
Query 2 LPNLLSLLSS-NRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
+P + LS + +Q EA W I+NI +GNQ+Q + ++GAVP L+ LLE D ++
Sbjct 132 VPIFVEFLSRYDAPELQFEAAWAITNIASGNQQQTKVATDNGAVPKLIALLEAPKEDVRE 191
Query 61 EAAWAVSNAVSGGSDMQVAELV-RLGCAPPLCQLLG 95
+A WA+ N G + +LV LG PL L+
Sbjct 192 QAIWALGNI--AGDSAECRDLVLSLGALKPLLYLMA 225
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 32/94 (34%), Positives = 52/94 (55%), Gaps = 1/94 (1%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
A+P L L+ V +ACW +S I+ G+++ IQ VL+SGA P L+ L++ +
Sbjct 258 AIPYLAKLIEHPDSEVLTDACWALSYISDGSEDHIQAVLDSGACPRLIQLMDHVLPVIQT 317
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLL 94
+ + N ++ G+D Q +V GC P L +LL
Sbjct 318 PSLRTIGN-IATGNDRQTQVIVDSGCIPVLYKLL 350
Score = 45.4 bits (106), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 32/113 (28%), Positives = 58/113 (51%), Gaps = 3/113 (2%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTK- 59
A+P L++LL + ++ V+++A W + NI + E VL GA+ LL+L+ + D+
Sbjct 174 AVPKLIALLEAPKEDVREQAIWALGNIAGDSAECRDLVLSLGALKPLLYLMANSQKDSVL 233
Query 60 KEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAI 112
+ A W +SN G ++ P L +L+ DS+++ + AL I
Sbjct 234 RNATWTISNLCRGKPKPYFDDI--RPAIPYLAKLIEHPDSEVLTDACWALSYI 284
> ath:AT4G16143 IMPA-2; IMPA-2 (IMPORTIN ALPHA ISOFORM 2); binding
/ protein transporter
Length=535
Score = 135 bits (339), Expect = 4e-32, Method: Compositional matrix adjust.
Identities = 68/130 (52%), Positives = 92/130 (70%), Gaps = 1/130 (0%)
Query 11 SNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAAWAVSNAV 70
+++K+++KEACWTISNITAGN++QIQ V E+G + L++LL+ A+FD KKEAAWA+SNA
Sbjct 343 NHKKSIKKEACWTISNITAGNRDQIQAVCEAGLICPLVNLLQNAEFDIKKEAAWAISNAT 402
Query 71 SGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIKETEGL-SINP 129
SGGS Q+ +V G PLC LL C D +I+ V LE L IL+ G+ K T +N
Sbjct 403 SGGSPDQIKYMVEQGVVKPLCDLLVCPDPRIITVCLEGLENILKVGEAEKVTGNTGDVNF 462
Query 130 YATLIEQADG 139
YA LI+ A+G
Sbjct 463 YAQLIDDAEG 472
Score = 51.6 bits (122), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 26/74 (35%), Positives = 43/74 (58%), Gaps = 2/74 (2%)
Query 2 LPNLLSLLS-SNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
+P + L+ + +Q EA W ++NI +G E + V+E GAVP + LL + D ++
Sbjct 122 VPRFVEFLTREDYPQLQFEAAWALTNIASGTSENTKVVIEHGAVPIFVQLLASQSDDVRE 181
Query 61 EAAWAVSNAVSGGS 74
+A WA+ N V+G S
Sbjct 182 QAVWALGN-VAGDS 194
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 36/122 (29%), Positives = 66/122 (54%), Gaps = 6/122 (4%)
Query 1 ALPNLLSLLSSNRK-TVQKEACWTISNITAGN-QEQIQEVLESGAVPALLHLLETADFDT 58
AL LLS L+ + K ++ + A WT+SN G Q +V A+PAL L+ + D +
Sbjct 206 ALIPLLSQLNEHAKLSMLRNATWTLSNFCRGKPQPPFDQV--RPALPALERLIHSTDEEV 263
Query 59 KKEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKK 118
+A WA+S +S G++ ++ ++ G P L +LL +++ +L ++ I+ TG
Sbjct 264 LTDACWALS-YLSDGTNDKIQSVIEAGVVPRLVELLQHQSPSVLIPALRSIGNIV-TGDD 321
Query 119 IK 120
++
Sbjct 322 LQ 323
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 28/103 (27%), Positives = 51/103 (49%), Gaps = 3/103 (2%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNI-TAGNQEQIQEVLESGAVPALLHLLETADFDT- 58
+LP ++ + S+ +++Q EA + + I+EV+++G VP + L D+
Sbjct 78 SLPAMVGGVWSDDRSLQLEATTQFRKLLSIERSPPIEEVIDAGVVPRFVEFLTREDYPQL 137
Query 59 KKEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKI 101
+ EAAWA++N SG S+ ++ G P QLL +
Sbjct 138 QFEAAWALTNIASGTSE-NTKVVIEHGAVPIFVQLLASQSDDV 179
Score = 38.1 bits (87), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 25/74 (33%), Positives = 40/74 (54%), Gaps = 3/74 (4%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQE-VLESGA-VPALLHLLETADFDT 58
A+P + LL+S V+++A W + N+ AG+ + ++ VL GA +P L L E A
Sbjct 164 AVPIFVQLLASQSDDVREQAVWALGNV-AGDSPRCRDLVLGQGALIPLLSQLNEHAKLSM 222
Query 59 KKEAAWAVSNAVSG 72
+ A W +SN G
Sbjct 223 LRNATWTLSNFCRG 236
> pfa:PF08_0087 importin alpha, putative
Length=545
Score = 130 bits (328), Expect = 9e-31, Method: Compositional matrix adjust.
Identities = 64/139 (46%), Positives = 91/139 (65%), Gaps = 0/139 (0%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
A+ L LL+S++K+++KEACW +SNITAGN QIQ V+++ +P L+++L DF+ +K
Sbjct 346 AVQKLSCLLNSSKKSIKKEACWALSNITAGNISQIQAVIDNNVIPQLINILMKEDFEVRK 405
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
EAAWA+SNA SGGS++Q+ LV G L LL D+ I+ V+LE L IL G+ K
Sbjct 406 EAAWAISNASSGGSELQIEYLVECGAIHSLSNLLDVEDANIISVTLEGLENILEMGENKK 465
Query 121 ETEGLSINPYATLIEQADG 139
+ L NPY L E+ D
Sbjct 466 LRDNLPTNPYVHLFEECDS 484
Score = 55.5 bits (132), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 29/74 (39%), Positives = 48/74 (64%), Gaps = 2/74 (2%)
Query 2 LPNLLSLLSSNRKT-VQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
+P ++ L + KT +Q EA W I+NI +G+QEQ + V+++ AVP L+ LL + D +
Sbjct 136 VPYIVEFLKYDDKTDLQFEAAWVITNIASGSQEQTKVVIDNNAVPHLVRLLSSEKEDVCE 195
Query 61 EAAWAVSNAVSGGS 74
+A WA+ N ++G S
Sbjct 196 QAVWALGN-IAGDS 208
Score = 55.5 bits (132), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 32/95 (33%), Positives = 52/95 (54%), Gaps = 1/95 (1%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
ALP L +L+ ++ + + +ACWT+S ++ G+ E I VL++G ++ LL F +
Sbjct 262 ALPTLAALIYNDDEEILTDACWTLSYLSDGSNENINSVLDAGVAERVVELLSHCSFLVQT 321
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLG 95
A V N V+ G D+Q +V+LG L LL
Sbjct 322 PALRTVGNIVT-GDDLQTDVVVKLGAVQKLSCLLN 355
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 27/73 (36%), Positives = 43/73 (58%), Gaps = 1/73 (1%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDT-K 59
A+P+L+ LLSS ++ V ++A W + NI + E + VL ++P LL +L T+ T
Sbjct 178 AVPHLVRLLSSEKEDVCEQAVWALGNIAGDSAECREYVLNQNSLPLLLKILRTSHKRTLI 237
Query 60 KEAAWAVSNAVSG 72
+ AAW +SN G
Sbjct 238 RNAAWTLSNLCRG 250
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 30/96 (31%), Positives = 54/96 (56%), Gaps = 3/96 (3%)
Query 1 ALPNLLSLL-SSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTK 59
+LP LL +L +S+++T+ + A WT+SN+ G E++ S A+P L L+ D +
Sbjct 220 SLPLLLKILRTSHKRTLIRNAAWTLSNLCRGKPAPKFEIV-SKALPTLAALIYNDDEEIL 278
Query 60 KEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLG 95
+A W +S +S GS+ + ++ G A + +LL
Sbjct 279 TDACWTLS-YLSDGSNENINSVLDAGVAERVVELLS 313
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 35/62 (56%), Gaps = 2/62 (3%)
Query 35 IQEVLESGAVPALLHLLETAD-FDTKKEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQL 93
IQEV+ SG VP ++ L+ D D + EAAW ++N ++ GS Q ++ P L +L
Sbjct 127 IQEVINSGVVPYIVEFLKYDDKTDLQFEAAWVITN-IASGSQEQTKVVIDNNAVPHLVRL 185
Query 94 LG 95
L
Sbjct 186 LS 187
> bbo:BBOV_III009880 17.m07856; armadillo/beta-catenin-like repeat
domain containing protein
Length=539
Score = 129 bits (325), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 59/138 (42%), Positives = 89/138 (64%), Gaps = 0/138 (0%)
Query 2 LPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKE 61
+P L LL S++KT++KEACWT+SNI AG ++Q++ L+S V LL ++ DFD K+E
Sbjct 345 IPILYKLLYSDKKTIKKEACWTLSNIAAGTRDQVEAFLQSDTVEKLLEMMTCNDFDIKRE 404
Query 62 AAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIKE 121
A+WA+ NA SGG Q + GC +C +L D+K++ V+L A+ IL+ G ++ E
Sbjct 405 ASWAICNAASGGDSKQAENMASRGCLRYICGILSTTDTKLIGVALRAMENILQVGHQLME 464
Query 122 TEGLSINPYATLIEQADG 139
LS NPY + +E+ DG
Sbjct 465 LNALSGNPYVSAVEECDG 482
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 37/94 (39%), Positives = 51/94 (54%), Gaps = 1/94 (1%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
A+P L LL V +ACW +S I+ GN+E IQ VL++GA L+ LLE +
Sbjct 260 AVPYLSKLLEHTDTEVLTDACWALSYISDGNEEHIQAVLDAGACGRLVQLLEHPQPVIQT 319
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLL 94
A V N ++ G+D Q +V GC P L +LL
Sbjct 320 PALRTVGN-IATGNDRQTQMIVDCGCIPILYKLL 352
Score = 55.1 bits (131), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 29/79 (36%), Positives = 49/79 (62%), Gaps = 1/79 (1%)
Query 16 VQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAAWAVSNAVSGGSD 75
+Q EA W I+N+ +GNQ+Q + ++GAVP L+ LL++ + +++A WA+ N ++G S
Sbjct 149 LQFEAAWAITNVASGNQQQTKVATDNGAVPKLIALLDSPKEEVREQAVWALGN-IAGDSP 207
Query 76 MQVAELVRLGCAPPLCQLL 94
++ LG PL LL
Sbjct 208 ECRDLVLGLGALKPLLYLL 226
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 36/113 (31%), Positives = 60/113 (53%), Gaps = 3/113 (2%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTK- 59
A+P L++LL S ++ V+++A W + NI + E VL GA+ LL+LL ++ D+
Sbjct 176 AVPKLIALLDSPKEEVREQAVWALGNIAGDSPECRDLVLGLGALKPLLYLLVHSEKDSVI 235
Query 60 KEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAI 112
+ A W VSN G ++ L P L +LL D++++ + AL I
Sbjct 236 RNATWTVSNLCRGKPKPVFHDV--LPAVPYLSKLLEHTDTEVLTDACWALSYI 286
> mmu:16647 Kpna2, 2410044B12Rik, IPOA1, MGC91246, Rch1; karyopherin
(importin) alpha 2; K15043 importin subunit alpha-2
Length=529
Score = 123 bits (309), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 66/139 (47%), Positives = 90/139 (64%), Gaps = 5/139 (3%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
AL SLL++ + +QKEA WT+SNITAG Q+QIQ+V+ G VP L+ +L ADF T+K
Sbjct 336 ALAVFPSLLTNPKTNIQKEATWTMSNITAGRQDQIQQVVNHGLVPFLVGVLSKADFKTQK 395
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
EAAWA++N SGG+ Q+ LV G PL LL D+KI+ V L+A+ I + +K+
Sbjct 396 EAAWAITNYTSGGTVEQIVYLVHCGIIEPLMNLLSAKDTKIIQVILDAISNIFQAAEKLG 455
Query 121 ETEGLSINPYATLIEQADG 139
ETE LSI +IE+ G
Sbjct 456 ETEKLSI-----MIEECGG 469
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 30/98 (30%), Positives = 52/98 (53%), Gaps = 2/98 (2%)
Query 2 LPNLLSLL-SSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
+P +S L ++ +Q E+ W ++NI +G EQ + V++ GA+PA + LL + +
Sbjct 121 IPKFVSFLGKTDCSPIQFESAWALTNIASGTSEQTKAVVDGGAIPAFISLLASPHAHISE 180
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMD 98
+A WA+ N GS + +++ G PL LL D
Sbjct 181 QAVWALGNIAGDGSAFRDL-VIKHGAIDPLLALLAVPD 217
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 48/93 (51%), Gaps = 1/93 (1%)
Query 2 LPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKE 61
LP L+ LL N V ++CW IS +T G E+I+ V++ G VP L+ LL +
Sbjct 253 LPTLVRLLHHNDPEVLADSCWAISYLTDGPNERIEMVVKKGVVPQLVKLLGATELPIVTP 312
Query 62 AAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLL 94
A A+ N V+ G+D Q +++ G LL
Sbjct 313 ALRAIGNIVT-GTDEQTQKVIDAGALAVFPSLL 344
Score = 39.3 bits (90), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 36/161 (22%), Positives = 66/161 (40%), Gaps = 50/161 (31%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQE-VLESGAV--------------- 44
A+P +SLL+S + ++A W + NI AG+ ++ V++ GA+
Sbjct 163 AIPAFISLLASPHAHISEQAVWALGNI-AGDGSAFRDLVIKHGAIDPLLALLAVPDLSTL 221
Query 45 --------------------------------PALLHLLETADFDTKKEAAWAVSNAVSG 72
P L+ LL D + ++ WA+S ++
Sbjct 222 ACGYLRNLTWTLSNLCRNKNPAPPLDAVEQILPTLVRLLHHNDPEVLADSCWAIS-YLTD 280
Query 73 GSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAIL 113
G + ++ +V+ G P L +LLG + IV +L A+ I+
Sbjct 281 GPNERIEMVVKKGVVPQLVKLLGATELPIVTPALRAIGNIV 321
> mmu:100039592 Gm10259; predicted pseudogene 10259; K15043 importin
subunit alpha-2
Length=529
Score = 123 bits (309), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 66/139 (47%), Positives = 90/139 (64%), Gaps = 5/139 (3%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
AL SLL++ + +QKEA WT+SNITAG Q+QIQ+V+ G VP L+ +L ADF T+K
Sbjct 336 ALAVFPSLLTNPKTNIQKEATWTMSNITAGRQDQIQQVVNHGLVPFLVGVLSKADFKTQK 395
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
EAAWA++N SGG+ Q+ LV G PL LL D+KI+ V L+A+ I + +K+
Sbjct 396 EAAWAITNYTSGGTVEQIVYLVHCGIIEPLMNLLSAKDTKIIQVILDAISNIFQAAEKLG 455
Query 121 ETEGLSINPYATLIEQADG 139
ETE LSI +IE+ G
Sbjct 456 ETEKLSI-----MIEECGG 469
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 30/98 (30%), Positives = 52/98 (53%), Gaps = 2/98 (2%)
Query 2 LPNLLSLL-SSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
+P +S L ++ +Q E+ W ++NI +G EQ + V++ GA+PA + LL + +
Sbjct 121 IPKFVSFLGKTDCSPIQFESAWALTNIASGTSEQTKAVVDGGAIPAFISLLASPHAHISE 180
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMD 98
+A WA+ N GS + +++ G PL LL D
Sbjct 181 QAVWALGNIAGDGSAFRDL-VIKHGAIDPLLALLAVPD 217
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 48/93 (51%), Gaps = 1/93 (1%)
Query 2 LPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKE 61
LP L+ LL N V ++CW IS +T G E+I+ V++ G VP L+ LL +
Sbjct 253 LPTLVRLLHHNDPEVLADSCWAISYLTDGPNERIEMVVKKGVVPQLVKLLGATELPIVTP 312
Query 62 AAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLL 94
A A+ N V+ G+D Q +++ G LL
Sbjct 313 ALRAIGNIVT-GTDEQTQKVIDAGALAVFPSLL 344
Score = 39.3 bits (90), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 36/161 (22%), Positives = 66/161 (40%), Gaps = 50/161 (31%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQE-VLESGAV--------------- 44
A+P +SLL+S + ++A W + NI AG+ ++ V++ GA+
Sbjct 163 AIPAFISLLASPHAHISEQAVWALGNI-AGDGSAFRDLVIKHGAIDPLLALLAVPDLSTL 221
Query 45 --------------------------------PALLHLLETADFDTKKEAAWAVSNAVSG 72
P L+ LL D + ++ WA+S ++
Sbjct 222 ACGYLRNLTWTLSNLCRNKNPAPPLDAVEQILPTLVRLLHHNDPEVLADSCWAIS-YLTD 280
Query 73 GSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAIL 113
G + ++ +V+ G P L +LLG + IV +L A+ I+
Sbjct 281 GPNERIEMVVKKGVVPQLVKLLGATELPIVTPALRAIGNIV 321
> mmu:100043906 Gm10184; predicted pseudogene 10184
Length=529
Score = 123 bits (309), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 66/139 (47%), Positives = 90/139 (64%), Gaps = 5/139 (3%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
AL SLL++ + +QKEA WT+SNITAG Q+QIQ+V+ G VP L+ +L ADF T+K
Sbjct 336 ALAVFPSLLTNPKTNIQKEATWTMSNITAGRQDQIQQVVNHGLVPFLVGVLSKADFKTQK 395
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
EAAWA++N SGG+ Q+ LV G PL LL D+KI+ V L+A+ I + +K+
Sbjct 396 EAAWAITNYTSGGTVEQIVYLVHCGIIEPLMNLLSAKDTKIIQVILDAISNIFQAAEKLG 455
Query 121 ETEGLSINPYATLIEQADG 139
ETE LSI +IE+ G
Sbjct 456 ETEKLSI-----MIEECGG 469
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 30/98 (30%), Positives = 52/98 (53%), Gaps = 2/98 (2%)
Query 2 LPNLLSLL-SSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
+P +S L ++ +Q E+ W ++NI +G EQ + V++ GA+PA + LL + +
Sbjct 121 IPKFVSFLGKTDCSPIQFESAWALTNIASGTSEQTKAVVDGGAIPAFISLLASPHAHISE 180
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMD 98
+A WA+ N GS + +++ G PL LL D
Sbjct 181 QAVWALGNIAGDGSAFRDL-VIKHGAIDPLLALLAVPD 217
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 48/93 (51%), Gaps = 1/93 (1%)
Query 2 LPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKE 61
LP L+ LL N V ++CW IS +T G E+I+ V++ G VP L+ LL +
Sbjct 253 LPTLVRLLHHNDPEVLADSCWAISYLTDGPNERIEMVVKKGVVPQLVKLLGATELPIVTP 312
Query 62 AAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLL 94
A A+ N V+ G+D Q +++ G LL
Sbjct 313 ALRAIGNIVT-GTDEQTQKVIDAGALAVFPSLL 344
Score = 39.3 bits (90), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 36/161 (22%), Positives = 66/161 (40%), Gaps = 50/161 (31%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQE-VLESGAV--------------- 44
A+P +SLL+S + ++A W + NI AG+ ++ V++ GA+
Sbjct 163 AIPAFISLLASPHAHISEQAVWALGNI-AGDGSAFRDLVIKHGAIDPLLALLAVPDLSTL 221
Query 45 --------------------------------PALLHLLETADFDTKKEAAWAVSNAVSG 72
P L+ LL D + ++ WA+S ++
Sbjct 222 ACGYLRNLTWTLSNLCRNKNPAPPLDAVEQILPTLVRLLHHNDPEVLADSCWAIS-YLTD 280
Query 73 GSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAIL 113
G + ++ +V+ G P L +LLG + IV +L A+ I+
Sbjct 281 GPNERIEMVVKKGVVPQLVKLLGATELPIVTPALRAIGNIV 321
> hsa:3838 KPNA2, IPOA1, QIP2, RCH1, SRP1alpha; karyopherin alpha
2 (RAG cohort 1, importin alpha 1); K15043 importin subunit
alpha-2
Length=529
Score = 123 bits (308), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 66/139 (47%), Positives = 90/139 (64%), Gaps = 5/139 (3%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
AL SLL++ + +QKEA WT+SNITAG Q+QIQ+V+ G VP L+ +L ADF T+K
Sbjct 336 ALAVFPSLLTNPKTNIQKEATWTMSNITAGRQDQIQQVVNHGLVPFLVSVLSKADFKTQK 395
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
EA WAV+N SGG+ Q+ LV G PL LL D+KI++V L+A+ I + +K+
Sbjct 396 EAVWAVTNYTSGGTVEQIVYLVHCGIIEPLMNLLTAKDTKIILVILDAISNIFQAAEKLG 455
Query 121 ETEGLSINPYATLIEQADG 139
ETE LSI +IE+ G
Sbjct 456 ETEKLSI-----MIEECGG 469
Score = 57.8 bits (138), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 30/98 (30%), Positives = 52/98 (53%), Gaps = 2/98 (2%)
Query 2 LPNLLSLLS-SNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
+P +S L ++ +Q E+ W ++NI +G EQ + V++ GA+PA + LL + +
Sbjct 121 IPKFVSFLGRTDCSPIQFESAWALTNIASGTSEQTKAVVDGGAIPAFISLLASPHAHISE 180
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMD 98
+A WA+ N GS + +++ G PL LL D
Sbjct 181 QAVWALGNIAGDGSVFRDL-VIKYGAVDPLLALLAVPD 217
Score = 39.3 bits (90), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 37/161 (22%), Positives = 65/161 (40%), Gaps = 50/161 (31%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQE-VLESGAV--------------- 44
A+P +SLL+S + ++A W + NI AG+ ++ V++ GAV
Sbjct 163 AIPAFISLLASPHAHISEQAVWALGNI-AGDGSVFRDLVIKYGAVDPLLALLAVPDMSSL 221
Query 45 --------------------------------PALLHLLETADFDTKKEAAWAVSNAVSG 72
P L+ LL D + + WA+S ++
Sbjct 222 ACGYLRNLTWTLSNLCRNKNPAPPIDAVEQILPTLVRLLHHDDPEVLADTCWAIS-YLTD 280
Query 73 GSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAIL 113
G + ++ +V+ G P L +LLG + IV +L A+ I+
Sbjct 281 GPNERIGMVVKTGVVPQLVKLLGASELPIVTPALRAIGNIV 321
> ath:AT5G49310 IMPA-5; IMPA-5 (IMPORTIN ALPHA ISOFORM 5); binding
/ protein transporter
Length=519
Score = 123 bits (308), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 65/140 (46%), Positives = 91/140 (65%), Gaps = 3/140 (2%)
Query 1 ALPNLLSLLSSNR-KTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTK 59
LP L LL+ N + +++EACWTISNITAG +EQIQ V+++ +P+L++L + A+FD K
Sbjct 325 VLPVLADLLTQNHMRGIRREACWTISNITAGLEEQIQSVIDANLIPSLVNLAQHAEFDIK 384
Query 60 KEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKI 119
KEA WA+SNA GGS Q+ LV C LC +L C D +I++VSL L IL G+
Sbjct 385 KEAIWAISNASVGGSPNQIKYLVEQNCIKALCDILVCPDLRIILVSLGGLEMILIAGEVD 444
Query 120 KETEGLSINPYATLIEQADG 139
K +N Y+ +IE A+G
Sbjct 445 KNLR--DVNCYSQMIEDAEG 462
Score = 55.1 bits (131), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 35/94 (37%), Positives = 50/94 (53%), Gaps = 1/94 (1%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
LP L L+ S+ + V +ACW +SN++ + E IQ V+E+G VP L+ LL+ A
Sbjct 241 VLPVLKRLVYSDDEQVLIDACWALSNLSDASNENIQSVIEAGVVPRLVELLQHASPVVLV 300
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLL 94
A + N VSG S Q ++ G P L LL
Sbjct 301 PALRCIGNIVSGNS-QQTHCVINCGVLPVLADLL 333
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 24/83 (28%), Positives = 42/83 (50%), Gaps = 3/83 (3%)
Query 1 ALPNLLSLLS-SNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTK 59
+P + L + +Q EA W ++NI +G E + V++ G VP + LL + D D +
Sbjct 114 VVPRFVEFLKKDDNPKLQFEAAWALTNIASGASEHTKVVIDHGVVPLFVQLLASPDDDVR 173
Query 60 KEAAWAVSNAVSGGSDMQVAELV 82
++A W + N G +Q + V
Sbjct 174 EQAIWGLGNV--AGDSIQCRDFV 194
Score = 38.9 bits (89), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 26/74 (35%), Positives = 39/74 (52%), Gaps = 3/74 (4%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQE-VLESGAVPALLHLLET-ADFDT 58
+P + LL+S V+++A W + N+ AG+ Q ++ VL SGA LLH L A
Sbjct 157 VVPLFVQLLASPDDDVREQAIWGLGNV-AGDSIQCRDFVLNSGAFIPLLHQLNNHATLSI 215
Query 59 KKEAAWAVSNAVSG 72
+ A W +SN G
Sbjct 216 LRNATWTLSNFFRG 229
> cel:F32E10.4 ima-3; IMportin Alpha family member (ima-3)
Length=514
Score = 118 bits (295), Expect = 7e-27, Method: Compositional matrix adjust.
Identities = 51/114 (44%), Positives = 78/114 (68%), Gaps = 0/114 (0%)
Query 8 LLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAAWAVS 67
LL+ ++ + KEA W +SNITAGNQ+Q+Q+V ++G +P ++HLL+ DF T+KEAAWA+S
Sbjct 327 LLAHYKEKINKEAVWFVSNITAGNQQQVQDVFDAGIMPMIIHLLDRGDFPTQKEAAWAIS 386
Query 68 NAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIKE 121
N G QV ++V+LG P C +L C DS+I+ V L+ ++ IL+ + E
Sbjct 387 NVTISGRPNQVEQMVKLGVLRPFCAMLSCTDSQIIQVVLDGINNILKMAGEAAE 440
Score = 62.8 bits (151), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 33/93 (35%), Positives = 50/93 (53%), Gaps = 1/93 (1%)
Query 2 LPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKE 61
LP L+ LSS +Q EA W ++NI +G EQ Q V+ +GAVP L LL + + ++
Sbjct 110 LPVLVQCLSSTDPNLQFEAAWALTNIASGTSEQTQAVVNAGAVPLFLQLLSCGNLNVCEQ 169
Query 62 AAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLL 94
+ WA+ N + G + + LG PL Q +
Sbjct 170 SVWALGNIIGDGPHFR-DYCLELGILQPLLQFI 201
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 29/87 (33%), Positives = 47/87 (54%), Gaps = 1/87 (1%)
Query 35 IQEVLESGAVPALLHLLETADFDTKKEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLL 94
I +++ SG +P L+ L + D + + EAAWA++N SG S+ Q +V G P QLL
Sbjct 101 IDDLIGSGILPVLVQCLSSTDPNLQFEAAWALTNIASGTSE-QTQAVVNAGAVPLFLQLL 159
Query 95 GCMDSKIVMVSLEALHAILRTGKKIKE 121
C + + S+ AL I+ G ++
Sbjct 160 SCGNLNVCEQSVWALGNIIGDGPHFRD 186
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 31/100 (31%), Positives = 45/100 (45%), Gaps = 1/100 (1%)
Query 2 LPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKE 61
LP L L+ + + W +S +T G E IQ V+E+ V L+ LL D +
Sbjct 237 LPALSLLIHHQDTNILIDTVWALSYLTDGGNEHIQMVIEAQVVTHLVPLLGHVDVKVQTA 296
Query 62 AAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKI 101
A AV N V+ G+D Q ++ G + LL KI
Sbjct 297 ALRAVGNIVT-GTDEQTQLVLDSGVLRFMPGLLAHYKEKI 335
> xla:380125 kpna2, MGC53789, ipoa1, pen, qip2, rch1, srp1alpha;
karyopherin alpha-2 subunit like; K15043 importin subunit
alpha-2
Length=526
Score = 117 bits (293), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 60/125 (48%), Positives = 78/125 (62%), Gaps = 0/125 (0%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
AL LL+ ++ +QKEA WT+SNITAG Q+QIQEV+ G +P L+ +L D+ T+K
Sbjct 333 ALSAFTELLTHHKNNIQKEAAWTLSNITAGRQDQIQEVVNCGLIPYLVEILRKGDYKTQK 392
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
EA WAV+N SGG+ Q+ LV+ G PL LL DSK V+V L+A I KI
Sbjct 393 EAIWAVTNYTSGGTIDQIIYLVQCGIIEPLSNLLSVKDSKTVLVMLDAFTNIFAAADKIG 452
Query 121 ETEGL 125
ETE L
Sbjct 453 ETEKL 457
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 32/98 (32%), Positives = 53/98 (54%), Gaps = 2/98 (2%)
Query 2 LPNLLSLLS-SNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
+P L++ L+ S+ +Q EA W ++NI +G +Q + V+E G VPA + LL + +
Sbjct 118 IPKLVTFLAHSDCSPIQFEAAWALTNIASGTSDQTKAVVEGGGVPAFISLLASPHPHISE 177
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMD 98
+A WA+ N GS + +++ G PL LL D
Sbjct 178 QAVWALGNIAGDGSAYRDL-VIKHGAVGPLLALLAGPD 214
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 32/118 (27%), Positives = 54/118 (45%), Gaps = 15/118 (12%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQ-----EQIQEVLESGAVPALLHLLETAD 55
A P+L +L + + V WT+SN+ + IQ++L P ++ LL D
Sbjct 211 AGPDLSTLATGYLRNV----TWTLSNLCRNKNPAPPLDAIQQIL-----PTIVRLLHHDD 261
Query 56 FDTKKEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAIL 113
+ + WAVS G +D ++ +VR G + QLL C + +V L + I+
Sbjct 262 REVLADTCWAVSYLTDGSND-RIDVVVRTGLVSRIVQLLACGELTVVTPCLRTIGNIV 318
Score = 36.2 bits (82), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 22/73 (30%), Positives = 33/73 (45%), Gaps = 5/73 (6%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDT-- 58
+P +SLL+S + ++A W + NI V++ GAV LL LL D T
Sbjct 160 GVPAFISLLASPHPHISEQAVWALGNIAGDGSAYRDLVIKHGAVGPLLALLAGPDLSTLA 219
Query 59 ---KKEAAWAVSN 68
+ W +SN
Sbjct 220 TGYLRNVTWTLSN 232
> xla:380151 kpna2, MGC52989; karyopherin alpha 2 (RAG cohort
1, importin alpha 1); K15043 importin subunit alpha-2
Length=526
Score = 115 bits (289), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 59/125 (47%), Positives = 78/125 (62%), Gaps = 0/125 (0%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
AL LL+ ++ +QKEA WT+SNITAG Q+QIQEV+ G +P L+ +L D+ T+K
Sbjct 333 ALSVFAELLTHHKNNIQKEAAWTLSNITAGRQDQIQEVVNHGLMPYLIEILRKGDYKTQK 392
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
EA WAV+N SGG+ Q+ LV+ G PL LL DSK V+V L+A I K+
Sbjct 393 EAIWAVTNYTSGGTIDQIIYLVQCGVIEPLSNLLSVKDSKTVLVILDAFTNIFAAADKLG 452
Query 121 ETEGL 125
ETE L
Sbjct 453 ETEKL 457
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 32/98 (32%), Positives = 53/98 (54%), Gaps = 2/98 (2%)
Query 2 LPNLLSLLS-SNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
+P L++ L+ S+ +Q EA W ++NI +G +Q + V+E G VPA + LL + +
Sbjct 118 IPKLVAFLAHSDCSPIQFEAAWALTNIASGTSDQTKAVVEGGGVPAFISLLASPHPHISE 177
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMD 98
+A WA+ N GS + +++ G PL LL D
Sbjct 178 QAVWALGNIAGDGSAYRDL-VIKHGAVGPLLALLAGPD 214
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 35/118 (29%), Positives = 55/118 (46%), Gaps = 15/118 (12%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQ-----EQIQEVLESGAVPALLHLLETAD 55
A P+L +L + + V WT+SN+ + IQ++L P L+ LL D
Sbjct 211 AGPDLSTLATGYLRNV----TWTLSNLCRNKNPAPPLDAIQQIL-----PTLVRLLHHDD 261
Query 56 FDTKKEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAIL 113
+ + WAVS G +D ++ +VR G + QLLGC + IV L + I+
Sbjct 262 REVLADTCWAVSYLTDGSND-RIDVVVRTGLVSRIVQLLGCGELTIVTPCLRTIGNIV 318
> mmu:16649 Kpna4, 1110058D08Rik, IPOA3; karyopherin (importin)
alpha 4
Length=521
Score = 115 bits (288), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 59/139 (42%), Positives = 85/139 (61%), Gaps = 7/139 (5%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
AL + +LL+ ++ + KEA W +SNITAGNQ+Q+Q V+++ VP ++HLL+ DF T+K
Sbjct 327 ALSHFPALLTHPKEKINKEAVWFLSNITAGNQQQVQAVIDANLVPMIIHLLDKGDFGTQK 386
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
EAAWA+SN G QVA L++ PP C LL D+++V V L+ L IL+ +
Sbjct 387 EAAWAISNLTISGRKDQVAYLIQQNVIPPFCNLLTVKDAQVVQVVLDGLSNILKMAEDQA 446
Query 121 ETEGLSINPYATLIEQADG 139
ET A LIE+ G
Sbjct 447 ET-------IANLIEECGG 458
Score = 54.7 bits (130), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 31/95 (32%), Positives = 50/95 (52%), Gaps = 2/95 (2%)
Query 2 LPNLLSLLS-SNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
LP L+ L + ++Q EA W ++NI +G EQ Q V++S AVP L LL + + +
Sbjct 116 LPILVHCLERDDNPSLQFEAAWALTNIASGTSEQTQAVVQSNAVPLFLRLLHSPHQNVCE 175
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLG 95
+A WA+ N + G + ++ LG PL +
Sbjct 176 QAVWALGNIIGDGPQCR-DYVISLGVVKPLLSFIS 209
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 36/71 (50%), Gaps = 0/71 (0%)
Query 2 LPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKE 61
LP L L+ + + W +S +T EQIQ V++SG VP L+ LL + +
Sbjct 244 LPALCVLIHHTDVNILVDTVWALSYLTDAGNEQIQMVIDSGIVPHLVPLLSHQEVKVQTA 303
Query 62 AAWAVSNAVSG 72
A AV N V+G
Sbjct 304 ALRAVGNIVTG 314
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/88 (30%), Positives = 48/88 (54%), Gaps = 2/88 (2%)
Query 35 IQEVLESGAVPALLHLLETADFDT-KKEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQL 93
I ++++SG +P L+H LE D + + EAAWA++N SG S+ Q +V+ P +L
Sbjct 107 IDDLIKSGILPILVHCLERDDNPSLQFEAAWALTNIASGTSE-QTQAVVQSNAVPLFLRL 165
Query 94 LGCMDSKIVMVSLEALHAILRTGKKIKE 121
L + ++ AL I+ G + ++
Sbjct 166 LHSPHQNVCEQAVWALGNIIGDGPQCRD 193
Score = 31.2 bits (69), Expect = 0.93, Method: Compositional matrix adjust.
Identities = 28/119 (23%), Positives = 48/119 (40%), Gaps = 2/119 (1%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETA-DFDTK 59
A+P L LL S + V ++A W + NI + V+ G V LL + +
Sbjct 158 AVPLFLRLLHSPHQNVCEQAVWALGNIIGDGPQCRDYVISLGVVKPLLSFISPSIPITFL 217
Query 60 KEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKK 118
+ W + N E ++ P LC L+ D I++ ++ AL + G +
Sbjct 218 RNVTWVMVNLCRHKDPPPPMETIQ-EILPALCVLIHHTDVNILVDTVWALSYLTDAGNE 275
> xla:432010 kpna4, MGC78839, ipoa3, qip1, srp3; karyopherin alpha
4 (importin alpha 3)
Length=520
Score = 115 bits (287), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 59/139 (42%), Positives = 84/139 (60%), Gaps = 7/139 (5%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
AL + SLL+ ++ + KEA W +SNITAGNQ+Q+Q V+++ VP ++HLL+ DF T+K
Sbjct 327 ALLHFPSLLTHAKEKINKEAVWFLSNITAGNQQQVQAVIDANLVPMIIHLLDKGDFGTQK 386
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
EAAWA+SN G QVA L++ PP C LL D +++ V L+ L IL+ +
Sbjct 387 EAAWAISNLTISGRKDQVAYLIQQNVIPPFCNLLTVKDPQVIQVVLDGLSNILKMADEDS 446
Query 121 ETEGLSINPYATLIEQADG 139
ET A LIE+ G
Sbjct 447 ET-------IANLIEECGG 458
Score = 55.1 bits (131), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 31/95 (32%), Positives = 50/95 (52%), Gaps = 2/95 (2%)
Query 2 LPNLLSLLS-SNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
LP L+ L + ++Q EA W ++NI +G EQ Q V++S AVP L LL + + +
Sbjct 116 LPILVHCLDRDDNPSLQFEAAWALTNIASGTSEQTQAVVQSNAVPLFLRLLHSPHQNVCE 175
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLG 95
+A WA+ N + G + ++ LG PL +
Sbjct 176 QAVWALGNIIGDGPQCR-DYVISLGVVKPLLSFIN 209
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 35/71 (49%), Gaps = 0/71 (0%)
Query 2 LPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKE 61
LP L L+ + + W +S +T EQIQ V+ESG V L+ LL + +
Sbjct 244 LPALCVLIHHTDVNILVDTVWALSYLTDAGNEQIQMVIESGIVTNLVPLLSNPEVKVQTA 303
Query 62 AAWAVSNAVSG 72
A AV N V+G
Sbjct 304 ALRAVGNIVTG 314
Score = 39.3 bits (90), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 30/114 (26%), Positives = 59/114 (51%), Gaps = 3/114 (2%)
Query 10 SSNRKTVQKEACWTISNITAGNQEQ-IQEVLESGAVPALLHLLETADFDT-KKEAAWAVS 67
SS+ + +Q A + + ++ I ++++SG +P L+H L+ D + + EAAWA++
Sbjct 81 SSDHQGIQLSAVQAARKLLSSDRNPPIDDLIKSGILPILVHCLDRDDNPSLQFEAAWALT 140
Query 68 NAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIKE 121
N SG S+ Q +V+ P +LL + ++ AL I+ G + ++
Sbjct 141 NIASGTSE-QTQAVVQSNAVPLFLRLLHSPHQNVCEQAVWALGNIIGDGPQCRD 193
Score = 31.2 bits (69), Expect = 0.91, Method: Compositional matrix adjust.
Identities = 28/119 (23%), Positives = 48/119 (40%), Gaps = 2/119 (1%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETA-DFDTK 59
A+P L LL S + V ++A W + NI + V+ G V LL + +
Sbjct 158 AVPLFLRLLHSPHQNVCEQAVWALGNIIGDGPQCRDYVISLGVVKPLLSFINPSIPITFL 217
Query 60 KEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKK 118
+ W + N E ++ P LC L+ D I++ ++ AL + G +
Sbjct 218 RNVTWVMVNLCRHKDPPPPMETIQ-EILPALCVLIHHTDVNILVDTVWALSYLTDAGNE 275
> hsa:3840 KPNA4, IPOA3, MGC12217, MGC26703, QIP1, SRP3; karyopherin
alpha 4 (importin alpha 3)
Length=521
Score = 115 bits (287), Expect = 6e-26, Method: Compositional matrix adjust.
Identities = 55/124 (44%), Positives = 80/124 (64%), Gaps = 0/124 (0%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
AL + +LL+ ++ + KEA W +SNITAGNQ+Q+Q V+++ VP ++HLL+ DF T+K
Sbjct 327 ALSHFPALLTHPKEKINKEAVWFLSNITAGNQQQVQAVIDANLVPMIIHLLDKGDFGTQK 386
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
EAAWA+SN G QVA L++ PP C LL D+++V V L+ L IL+ +
Sbjct 387 EAAWAISNLTISGRKDQVAYLIQQNVIPPFCNLLTVKDAQVVQVVLDGLSNILKMAEDEA 446
Query 121 ETEG 124
ET G
Sbjct 447 ETIG 450
Score = 54.7 bits (130), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 31/94 (32%), Positives = 50/94 (53%), Gaps = 2/94 (2%)
Query 2 LPNLLSLLS-SNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
LP L+ L + ++Q EA W ++NI +G EQ Q V++S AVP L LL + + +
Sbjct 116 LPILVHCLERDDNPSLQFEAAWALTNIASGTSEQTQAVVQSNAVPLFLRLLHSPHQNVCE 175
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLL 94
+A WA+ N + G + ++ LG PL +
Sbjct 176 QAVWALGNIIGDGPQCR-DYVISLGVVKPLLSFI 208
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 36/71 (50%), Gaps = 0/71 (0%)
Query 2 LPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKE 61
LP L L+ + + W +S +T EQIQ V++SG VP L+ LL + +
Sbjct 244 LPALCVLIHHTDVNILVDTVWALSYLTDAGNEQIQMVIDSGIVPHLVPLLSHQEVKVQTA 303
Query 62 AAWAVSNAVSG 72
A AV N V+G
Sbjct 304 ALRAVGNIVTG 314
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 31/114 (27%), Positives = 59/114 (51%), Gaps = 3/114 (2%)
Query 10 SSNRKTVQKEACWTISNITAGNQEQ-IQEVLESGAVPALLHLLETADFDT-KKEAAWAVS 67
SS+ + +Q A + + ++ I ++++SG +P L+H LE D + + EAAWA++
Sbjct 81 SSDNQGIQLSAVQAARKLLSSDRNPPIDDLIKSGILPILVHCLERDDNPSLQFEAAWALT 140
Query 68 NAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIKE 121
N SG S+ Q +V+ P +LL + ++ AL I+ G + ++
Sbjct 141 NIASGTSE-QTQAVVQSNAVPLFLRLLHSPHQNVCEQAVWALGNIIGDGPQCRD 193
Score = 31.6 bits (70), Expect = 0.86, Method: Compositional matrix adjust.
Identities = 28/119 (23%), Positives = 48/119 (40%), Gaps = 2/119 (1%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETA-DFDTK 59
A+P L LL S + V ++A W + NI + V+ G V LL + +
Sbjct 158 AVPLFLRLLHSPHQNVCEQAVWALGNIIGDGPQCRDYVISLGVVKPLLSFISPSIPITFL 217
Query 60 KEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKK 118
+ W + N E ++ P LC L+ D I++ ++ AL + G +
Sbjct 218 RNVTWVMVNLCRHKDPPPPMETIQ-EILPALCVLIHHTDVNILVDTVWALSYLTDAGNE 275
> dre:436607 kpna2, MGC113902, hm:zeh0389, hm:zeh0389r, zgc:113902,
zgc:86945; karyopherin alpha 2 (RAG cohort 1, importin
alpha 1); K15043 importin subunit alpha-2
Length=525
Score = 114 bits (286), Expect = 7e-26, Method: Compositional matrix adjust.
Identities = 59/125 (47%), Positives = 77/125 (61%), Gaps = 0/125 (0%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
ALP +LL + +QKEA WT+SNITAG QIQEV+ +G VP L+ +L D+ T+K
Sbjct 333 ALPMFPALLRHQKSNIQKEASWTLSNITAGRDYQIQEVINAGIVPYLVEVLRRGDYKTQK 392
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
EA WAV+N SGG+ QV LV+ PL LL DS ++V L+A+ I G+KI
Sbjct 393 EAVWAVTNYTSGGTVEQVVYLVQANVLEPLLNLLSTKDSTTILVILDAITNIFLAGEKIG 452
Query 121 ETEGL 125
E E L
Sbjct 453 EVEKL 457
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 30/98 (30%), Positives = 51/98 (52%), Gaps = 2/98 (2%)
Query 2 LPNLLSLLS-SNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
+P + LS + +Q EA W ++NI +G +Q V++ GA+PA + L+ + +
Sbjct 118 IPKFVGFLSLVDSPPIQFEAAWALTNIASGTSDQTSAVVQGGAIPAFISLISSPHAHISE 177
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMD 98
+A WA+ N GS + +++ G PPL LL D
Sbjct 178 QAVWALGNIAGDGSGYR-DRVIKHGAIPPLLALLAVPD 214
Score = 42.0 bits (97), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 22/73 (30%), Positives = 34/73 (46%), Gaps = 5/73 (6%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTK- 59
A+P +SL+SS + ++A W + NI V++ GA+P LL LL D
Sbjct 160 AIPAFISLISSPHAHISEQAVWALGNIAGDGSGYRDRVIKHGAIPPLLALLAVPDLSVFP 219
Query 60 ----KEAAWAVSN 68
+ W +SN
Sbjct 220 AGYLRNVTWTLSN 232
> mmu:16648 Kpna3, IPOA4; karyopherin (importin) alpha 3
Length=521
Score = 109 bits (272), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 52/112 (46%), Positives = 72/112 (64%), Gaps = 0/112 (0%)
Query 2 LPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKE 61
L + +LLS ++ + KEA W +SNITAGNQ+Q+Q V+++G +P ++H L DF T+KE
Sbjct 328 LSHFPNLLSHPKEKINKEAVWFLSNITAGNQQQVQAVIDAGLIPMIIHQLAKGDFGTQKE 387
Query 62 AAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAIL 113
AAWA+SN G QV LV+ PP C LL DS++V V L+ L IL
Sbjct 388 AAWAISNLTISGRKDQVEYLVQQNVIPPFCNLLSVKDSQVVQVVLDGLKNIL 439
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 30/95 (31%), Positives = 49/95 (51%), Gaps = 2/95 (2%)
Query 2 LPNLLSLLS-SNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
LP L+ L + ++Q EA W ++NI +G Q Q V++S AVP L LL + + +
Sbjct 116 LPILVKCLERDDNPSLQFEAAWALTNIASGTSAQTQAVVQSNAVPLFLRLLHSPHQNVCE 175
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLG 95
+A WA+ N + G + ++ LG PL +
Sbjct 176 QAVWALGNIIGDGPQCR-DYVISLGVVKPLLSFIN 209
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 26/71 (36%), Positives = 37/71 (52%), Gaps = 0/71 (0%)
Query 2 LPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKE 61
LP L L+ + + W +S +T G EQIQ V++SG VP L+ LL + +
Sbjct 244 LPALCVLIYHTDINILVDTVWALSYLTDGGNEQIQMVIDSGVVPFLVPLLSHQEVKVQTA 303
Query 62 AAWAVSNAVSG 72
A AV N V+G
Sbjct 304 ALRAVGNIVTG 314
Score = 32.3 bits (72), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 29/119 (24%), Positives = 48/119 (40%), Gaps = 2/119 (1%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETA-DFDTK 59
A+P L LL S + V ++A W + NI + V+ G V LL + +
Sbjct 158 AVPLFLRLLHSPHQNVCEQAVWALGNIIGDGPQCRDYVISLGVVKPLLSFINPSIPITFL 217
Query 60 KEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKK 118
+ W + N E V+ P LC L+ D I++ ++ AL + G +
Sbjct 218 RNVTWVIVNLCRNKDPPPPMETVQ-EILPALCVLIYHTDINILVDTVWALSYLTDGGNE 275
> hsa:3839 KPNA3, IPOA4, SRP1, SRP1gamma, SRP4, hSRP1; karyopherin
alpha 3 (importin alpha 4)
Length=521
Score = 109 bits (272), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 52/112 (46%), Positives = 72/112 (64%), Gaps = 0/112 (0%)
Query 2 LPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKE 61
L + +LLS ++ + KEA W +SNITAGNQ+Q+Q V+++G +P ++H L DF T+KE
Sbjct 328 LSHFPNLLSHPKEKINKEAVWFLSNITAGNQQQVQAVIDAGLIPMIIHQLAKGDFGTQKE 387
Query 62 AAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAIL 113
AAWA+SN G QV LV+ PP C LL DS++V V L+ L IL
Sbjct 388 AAWAISNLTISGRKDQVEYLVQQNVIPPFCNLLSVKDSQVVQVVLDGLKNIL 439
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 30/95 (31%), Positives = 49/95 (51%), Gaps = 2/95 (2%)
Query 2 LPNLLSLLS-SNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
LP L+ L + ++Q EA W ++NI +G Q Q V++S AVP L LL + + +
Sbjct 116 LPILVKCLERDDNPSLQFEAAWALTNIASGTSAQTQAVVQSNAVPLFLRLLRSPHQNVCE 175
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLG 95
+A WA+ N + G + ++ LG PL +
Sbjct 176 QAVWALGNIIGDGPQCR-DYVISLGVVKPLLSFIS 209
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 26/71 (36%), Positives = 37/71 (52%), Gaps = 0/71 (0%)
Query 2 LPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKE 61
LP L L+ + + W +S +T G EQIQ V++SG VP L+ LL + +
Sbjct 244 LPALCVLIYHTDINILVDTVWALSYLTDGGNEQIQMVIDSGVVPFLVPLLSHQEVKVQTA 303
Query 62 AAWAVSNAVSG 72
A AV N V+G
Sbjct 304 ALRAVGNIVTG 314
Score = 32.3 bits (72), Expect = 0.44, Method: Compositional matrix adjust.
Identities = 29/119 (24%), Positives = 48/119 (40%), Gaps = 2/119 (1%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETA-DFDTK 59
A+P L LL S + V ++A W + NI + V+ G V LL + +
Sbjct 158 AVPLFLRLLRSPHQNVCEQAVWALGNIIGDGPQCRDYVISLGVVKPLLSFISPSIPITFL 217
Query 60 KEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKK 118
+ W + N E V+ P LC L+ D I++ ++ AL + G +
Sbjct 218 RNVTWVIVNLCRNKDPPPPMETVQ-EILPALCVLIYHTDINILVDTVWALSYLTDGGNE 275
> dre:327373 kpna4, impa3, wu:fa56d07, wu:fa66g10, wu:fe14c04,
wu:fi04h11, wu:fi19b05; karyopherin alpha 4 (importin alpha
3)
Length=521
Score = 108 bits (270), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 58/139 (41%), Positives = 83/139 (59%), Gaps = 7/139 (5%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
AL + +LL+ ++ + KEA W +SNITAGNQ+Q+Q V+++ VP ++ LL+ DF T+K
Sbjct 327 ALGHFPALLTHPKEKINKEAVWFLSNITAGNQQQVQAVIDANLVPMIILLLDKGDFGTQK 386
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
EAAWA+SN G QVA L++ PP C LL D+++V V L+ L IL+
Sbjct 387 EAAWAISNLTISGRKDQVAYLIQQQVIPPFCNLLTVKDAQVVQVVLDGLSNILKMADDEA 446
Query 121 ETEGLSINPYATLIEQADG 139
ET A LIE+ G
Sbjct 447 ET-------VANLIEECGG 458
Score = 55.1 bits (131), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 31/95 (32%), Positives = 50/95 (52%), Gaps = 2/95 (2%)
Query 2 LPNLLSLLS-SNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
LP L+ L + ++Q EA W ++NI +G EQ Q V++S AVP L LL + + +
Sbjct 116 LPILVHCLDRDDNPSLQFEAAWALTNIASGTSEQTQAVVQSNAVPLFLRLLHSPHQNVCE 175
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLG 95
+A WA+ N + G + ++ LG PL +
Sbjct 176 QAVWALGNIIGDGPQCR-DYVISLGVVKPLLSFIS 209
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 36/71 (50%), Gaps = 0/71 (0%)
Query 2 LPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKE 61
LP L L+ + + W +S +T EQIQ V++SG VP L+ LL + +
Sbjct 244 LPALCVLIHHTDVNILVDTVWALSYLTDAGNEQIQMVIDSGIVPYLVPLLSHQEVKVQTA 303
Query 62 AAWAVSNAVSG 72
A AV N V+G
Sbjct 304 ALRAVGNIVTG 314
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 26/88 (29%), Positives = 48/88 (54%), Gaps = 2/88 (2%)
Query 35 IQEVLESGAVPALLHLLETADFDT-KKEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQL 93
I ++++SG +P L+H L+ D + + EAAWA++N SG S+ Q +V+ P +L
Sbjct 107 IDDLIKSGILPILVHCLDRDDNPSLQFEAAWALTNIASGTSE-QTQAVVQSNAVPLFLRL 165
Query 94 LGCMDSKIVMVSLEALHAILRTGKKIKE 121
L + ++ AL I+ G + ++
Sbjct 166 LHSPHQNVCEQAVWALGNIIGDGPQCRD 193
Score = 32.0 bits (71), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 32/132 (24%), Positives = 54/132 (40%), Gaps = 3/132 (2%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETA-DFDTK 59
A+P L LL S + V ++A W + NI + V+ G V LL + +
Sbjct 158 AVPLFLRLLHSPHQNVCEQAVWALGNIIGDGPQCRDYVISLGVVKPLLSFISPSIPITFL 217
Query 60 KEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTG-KK 118
+ W + N E ++ P LC L+ D I++ ++ AL + G ++
Sbjct 218 RNVTWVMVNLCRHKDPPPPMETIQ-EILPALCVLIHHTDVNILVDTVWALSYLTDAGNEQ 276
Query 119 IKETEGLSINPY 130
I+ I PY
Sbjct 277 IQMVIDSGIVPY 288
> xla:432069 kpna3, MGC78841, ipoa4, srp1, srp1gamma, srp4; karyopherin
alpha 3 (importin alpha 4)
Length=521
Score = 108 bits (270), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 51/112 (45%), Positives = 72/112 (64%), Gaps = 0/112 (0%)
Query 2 LPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKE 61
L + +LL+ ++ + KEA W +SNITAGNQ+Q+Q V+++G +P ++H L DF T+KE
Sbjct 328 LSHFHNLLTHPKEKINKEAVWFLSNITAGNQQQVQAVIDAGLIPMIIHQLAKGDFGTQKE 387
Query 62 AAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAIL 113
AAWA+SN G QV LV+ PP C LL DS++V V L+ L IL
Sbjct 388 AAWAISNLTISGRKDQVEYLVQQNVIPPFCSLLSVKDSQVVQVVLDGLKNIL 439
Score = 52.8 bits (125), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 30/95 (31%), Positives = 50/95 (52%), Gaps = 2/95 (2%)
Query 2 LPNLLSLL-SSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
LP L+ L + + ++Q EA W ++NI +G Q Q V++S AVP L LL + + +
Sbjct 116 LPILVKCLGADDNPSLQFEAAWALTNIASGTSAQTQAVVQSNAVPLFLRLLHSLHQNVCE 175
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLG 95
+A WA+ N + G + ++ LG PL +
Sbjct 176 QAVWALGNIIGDGPQCR-DYVISLGVVKPLLSFIN 209
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 26/71 (36%), Positives = 37/71 (52%), Gaps = 0/71 (0%)
Query 2 LPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKE 61
LP L L+ + + W +S +T G EQIQ V++SG VP L+ LL + +
Sbjct 244 LPALCVLIYHTDINILVDTVWALSYLTDGGNEQIQMVIDSGVVPFLVPLLSHQEVKVQTA 303
Query 62 AAWAVSNAVSG 72
A AV N V+G
Sbjct 304 ALRAVGNIVTG 314
Score = 31.6 bits (70), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 29/119 (24%), Positives = 48/119 (40%), Gaps = 2/119 (1%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETA-DFDTK 59
A+P L LL S + V ++A W + NI + V+ G V LL + +
Sbjct 158 AVPLFLRLLHSLHQNVCEQAVWALGNIIGDGPQCRDYVISLGVVKPLLSFINPSIPITFL 217
Query 60 KEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKK 118
+ W + N E V+ P LC L+ D I++ ++ AL + G +
Sbjct 218 RNVTWVIVNLCRNKDPPPPMETVQ-EILPALCVLIYHTDINILVDTVWALSYLTDGGNE 275
> dre:368640 kpna3, si:dz79f24.2; karyopherin (importin) alpha
3
Length=496
Score = 108 bits (269), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 51/113 (45%), Positives = 72/113 (63%), Gaps = 0/113 (0%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
L + +LL+ ++ + KEA W +SNITAGNQ+Q+Q V+++G +P ++H L DF T+K
Sbjct 303 VLTHFSTLLTHPKEKINKEAVWFLSNITAGNQQQVQAVIDAGLIPMIIHQLAKGDFGTQK 362
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAIL 113
EAAWA+SN G QV LV+ PP C LL DS++V V L+ L IL
Sbjct 363 EAAWAISNLTISGRKDQVEYLVQQNVIPPFCNLLSVKDSQVVQVVLDGLKNIL 415
Score = 52.4 bits (124), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 30/95 (31%), Positives = 49/95 (51%), Gaps = 2/95 (2%)
Query 2 LPNLLSLLS-SNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
LP L+ L + ++Q EA W ++NI +G Q Q V++S AVP L LL + + +
Sbjct 92 LPILVKCLERDDNPSLQFEAAWALTNIASGTSAQTQAVVKSNAVPLFLRLLHSPHQNVCE 151
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLG 95
+A WA+ N + G + ++ LG PL +
Sbjct 152 QAVWALGNIIGDGPQCR-DYVISLGVVKPLLSFIN 185
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 26/71 (36%), Positives = 37/71 (52%), Gaps = 0/71 (0%)
Query 2 LPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKE 61
LP L L+ + + W +S +T G EQIQ V++SG VP L+ LL + +
Sbjct 220 LPALCVLIYHTDINILVDTVWALSYLTDGGNEQIQMVIDSGVVPYLVPLLSHQEVKVQTA 279
Query 62 AAWAVSNAVSG 72
A AV N V+G
Sbjct 280 ALRAVGNIVTG 290
Score = 32.7 bits (73), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 32/132 (24%), Positives = 54/132 (40%), Gaps = 3/132 (2%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETA-DFDTK 59
A+P L LL S + V ++A W + NI + V+ G V LL + +
Sbjct 134 AVPLFLRLLHSPHQNVCEQAVWALGNIIGDGPQCRDYVISLGVVKPLLSFINPSIPITFL 193
Query 60 KEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTG-KK 118
+ W + N E V+ P LC L+ D I++ ++ AL + G ++
Sbjct 194 RNVTWVIVNLCRNKDPPPPMETVQ-EILPALCVLIYHTDINILVDTVWALSYLTDGGNEQ 252
Query 119 IKETEGLSINPY 130
I+ + PY
Sbjct 253 IQMVIDSGVVPY 264
> xla:407837 imp_alpha_4; importin alpha 4 protein
Length=521
Score = 107 bits (268), Expect = 9e-24, Method: Compositional matrix adjust.
Identities = 55/127 (43%), Positives = 77/127 (60%), Gaps = 8/127 (6%)
Query 7 SLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAAWAV 66
+LL+ ++ + KEA W +SNITAGNQ+Q+Q V+++G +P ++H L DF T+KEAAWA+
Sbjct 333 NLLTHPKEKINKEAVWFLSNITAGNQQQVQAVIDAGLIPMIIHQLAKGDFGTQKEAAWAI 392
Query 67 SNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAIL--------RTGKK 118
SN G QV LV+ PP C LL DS++V V L+ L IL +
Sbjct 393 SNLTISGRKDQVEYLVQQNVIPPFCSLLSVKDSQVVQVVLDGLKNILIMAGDEASTIAEI 452
Query 119 IKETEGL 125
I+E EGL
Sbjct 453 IEECEGL 459
Score = 51.6 bits (122), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 30/95 (31%), Positives = 49/95 (51%), Gaps = 2/95 (2%)
Query 2 LPNLLSLL-SSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
LP L+ L + + +Q EA W ++NI +G Q Q V++S AVP L LL + + +
Sbjct 116 LPILVKCLEADDNPPLQFEAAWALTNIASGTSAQTQAVVQSNAVPLFLRLLHSLHQNVCE 175
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLG 95
+A WA+ N + G + ++ LG PL +
Sbjct 176 QAVWALGNIIGDGPQCR-DYVISLGVVKPLLSFIN 209
Score = 45.1 bits (105), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 31/88 (35%), Positives = 43/88 (48%), Gaps = 3/88 (3%)
Query 2 LPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKE 61
LP L L+ + + W +S +T G EQIQ V++SG P L+ LL + +
Sbjct 244 LPALCVLIYHTDFNILVDTVWALSYLTDGGNEQIQMVIDSGVAPFLVPLLSHQEVKVQTA 303
Query 62 AAWAVSNAVSGGSDMQVAELVRLGCAPP 89
A AV N V+ G+D Q V L C P
Sbjct 304 ALRAVGNIVT-GTDEQTQ--VVLNCDVP 328
Score = 37.0 bits (84), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 31/55 (56%), Gaps = 1/55 (1%)
Query 2 LPNLLSLLSSNRKTVQKEACWTISNIT-AGNQEQIQEVLESGAVPALLHLLETAD 55
+P ++ L+ QKEA W ISN+T +G ++Q++ +++ +P LL D
Sbjct 370 IPMIIHQLAKGDFGTQKEAAWAISNLTISGRKDQVEYLVQQNVIPPFCSLLSVKD 424
Score = 35.0 bits (79), Expect = 0.069, Method: Compositional matrix adjust.
Identities = 36/156 (23%), Positives = 57/156 (36%), Gaps = 44/156 (28%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAV---------------- 44
A+P L LL S + V ++A W + NI + V+ G V
Sbjct 158 AVPLFLRLLHSLHQNVCEQAVWALGNIIGDGPQCRDYVISLGVVKPLLSFINPSIPITFL 217
Query 45 ---------------------------PALLHLLETADFDTKKEAAWAVSNAVSGGSDMQ 77
PAL L+ DF+ + WA+S GG++ Q
Sbjct 218 RNVTWVIVNLCRNKDPPPPMETVQEILPALCVLIYHTDFNILVDTVWALSYLTDGGNE-Q 276
Query 78 VAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAIL 113
+ ++ G AP L LL + K+ +L A+ I+
Sbjct 277 IQMVIDSGVAPFLVPLLSHQEVKVQTAALRAVGNIV 312
> ath:AT5G52000 IMPA-8; IMPA-8 (IMPORTIN ALPHA ISOFORM 8); binding
/ protein transporter
Length=441
Score = 104 bits (260), Expect = 7e-23, Method: Compositional matrix adjust.
Identities = 56/141 (39%), Positives = 85/141 (60%), Gaps = 4/141 (2%)
Query 1 ALPNLLSLLSSNRKT-VQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTK 59
ALP + ++L+ N + ++K ACW ISNITAG +EQIQ V+++ +P L++L + DF K
Sbjct 255 ALPIISNMLTRNHENKIKKCACWVISNITAGTKEQIQSVIDANLIPILVNLAQDTDFYMK 314
Query 60 KEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVS-LEALHAILRTGKK 118
KEA WA+SN GS Q+ + C LC +L D + ++ L+ L +L+ G+
Sbjct 315 KEAVWAISNMALNGSHDQIKYMAEQSCIKQLCDILVYSDERTTILKCLDGLENMLKAGEA 374
Query 119 IKETEGLSINPYATLIEQADG 139
K +E +NPY LIE A+G
Sbjct 375 EKNSE--DVNPYCLLIEDAEG 393
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 21/52 (40%), Positives = 35/52 (67%), Gaps = 0/52 (0%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLE 52
ALP L LL S+ + V K AC + +++ G+++ IQ V+E+G VP L+ +L+
Sbjct 171 ALPALEILLHSHDEDVLKNACMALCHLSEGSEDGIQSVIEAGFVPKLVQILQ 222
Score = 38.1 bits (87), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 36/68 (52%), Gaps = 2/68 (2%)
Query 5 LLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAAW 64
L+ L++S + V+++A WT+SN+ + VL SG + LL LL T + A W
Sbjct 94 LIQLIASPKDYVREQAIWTLSNVAGHSIHYRDFVLNSGVLMPLLRLLYKD--TTLRIATW 151
Query 65 AVSNAVSG 72
A+ N G
Sbjct 152 ALRNLCRG 159
> xla:398026 kpna7, kpna1; karyopherin alpha 7 (importin alpha
8)
Length=522
Score = 99.4 bits (246), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 53/121 (43%), Positives = 72/121 (59%), Gaps = 0/121 (0%)
Query 5 LLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAAW 64
L LL + ++QKEA W ISNI AG QIQ+++ G + L+ LL DF +KEA W
Sbjct 333 LPQLLRHQKPSIQKEAAWAISNIAAGPAPQIQQMITCGLLSPLVDLLNKGDFKAQKEAVW 392
Query 65 AVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIKETEG 124
AV+N SGG+ QV +LV+ G PL LL DSK ++V L+A+ I +K+ E E
Sbjct 393 AVTNYTSGGTVEQVVQLVQCGVLEPLLNLLTIKDSKTILVILDAISNIFLAAEKLGEQEK 452
Query 125 L 125
L
Sbjct 453 L 453
Score = 59.7 bits (143), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 29/95 (30%), Positives = 52/95 (54%), Gaps = 2/95 (2%)
Query 2 LPNLLSLLSS-NRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
+P L+ LS + T+Q EA W ++NI +G +Q + V++ GA+PA + L+ + +
Sbjct 118 IPKLVEFLSRHDNSTLQFEAAWALTNIASGTSDQTKSVVDGGAIPAFISLISSPHLHISE 177
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLG 95
+A WA+ N ++G + L+ PPL L+
Sbjct 178 QAVWALGN-IAGDGPLYRDALINCNVIPPLLALVN 211
Score = 31.2 bits (69), Expect = 0.97, Method: Compositional matrix adjust.
Identities = 25/103 (24%), Positives = 41/103 (39%), Gaps = 2/103 (1%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLET-ADFDTK 59
A+P +SL+SS + ++A W + NI ++ +P LL L+
Sbjct 160 AIPAFISLISSPHLHISEQAVWALGNIAGDGPLYRDALINCNVIPPLLALVNPQTPLGYL 219
Query 60 KEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIV 102
+ W +SN + V L P L QL+ D I+
Sbjct 220 RNITWTLSNLCRNKNPYPPMSAV-LQILPVLTQLMHHDDKDIL 261
> xla:398027 kpna2, MGC154872, MGC86340; importin alpha 1b; K15043
importin subunit alpha-2
Length=523
Score = 98.6 bits (244), Expect = 5e-21, Method: Compositional matrix adjust.
Identities = 52/121 (42%), Positives = 73/121 (60%), Gaps = 0/121 (0%)
Query 5 LLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAAW 64
L LL + ++QKEA W +SNI AG QIQ+++ G + L+ LL+ DF +KEA W
Sbjct 333 LPQLLRHQKPSIQKEAAWALSNIAAGPAPQIQQMITCGLLSPLVDLLKKGDFKAQKEAVW 392
Query 65 AVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIKETEG 124
AV+N SGG+ QV +LV+ G PL LL DSK ++V L+A+ I +K+ E E
Sbjct 393 AVTNYTSGGTVEQVVQLVQCGVLEPLLNLLTIKDSKTILVILDAISNIFLAAEKLGEQEK 452
Query 125 L 125
L
Sbjct 453 L 453
Score = 61.2 bits (147), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 30/95 (31%), Positives = 52/95 (54%), Gaps = 2/95 (2%)
Query 2 LPNLLSLLS-SNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
+P L+ LS N T+Q EA W ++NI +G +Q + V++ GA+PA + L+ + +
Sbjct 118 IPKLVEFLSYHNNSTLQFEAAWALTNIASGTSDQTKSVVDGGAIPAFISLISSPHLHISE 177
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLG 95
+A WA+ N ++G + L+ PPL L+
Sbjct 178 QAVWALGN-IAGDGPLYRDALISCNVIPPLLTLVN 211
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 27/93 (29%), Positives = 46/93 (49%), Gaps = 1/93 (1%)
Query 2 LPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKE 61
LP L L+ K + + CW +S +T G+ ++I V+++G V L+ L+ + +
Sbjct 246 LPVLTQLMLHEDKDILSDTCWAMSYLTDGSNDRIDVVVKTGLVERLIQLMYSPELSILTP 305
Query 62 AAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLL 94
+ V N V+ G+D Q + G L QLL
Sbjct 306 SLRTVGNIVT-GTDKQTQAAIDAGVLSVLPQLL 337
Score = 30.4 bits (67), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 29/104 (27%), Positives = 46/104 (44%), Gaps = 4/104 (3%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLES-GAVPALLHLLET-ADFDT 58
A+P +SL+SS + ++A W + NI AG+ ++ L S +P LL L+
Sbjct 160 AIPAFISLISSPHLHISEQAVWALGNI-AGDGPLYRDALISCNVIPPLLTLVNPQTPLGY 218
Query 59 KKEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIV 102
+ W +SN + V L P L QL+ D I+
Sbjct 219 LRNITWTLSNLCRNKNPYPPMSAV-LQILPVLTQLMLHEDKDIL 261
> dre:406343 fi21b05, wu:fi21b05, wu:fj93d06; zgc:55877
Length=520
Score = 94.0 bits (232), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 54/139 (38%), Positives = 79/139 (56%), Gaps = 8/139 (5%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
ALP L+ + +VQKEA W +SNI AG +QIQ+++ G L+ LL DF T++
Sbjct 332 ALP---KLMRHQKSSVQKEAAWAVSNIAAGPCQQIQQLITCGLSAPLVDLLRNGDFKTQR 388
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIK 120
EA WAV+N SGG+ QV LV+ G + LL D+K V+V L+A+ I +K+
Sbjct 389 EAVWAVTNYTSGGTVEQVVHLVKCGALEAILNLLQVKDTKTVLVILDAISNIFLAAEKLG 448
Query 121 ETEGLSINPYATLIEQADG 139
E + L + L+E+ G
Sbjct 449 EVDKLCL-----LVEEVGG 462
Score = 55.8 bits (133), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 31/90 (34%), Positives = 48/90 (53%), Gaps = 2/90 (2%)
Query 2 LPNLLSLLSSNRK-TVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
L + L N T+Q EA W+++N+ +G Q+V+E GAVPA + LL + + +
Sbjct 118 LTRFVEFLGRNDDPTLQFEAAWSLTNVASGTSWHTQQVVEHGAVPAFIALLASPMLNISE 177
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPL 90
+A WA+ N GS + A L+ G P L
Sbjct 178 QAVWALGNIAGDGSSYRDA-LIDCGVIPAL 206
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 35/99 (35%), Positives = 51/99 (51%), Gaps = 1/99 (1%)
Query 2 LPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKE 61
L L+ LL V A +I NI +G+ Q Q +++G + AL L+ +KE
Sbjct 288 LTRLVELLGFEELAVVTPALRSIENIVSGSDLQTQAAIDAGVLSALPKLMRHQKSSVQKE 347
Query 62 AAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSK 100
AAWAVSN ++ G Q+ +L+ G + PL LL D K
Sbjct 348 AAWAVSN-IAAGPCQQIQQLITCGLSAPLVDLLRNGDFK 385
Score = 35.4 bits (80), Expect = 0.056, Method: Compositional matrix adjust.
Identities = 26/103 (25%), Positives = 44/103 (42%), Gaps = 2/103 (1%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALL-HLLETADFDTK 59
A+P ++LL+S + ++A W + NI +++ G +PALL L A
Sbjct 160 AVPAFIALLASPMLNISEQAVWALGNIAGDGSSYRDALIDCGVIPALLARLTPDAPVGYL 219
Query 60 KEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIV 102
+ W +SN + V+ P + QLL D I+
Sbjct 220 RNLTWTLSNLCRNKNPFPRFSAVQ-QMLPSIIQLLHHSDKSIL 261
> hsa:402569 KPNA7; karyopherin alpha 7 (importin alpha 8)
Length=516
Score = 90.9 bits (224), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 51/135 (37%), Positives = 76/135 (56%), Gaps = 5/135 (3%)
Query 5 LLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAAW 64
L LL N+ ++QKEA W +SN+ AG IQ++L +P L+ LL+ +F +KEA W
Sbjct 330 LPQLLQHNKPSIQKEAAWALSNVAAGPCHHIQQLLAYDVLPPLVALLKNGEFKVQKEAVW 389
Query 65 AVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGKKIKETEG 124
V+N +G + Q+ +LV G PL LL D KIV++ L+ + IL+ +K E E
Sbjct 390 MVANFATGATMDQLIQLVHSGVLEPLVNLLTAPDVKIVLIILDVISCILQAAEKRSEKEN 449
Query 125 LSINPYATLIEQADG 139
L + LIE+ G
Sbjct 450 LCL-----LIEELGG 459
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 44/77 (57%), Gaps = 1/77 (1%)
Query 2 LPNLLSLLSSN-RKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
+P ++ L S+ +Q EA W ++NI +G EQ + V+E GA+ L+ LL +++ +
Sbjct 115 IPRMVEFLKSSLYPCLQFEAAWALTNIASGTSEQTRAVVEGGAIQPLIELLSSSNVAVCE 174
Query 61 EAAWAVSNAVSGGSDMQ 77
+A WA+ N G + +
Sbjct 175 QAVWALGNIAGDGPEFR 191
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 33/114 (28%), Positives = 59/114 (51%), Gaps = 2/114 (1%)
Query 1 ALPNLLSLLSSNRK-TVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTK 59
A+P+LL+L+S T + WT+SN+ + +PALLHLL+ D +
Sbjct 199 AIPHLLALISPTLPITFLRNITWTLSNLCRNKNPYPCDTAVKQILPALLHLLQHQDSEVL 258
Query 60 KEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAIL 113
+A WA+S ++ GS+ ++ ++V G P L L+ + ++ SL + I+
Sbjct 259 SDACWALS-YLTDGSNKRIGQVVNTGVLPRLVVLMTSSELNVLTPSLRTVGNIV 311
Score = 48.5 bits (114), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 30/93 (32%), Positives = 50/93 (53%), Gaps = 1/93 (1%)
Query 2 LPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKE 61
LP LL LL V +ACW +S +T G+ ++I +V+ +G +P L+ L+ +++ +
Sbjct 243 LPALLHLLQHQDSEVLSDACWALSYLTDGSNKRIGQVVNTGVLPRLVVLMTSSELNVLTP 302
Query 62 AAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLL 94
+ V N V+ G+D Q + G L QLL
Sbjct 303 SLRTVGNIVT-GTDEQTQMAIDAGMLNVLPQLL 334
Score = 38.9 bits (89), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 33/55 (60%), Gaps = 1/55 (1%)
Query 2 LPNLLSLLSSNRKTVQKEACWTISNITAG-NQEQIQEVLESGAVPALLHLLETAD 55
LP L++LL + VQKEA W ++N G +Q+ +++ SG + L++LL D
Sbjct 369 LPPLVALLKNGEFKVQKEAVWMVANFATGATMDQLIQLVHSGVLEPLVNLLTAPD 423
> cel:F26B1.3 ima-2; IMportin Alpha family member (ima-2)
Length=531
Score = 84.0 bits (206), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 42/116 (36%), Positives = 72/116 (62%), Gaps = 1/116 (0%)
Query 1 ALPNLLSLLSSNRKT-VQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTK 59
+L +L L+ R + + KE CW +SNI AG Q+QIQ VL++ +P L+++L++ D +
Sbjct 345 SLDEILPLMEKTRSSSIVKECCWLVSNIIAGTQKQIQAVLDANLLPVLINVLKSGDHKCQ 404
Query 60 KEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRT 115
EA+WA+SN GG++ QV ++ P LCQ L ++ ++ +LE L+ ++ T
Sbjct 405 FEASWALSNLAQGGTNRQVVAMLEDNVVPALCQALLQTNTDMLNNTLETLYTLMLT 460
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 25/76 (32%), Positives = 39/76 (51%), Gaps = 0/76 (0%)
Query 2 LPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKE 61
L L+ LS + VQ EA W ++NI +G+ EQ +E+G L+HL ++
Sbjct 134 LQALVQALSVENERVQYEAAWALTNIVSGSTEQTIAAVEAGVTIPLIHLSVHQSAQISEQ 193
Query 62 AAWAVSNAVSGGSDMQ 77
A WAV+N S ++
Sbjct 194 ALWAVANIAGDSSQLR 209
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 31/51 (60%), Gaps = 0/51 (0%)
Query 5 LLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETAD 55
L+ L+ + V+++ACW +S +T G EQI+ ESG +P ++ + A+
Sbjct 266 LVKLVQHTDRQVRQDACWAVSYLTDGPDEQIELARESGVLPHVVAFFKEAE 316
Score = 37.4 bits (85), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 22/59 (37%), Positives = 32/59 (54%), Gaps = 1/59 (1%)
Query 35 IQEVLESGAVPALLHLLETADFDTKKEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQL 93
I EV+ G + AL+ L + + EAAWA++N VSG ++ +A V G PL L
Sbjct 125 IDEVIHCGLLQALVQALSVENERVQYEAAWALTNIVSGSTEQTIA-AVEAGVTIPLIHL 182
Score = 34.3 bits (77), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 21/57 (36%), Positives = 36/57 (63%), Gaps = 5/57 (8%)
Query 16 VQKEACWTISNITAGNQEQIQE-VLESGAVPALLHLLETADF--DTK-KEAAWAVSN 68
+ ++A W ++NI AG+ Q+++ V++ V AL+HL+E D D+ + AWA SN
Sbjct 190 ISEQALWAVANI-AGDSSQLRDYVIKCHGVEALMHLMEKVDQLGDSHVRTIAWAFSN 245
> ath:AT1G32880 importin alpha-1 subunit, putative
Length=183
Score = 69.7 bits (169), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 36/102 (35%), Positives = 58/102 (56%), Gaps = 1/102 (0%)
Query 34 QIQEVLESGAVPALLHLLETADFDTKKEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQL 93
++Q V+++ +P L+ L + A+FD KKE+ A+SNA GS Q+ +V C PLC +
Sbjct 40 KLQSVIDANLIPTLVKLTQNAEFDMKKESVCAISNATLLGSHDQIKYMVEQSCIKPLCDI 99
Query 94 LGCMDSKIVMVSLEALHAILRTGKKIKETEGLSINPYATLIE 135
L C D K ++ L+ + L+ G+ K G ++ Y LIE
Sbjct 100 LFCPDVKTILKCLDGMENTLKVGEAEKNA-GDDVSWYTRLIE 140
> cel:T19B10.7 ima-1; IMportin Alpha family member (ima-1)
Length=524
Score = 58.5 bits (140), Expect = 5e-09, Method: Composition-based stats.
Identities = 27/106 (25%), Positives = 59/106 (55%), Gaps = 0/106 (0%)
Query 12 NRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAAWAVSNAVS 71
N + +E C+ ISNI ++ I +++ SG + + ++E +++ +++EAA+ + + +
Sbjct 333 NVSYMSQEVCFIISNICVEGEQTIDKLISSGVLREVARVMEASEYRSRREAAFVICHCCA 392
Query 72 GGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILRTGK 117
+ + +V LG LL CMD +V L+A++ +L+ G+
Sbjct 393 SANQKHLEYVVELGMLSAFTDLLTCMDVSLVSYILDAIYLLLQFGE 438
> ath:AT5G03070 IMPA-9; IMPA-9 (IMPORTIN ALPHA ISOFORM 9); binding
Length=519
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 36/128 (28%), Positives = 63/128 (49%), Gaps = 12/128 (9%)
Query 9 LSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAAWAVSN 68
L S + ++KEA W +SNI AG+ E + + + +P LL +L T+ FD +KE A+ + N
Sbjct 354 LRSEHRVLKKEAAWVLSNIAAGSIEHKRMIHSTEVMPLLLRILSTSPFDIRKEVAYVLGN 413
Query 69 ----AVSGGSDMQVAE-----LVRLGCAPPLCQLLGCMDSKIVMVSLEALHAILR---TG 116
+ G ++ + +V GC +L+ D + + L+ + +LR G
Sbjct 414 LCVESAEGDRKPRIIQEHLVSIVSGGCLRGFIELVRSPDIEAARLGLQFIELVLRGMPNG 473
Query 117 KKIKETEG 124
+ K EG
Sbjct 474 EGPKLVEG 481
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/70 (35%), Positives = 39/70 (55%), Gaps = 2/70 (2%)
Query 16 VQKEACWTISNITAGNQEQIQEVLES-GAVPALLHLLETADFDTKKEAAWAVSNAVSGGS 74
V ++ W I N+ AG E ++ VL S GA+P L ++ T + AAWA+SN + G
Sbjct 184 VAEQCAWAIGNV-AGEGEDLRNVLLSQGALPPLARMIFPDKGSTVRTAAWALSNLIKGPE 242
Query 75 DMQVAELVRL 84
A+LV++
Sbjct 243 SKAAAQLVKI 252
Score = 38.5 bits (88), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 28/95 (29%), Positives = 49/95 (51%), Gaps = 3/95 (3%)
Query 1 ALPNLLSLLSSNRKTVQK-EACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTK 59
A+P L+ LS Q E+ W ++NI AG E+ + +L + + + HL E +
Sbjct 127 AIPLLVQCLSFGSPDEQLLESAWCLTNIAAGKPEETKALLPALPL-LIAHLGEKSSAPVA 185
Query 60 KEAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLL 94
++ AWA+ N G D++ L+ G PPL +++
Sbjct 186 EQCAWAIGNVAGEGEDLRNV-LLSQGALPPLARMI 219
> ath:AT5G13060 ABAP1; ABAP1 (ARMADILLO BTB ARABIDOPSIS PROTEIN
1); protein binding
Length=737
Score = 52.4 bits (124), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 34/109 (31%), Positives = 59/109 (54%), Gaps = 1/109 (0%)
Query 1 ALPNLLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKK 60
ALP L+ +L S TV EA I N+ + + +EV+ +GA+ ++ LL + +T++
Sbjct 268 ALPTLVLMLQSQDSTVHGEAIGAIGNLVHSSPDIKKEVIRAGALQPVIGLLSSTCLETQR 327
Query 61 EAAWAVSNAVSGGSDMQVAELVRLGCAPPLCQLLGCMDSKIVMVSLEAL 109
EAA + + SD +V + + G PL ++L D ++V +S AL
Sbjct 328 EAALLIGQFAAPDSDCKV-HIAQRGAITPLIKMLESSDEQVVEMSAFAL 375
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 34/136 (25%), Positives = 65/136 (47%), Gaps = 3/136 (2%)
Query 5 LLSLLSSNRKTVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLETADFDTKKEAAW 64
L+ LL+ VQ+ A + ++ N E +++E A+P L+ +L++ D EA
Sbjct 230 LVELLNFPDVKVQRAAAGALRTVSFRNDENKSQIVELNALPTLVLMLQSQDSTVHGEAIG 289
Query 65 AVSNAVSGGSDMQVAELVRLGCAPPLCQLLG--CMDSKIVMVSLEALHAILRTGKKIKET 122
A+ N V D++ E++R G P+ LL C++++ L A + K+
Sbjct 290 AIGNLVHSSPDIK-KEVIRAGALQPVIGLLSSTCLETQREAALLIGQFAAPDSDCKVHIA 348
Query 123 EGLSINPYATLIEQAD 138
+ +I P ++E +D
Sbjct 349 QRGAITPLIKMLESSD 364
> tgo:TGME49_094770 hypothetical protein
Length=710
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 41/76 (53%), Gaps = 1/76 (1%)
Query 15 TVQKEACWTISNITAGNQEQIQEVLESGAVPALLHLLET-ADFDTKKEAAWAVSNAVSGG 73
T + AC T+ N+ +Q+Q ++E+ A P L+ + + D++T+ EAA+AV +S
Sbjct 464 TTRARACNTLGNLGCETPKQVQPIIETRAFPLLVEIFQMDPDYNTRIEAAYAVCACLSRA 523
Query 74 SDMQVAELVRLGCAPP 89
QV ++ PP
Sbjct 524 DSQQVGYIISCTARPP 539
Lambda K H
0.314 0.129 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2487377096
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40