bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0260_orf2
Length=261
Score E
Sequences producing significant alignments: (Bits) Value
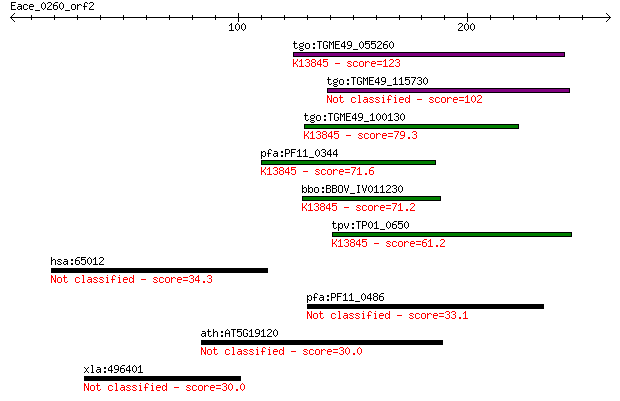
tgo:TGME49_055260 apical membrane antigen 1, putative ; K13845... 123 8e-28
tgo:TGME49_115730 apical membrane antigen, putative 102 2e-21
tgo:TGME49_100130 apical membrane antigen, putative ; K13845 a... 79.3 1e-14
pfa:PF11_0344 AMA1, AMA-1, Pf83, RMA-1, RMA1; apical membrane ... 71.6 3e-12
bbo:BBOV_IV011230 23.m06405; apical membrane antigen 1; K13845... 71.2 4e-12
tpv:TP01_0650 apical membrane antigen 1; K13845 apical merozoi... 61.2 3e-09
hsa:65012 SLC26A10, FLJ32854; solute carrier family 26, member... 34.3 0.53
pfa:PF11_0486 MAEBL, putative 33.1 1.1
ath:AT5G19120 aspartic-type endopeptidase 30.0 8.0
xla:496401 SLC26A3 anion exchanger 30.0 8.7
> tgo:TGME49_055260 apical membrane antigen 1, putative ; K13845
apical merozoite antigen 1
Length=569
Score = 123 bits (308), Expect = 8e-28, Method: Compositional matrix adjust.
Identities = 59/119 (49%), Positives = 81/119 (68%), Gaps = 2/119 (1%)
Query 124 TLQVSGNSNPFL-EPPLADFMSRFDIPKNHGSGIYVDLGSEKEVDGQMYREPSGRCPVFG 182
TL S + NPF + FM RF++ +H SGIYVDLG +KEVDG +YREP+G CP++G
Sbjct 62 TLSASTSGNPFQANVEMKTFMERFNLTHHHQSGIYVDLGQDKEVDGTLYREPAGLCPIWG 121
Query 183 KNIQLHQPPNNPHYKNDFLVDIPSKAESDAAGNPLPGGFNNSFKLPDKTPYSPMTAKIL 241
K+I+L Q P+ P Y+N+FL D+P++ E +GNPLPGGFN +F P SP ++L
Sbjct 122 KHIELQQ-PDRPPYRNNFLEDVPTEKEYKQSGNPLPGGFNLNFVTPSGQRISPFPMELL 179
> tgo:TGME49_115730 apical membrane antigen, putative
Length=388
Score = 102 bits (253), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 52/105 (49%), Positives = 66/105 (62%), Gaps = 9/105 (8%)
Query 139 LADFMSRFDIPKNHGSGIYVDLGSEKEVDGQMYREPSGRCPVFGKNIQLHQPPNNPHYKN 198
ADFM RF+IP+ HGSGI+VDLG D + YRE G+CPVFGK IQ+HQP Y N
Sbjct 106 FADFMKRFNIPQVHGSGIFVDLGR----DTEGYREVGGKCPVFGKAIQMHQP---AEYSN 158
Query 199 DFLVDIPSKAESDAAGNPLPGGFNNSFKLPDKTPYSPMTAKILAD 243
+FL D P+ +DA+ PLPGGFNN +SP+ +L +
Sbjct 159 NFLDDAPT--SNDASKKPLPGGFNNPQVYTSGQKFSPIDDSLLQE 201
> tgo:TGME49_100130 apical membrane antigen, putative ; K13845
apical merozoite antigen 1
Length=493
Score = 79.3 bits (194), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 41/94 (43%), Positives = 59/94 (62%), Gaps = 7/94 (7%)
Query 129 GNSNPFLEPPLADFMSRFDIPKNHGSGIYVDLGSEKEVDGQMYREPSGRCPVFGKNIQLH 188
G++NP+ A + R+++P HGSG+YVDLG+ K + + YREP G+CP +GK I+ +
Sbjct 52 GDANPW-----AKILERYNVPLVHGSGVYVDLGNTKILSKKKYREPGGKCPNYGKYIKTY 106
Query 189 QPPNNPH-YKNDFLVDIPSKAESDAAGNPLPGGF 221
QP NP + NDFL +P A + PL GGF
Sbjct 107 QPTTNPEIWPNDFLKPVP-YANTPQDTMPLGGGF 139
> pfa:PF11_0344 AMA1, AMA-1, Pf83, RMA-1, RMA1; apical membrane
antigen 1, AMA1; K13845 apical merozoite antigen 1
Length=622
Score = 71.6 bits (174), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 33/76 (43%), Positives = 48/76 (63%), Gaps = 1/76 (1%)
Query 110 AAAADVPPTAALVATLQVSGNSNPFLEPPLADFMSRFDIPKNHGSGIYVDLGSEKEVDGQ 169
A P L +++++ SN ++ P ++M+++DI + HGSGI VDLG + EV G
Sbjct 82 EGAEPAPQEQNLFSSIEIVERSN-YMGNPWTEYMAKYDIEEVHGSGIRVDLGEDAEVAGT 140
Query 170 MYREPSGRCPVFGKNI 185
YR PSG+CPVFGK I
Sbjct 141 QYRLPSGKCPVFGKGI 156
> bbo:BBOV_IV011230 23.m06405; apical membrane antigen 1; K13845
apical merozoite antigen 1
Length=605
Score = 71.2 bits (173), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 32/60 (53%), Positives = 39/60 (65%), Gaps = 0/60 (0%)
Query 128 SGNSNPFLEPPLADFMSRFDIPKNHGSGIYVDLGSEKEVDGQMYREPSGRCPVFGKNIQL 187
SG S + P +M +FDIP+NHGSGIYVDLG + V + YR P G+CPV GK I L
Sbjct 86 SGGSKNSGQSPWIKYMQKFDIPRNHGSGIYVDLGGYESVGSKSYRMPVGKCPVVGKIIDL 145
> tpv:TP01_0650 apical membrane antigen 1; K13845 apical merozoite
antigen 1
Length=785
Score = 61.2 bits (147), Expect = 3e-09, Method: Composition-based stats.
Identities = 41/106 (38%), Positives = 53/106 (50%), Gaps = 9/106 (8%)
Query 141 DFMSRFDIPKNHGSGIYVDLGSEKEVDGQMYREPSGRCPVFGKNIQLHQPPNNPHYKNDF 200
+FM++FDI K HGSG+YVDLG V YR P G+CPV GK I L DF
Sbjct 252 EFMAKFDIAKVHGSGVYVDLGESATVGIYDYRMPIGKCPVVGKAIILENGA-------DF 304
Query 201 LVDIPSKAESDAA-GNPLPGGFNNSFKLP-DKTPYSPMTAKILADW 244
L I + G P +NS K + SP++A++L W
Sbjct 305 LSSITHHDPKERGLGFPATKVASNSSKQDMENQLLSPISAQVLRSW 350
> hsa:65012 SLC26A10, FLJ32854; solute carrier family 26, member
10; K14707 solute carrier family 26, member 10
Length=563
Score = 34.3 bits (77), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 30/101 (29%), Positives = 47/101 (46%), Gaps = 7/101 (6%)
Query 19 PSPTRHLSSSSLPA-AFTSLAHLPSVLYLFTSFF-VFSFAFISTMRPLRSAAFPWAAPLL 76
PSP S+S+P AF LA +P V L+TSFF V ++ + T R L + F + +
Sbjct 28 PSPQHTFPSTSIPGMAFALLASVPPVFGLYTSFFPVLIYSLLGTGRHLSTGTFAILSLMT 87
Query 77 G-----FLSVFLLATVQGVVVKDKNLHRTNSEAAAAPAAAA 112
G + L+ + G+ + + R AA A + A
Sbjct 88 GSAVERLVPEPLVGNLSGIEKEQLDAQRVGVAAAVAFGSGA 128
> pfa:PF11_0486 MAEBL, putative
Length=2055
Score = 33.1 bits (74), Expect = 1.1, Method: Composition-based stats.
Identities = 33/116 (28%), Positives = 46/116 (39%), Gaps = 15/116 (12%)
Query 130 NSNPFLEPPLADFMSRFDIPKNH------GSGIYVDLGSEKEVDGQ------MYREPSGR 177
N+ + P +FM RFDI KNH SGI+ + E++ + E S
Sbjct 16 NNTSAINNPQEEFMDRFDINKNHVNIKWSNSGIHGKGKFKYEIEERDVLSEGNESEKSTI 75
Query 178 CPVFGKNIQLHQPPNNPHYKNDFLVDIPSKAESDAAGNPLPGGF-NNSFKLPDKTP 232
CP K P Y FL+ S+ N + GF N +KLP + P
Sbjct 76 CPNHVKEGTYKL--GCPDYGKTFLMGFEDNKYSEEFLNEISFGFLNKKYKLPIEIP 129
> ath:AT5G19120 aspartic-type endopeptidase
Length=386
Score = 30.0 bits (66), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 30/107 (28%), Positives = 46/107 (42%), Gaps = 8/107 (7%)
Query 84 LATVQGVVVKDKNLHRTNSEAAAAPAAAAADVPPTA--ALVATLQVSGNSNPFLEPPLAD 141
L+TV + + ++++ +EA A A A VPP A L T V P ++ L
Sbjct 260 LSTVVPYTILESSIYKVFAEAYAKAAGEATSVPPVAPFGLCFTSDV---DFPAVDLALQS 316
Query 142 FMSRFDIPKNHGSGIYVDLGSEKEVDGQMYREPSGRCPVFGKNIQLH 188
M R+ I HG + VD+G G + S P+ +QL
Sbjct 317 EMVRWRI---HGKNLMVDVGGGVRCSGIVDGGSSRVNPIVMGGLQLE 360
> xla:496401 SLC26A3 anion exchanger
Length=788
Score = 30.0 bits (66), Expect = 8.7, Method: Composition-based stats.
Identities = 17/69 (24%), Positives = 35/69 (50%), Gaps = 1/69 (1%)
Query 33 AFTSLAHLPSVLYLFTSFF-VFSFAFISTMRPLRSAAFPWAAPLLGFLSVFLLATVQGVV 91
AF LA +P +++SFF + ++ F+ T + + FP + ++G + + + +V
Sbjct 108 AFALLAEVPVGYGIYSSFFPILTYFFLGTSKHISVGPFPVISLMVGSVVLSMAPNENFIV 167
Query 92 VKDKNLHRT 100
NL+ T
Sbjct 168 YNTTNLNET 176
Lambda K H
0.321 0.135 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 9660501028
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40