bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
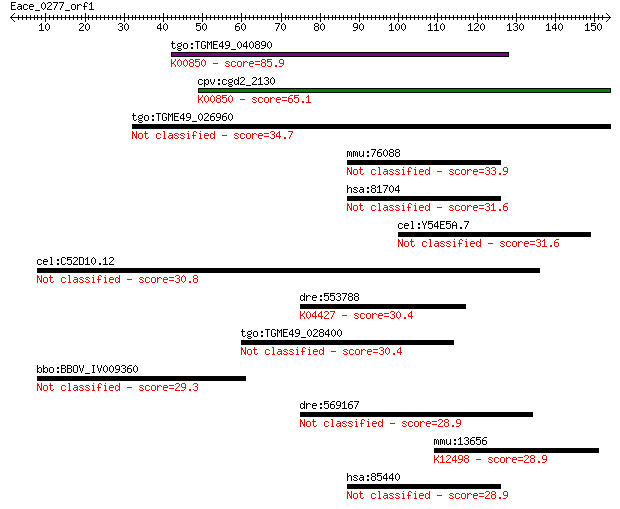
Query= Eace_0277_orf1
Length=153
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_040890 phosphofructokinase, putative (EC:2.7.1.90);... 85.9 5e-17
cpv:cgd2_2130 pyrophosphate-dependent phosphofructokinase (EC:... 65.1 9e-11
tgo:TGME49_026960 phosphofructokinase, putative (EC:2.7.1.90) 34.7 0.13
mmu:76088 Dock8, 1200017A24Rik, 5830472H07Rik, A130095G14Rik, ... 33.9 0.22
hsa:81704 DOCK8, FLJ00026, FLJ00152, FLJ00346, MRD2, ZIR8; ded... 31.6 1.0
cel:Y54E5A.7 hypothetical protein 31.6 1.2
cel:C52D10.12 hypothetical protein 30.8 1.8
dre:553788 map3k7, MGC110612, si:ch211-225h9.1, zgc:110612; mi... 30.4 2.1
tgo:TGME49_028400 hypothetical protein 30.4 2.3
bbo:BBOV_IV009360 23.m06219; hypothetical protein 29.3 5.1
dre:569167 pkhd-l2; si:ch211-244a23.1 28.9 5.9
mmu:13656 Egr4, NGF1-C, NGFI-C, NGFIC, pAT133; early growth re... 28.9 6.1
hsa:85440 DOCK7, KIAA1771, ZIR2; dedicator of cytokinesis 7 28.9
> tgo:TGME49_040890 phosphofructokinase, putative (EC:2.7.1.90);
K00850 6-phosphofructokinase [EC:2.7.1.11]
Length=1399
Score = 85.9 bits (211), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 44/93 (47%), Positives = 60/93 (64%), Gaps = 7/93 (7%)
Query 42 LKEQETNGKVHLHSADLFERSSPLQRLRRAWRCTLPSALESTQLRLVEA---DCPV---- 94
+ ++E + HLHSADLF+R+SP+Q+ R+AW CTLP+AL++ LV D V
Sbjct 119 VHQREQRARTHLHSADLFDRTSPVQKARQAWTCTLPAALDADTHTLVSGPPLDAKVKEAF 178
Query 95 EHPSLRQTPEVQQQLRGFFPATFGKKYVEVVPG 127
E L Q PEVQ++L FPAT+G +Y EV PG
Sbjct 179 EAGPLAQPPEVQKKLEALFPATWGFEYTEVAPG 211
> cpv:cgd2_2130 pyrophosphate-dependent phosphofructokinase (EC:2.7.1.11);
K00850 6-phosphofructokinase [EC:2.7.1.11]
Length=1426
Score = 65.1 bits (157), Expect = 9e-11, Method: Composition-based stats.
Identities = 43/105 (40%), Positives = 57/105 (54%), Gaps = 14/105 (13%)
Query 49 GKVHLHSADLFERSSPLQRLRRAWRCTLPSALESTQLRLVEADCPVEHPSLRQTPEVQQQ 108
GK HLHSADLFE SP+Q R+ W TLP +L S +++ E +E P L Q E +
Sbjct 72 GKPHLHSADLFENFSPVQLERQKWVPTLPYSLSSNCIKIKE----IEIPDLIQKDE--NK 125
Query 109 LRGFFPATFGKKYVEVVPGMGSGVASPKEPLDHPLSKALRIGIVL 153
L+ P TFG+ +E+ + E DH ALR+GIVL
Sbjct 126 LKELLPNTFGRPCIEIT----RDESKNSENSDH----ALRVGIVL 162
> tgo:TGME49_026960 phosphofructokinase, putative (EC:2.7.1.90)
Length=1225
Score = 34.7 bits (78), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 36/133 (27%), Positives = 55/133 (41%), Gaps = 20/133 (15%)
Query 32 SEAPPSVDAV--LKEQETNGKVHLHSADLFERSSPLQRLRRAWRCTLPSALES-TQLRLV 88
+ A P DAV E + + + HL S L + ++P + RR ++ LP S T + V
Sbjct 14 TSAHPEADAVETFFESQASSQSHLFSPQLADEATPNEVARRHFKPVLPPVFSSPTGVVTV 73
Query 89 ---EADCPVEHPSLRQTPEV-----QQQLRGFFPATFGKKYVEVVPGMGSGVASPKEPLD 140
+ D + + P V Q L FP T + + P G AS
Sbjct 74 PCNDTDLVNKQDEVNNAPHVLSAQDQDILASLFPNTINTNFCLLAPASGDRQAS------ 127
Query 141 HPLSKALRIGIVL 153
S+ LR+G+VL
Sbjct 128 ---SEPLRVGVVL 137
> mmu:76088 Dock8, 1200017A24Rik, 5830472H07Rik, A130095G14Rik,
AI461977; dedicator of cytokinesis 8
Length=2100
Score = 33.9 bits (76), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 13/39 (33%), Positives = 24/39 (61%), Gaps = 0/39 (0%)
Query 87 LVEADCPVEHPSLRQTPEVQQQLRGFFPATFGKKYVEVV 125
L E + + P++ + PE+ +L GF+ FG ++VEV+
Sbjct 1808 LDEQEFVYKEPAITKLPEISHRLEGFYGQCFGAEFVEVI 1846
> hsa:81704 DOCK8, FLJ00026, FLJ00152, FLJ00346, MRD2, ZIR8; dedicator
of cytokinesis 8
Length=1999
Score = 31.6 bits (70), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 12/39 (30%), Positives = 23/39 (58%), Gaps = 0/39 (0%)
Query 87 LVEADCPVEHPSLRQTPEVQQQLRGFFPATFGKKYVEVV 125
L E + + P++ + PE+ +L F+ FG ++VEV+
Sbjct 1707 LDEQEFVYKEPAITKLPEISHRLEAFYGQCFGAEFVEVI 1745
> cel:Y54E5A.7 hypothetical protein
Length=622
Score = 31.6 bits (70), Expect = 1.2, Method: Composition-based stats.
Identities = 20/49 (40%), Positives = 28/49 (57%), Gaps = 4/49 (8%)
Query 100 RQTPEVQQQLRGFFPATFGKKYVEVVPGMGSGVASPKEPLDHPLSKALR 148
R+ +V ++RG A +K VE+ M V+SP+ P HPLS ALR
Sbjct 458 REIMKVAGEMRGNI-APEARKVVEIATSM---VSSPERPAKHPLSFALR 502
> cel:C52D10.12 hypothetical protein
Length=811
Score = 30.8 bits (68), Expect = 1.8, Method: Composition-based stats.
Identities = 33/135 (24%), Positives = 55/135 (40%), Gaps = 21/135 (15%)
Query 8 LSRRLSSFGMTEGNVFGSLATNAPSEA-PPSVDAVLKEQET------NGKVHLHSADLFE 60
L R +S G +E L+ P P AV+KE T + ++ + A L +
Sbjct 678 LGRSMSIVGTSE------LSARRPDLMFKPDYSAVVKEDCTYLEISVSAYINAYKASLMQ 731
Query 61 RSSPLQRLRRAWRCTLPSALESTQLRLVEADCPVEHPSLRQTPEVQQQLRGFFPATFGKK 120
R PL L + S+ ++ L LVE P+ PS PE ++ P+
Sbjct 732 RERPLNDLSDV---SHNSSAHNSNLSLVEKPGPITDPSAMLVPENVRK-----PSVVSMD 783
Query 121 YVEVVPGMGSGVASP 135
+++ G+G +P
Sbjct 784 SPKILVGLGQHPVAP 798
> dre:553788 map3k7, MGC110612, si:ch211-225h9.1, zgc:110612;
mitogen activated protein kinase kinase kinase 7 (EC:2.7.11.25);
K04427 mitogen-activated protein kinase kinase kinase 7
[EC:2.7.11.25]
Length=544
Score = 30.4 bits (67), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 14/42 (33%), Positives = 22/42 (52%), Gaps = 1/42 (2%)
Query 75 TLPSALESTQLRLVEADCPVEHPSLRQTPEVQQQLRGFFPAT 116
LP A+ES R D P + PS+ + ++ L G+FP +
Sbjct 236 NLPKAIESLMTRCWSKD-PSQRPSMEEIVKIMSHLMGYFPGS 276
> tgo:TGME49_028400 hypothetical protein
Length=632
Score = 30.4 bits (67), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 17/54 (31%), Positives = 25/54 (46%), Gaps = 2/54 (3%)
Query 60 ERSSPLQRLRRAWRCTLPSALESTQLRLVEADCPVEHPSLRQTPEVQQQLRGFF 113
E + L RL R W P E + + DC ++ PSL Q + Q ++R F
Sbjct 152 EDKTGLSRLGRLWSLVEPD--EFGMINVWAVDCNMDSPSLMQCIKTQPKVRCMF 203
> bbo:BBOV_IV009360 23.m06219; hypothetical protein
Length=1007
Score = 29.3 bits (64), Expect = 5.1, Method: Composition-based stats.
Identities = 20/58 (34%), Positives = 30/58 (51%), Gaps = 5/58 (8%)
Query 8 LSRRLSSFGMT-----EGNVFGSLATNAPSEAPPSVDAVLKEQETNGKVHLHSADLFE 60
R+L + MT E + SL+ A +E PS+ V+K+ T V L+S DL+E
Sbjct 613 FQRQLDEYVMTLRTEYEAAIESSLSGEATAEQKPSLLNVVKKFLTPSVVFLYSIDLYE 670
> dre:569167 pkhd-l2; si:ch211-244a23.1
Length=3895
Score = 28.9 bits (63), Expect = 5.9, Method: Composition-based stats.
Identities = 17/62 (27%), Positives = 30/62 (48%), Gaps = 6/62 (9%)
Query 75 TLPSALESTQLRLVEADCPVEHPSLRQTPEVQQQLRGFFPAT---FGKKYVEVVPGMGSG 131
TLP A+ S +R+ P+ P + T E ++ GF P+ +G + ++ GSG
Sbjct 1115 TLPPAVYSVNIRVANQGYPLTSPDINTTIEYVLEVTGFSPSVGSLYGGTTITII---GSG 1171
Query 132 VA 133
+
Sbjct 1172 FS 1173
> mmu:13656 Egr4, NGF1-C, NGFI-C, NGFIC, pAT133; early growth
response 4; K12498 early growth response protein 4
Length=478
Score = 28.9 bits (63), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 21/43 (48%), Positives = 24/43 (55%), Gaps = 2/43 (4%)
Query 109 LRGFFPATFGKKYVEVVPGMGSGVASPKEP-LDHPLSKALRIG 150
LRG A F K V +PG GSGVA+P P P +KA R G
Sbjct 311 LRGAATADFSKPLVADLPG-GSGVAAPSSPAASFPAAKARRKG 352
> hsa:85440 DOCK7, KIAA1771, ZIR2; dedicator of cytokinesis 7
Length=2109
Score = 28.9 bits (63), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 12/39 (30%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 87 LVEADCPVEHPSLRQTPEVQQQLRGFFPATFGKKYVEVV 125
L E + + P++ + E+ +L GF+ FG+ VEV+
Sbjct 1824 LDEQEFVYKEPAITKLAEISHRLEGFYGERFGEDVVEVI 1862
Lambda K H
0.316 0.132 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3256415000
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40