bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0281_orf1
Length=83
Score E
Sequences producing significant alignments: (Bits) Value
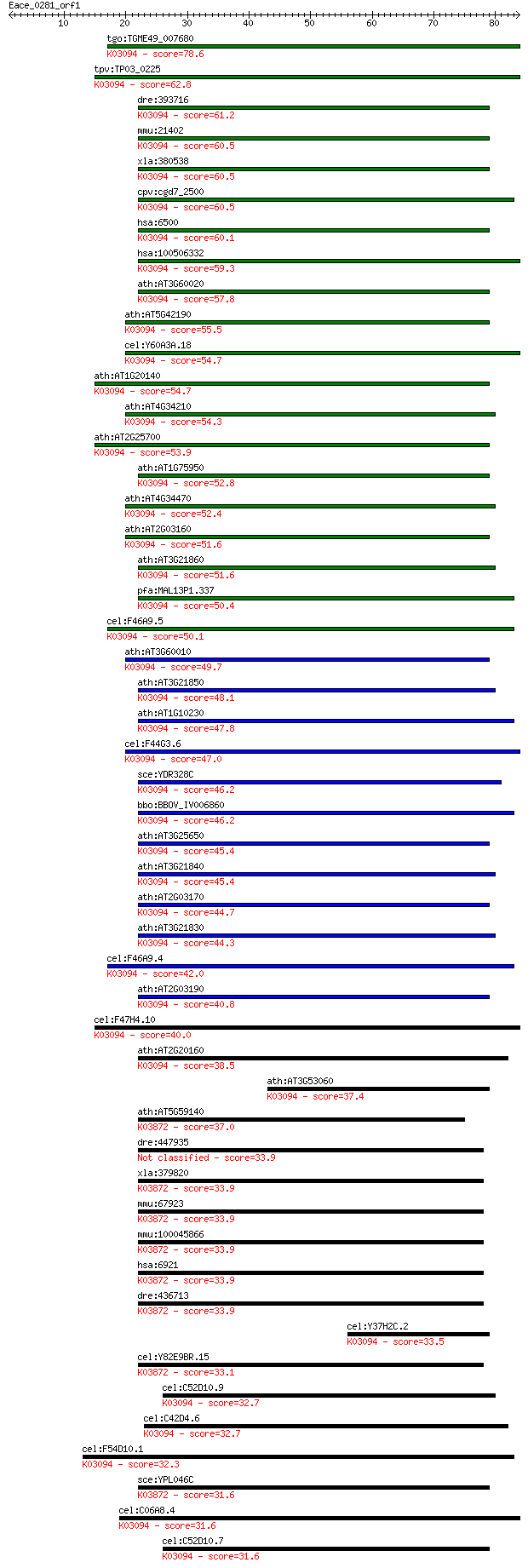
tgo:TGME49_007680 S-phase kinase-associated protein, putative ... 78.6 4e-15
tpv:TP03_0225 hypothetical protein; K03094 S-phase kinase-asso... 62.8 2e-10
dre:393716 skp1, MGC73186, skp1a, zgc:73186; S-phase kinase-as... 61.2 8e-10
mmu:21402 Skp1a, 15kDa, 2610043E24Rik, 2610206H23Rik, EMC19, O... 60.5 1e-09
xla:380538 skp1, MGC64317, skp1a; S-phase kinase-associated pr... 60.5 1e-09
cpv:cgd7_2500 Skp1 family protein ; K03094 S-phase kinase-asso... 60.5 1e-09
hsa:6500 SKP1, EMC19, MGC34403, OCP-II, OCP2, SKP1A, TCEB1L, p... 60.1 1e-09
hsa:100506332 s-phase kinase-associated protein 1-like; K03094... 59.3 3e-09
ath:AT3G60020 ASK5; ASK5 (ARABIDOPSIS SKP1-LIKE 5); protein bi... 57.8 9e-09
ath:AT5G42190 ASK2; ASK2 (ARABIDOPSIS SKP1-LIKE 2); protein bi... 55.5 5e-08
cel:Y60A3A.18 skr-4; SKp1 Related (ubiquitin ligase complex co... 54.7 7e-08
ath:AT1G20140 ASK4; ASK4 (ARABIDOPSIS SKP1-LIKE 4); protein bi... 54.7 8e-08
ath:AT4G34210 ASK11; ASK11 (ARABIDOPSIS SKP1-LIKE 11); protein... 54.3 8e-08
ath:AT2G25700 ASK3; ASK3 (ARABIDOPSIS SKP1-LIKE 3); protein bi... 53.9 1e-07
ath:AT1G75950 SKP1; SKP1 (S PHASE KINASE-ASSOCIATED PROTEIN 1)... 52.8 2e-07
ath:AT4G34470 ASK12; ASK12 (ARABIDOPSIS SKP1-LIKE 12); protein... 52.4 4e-07
ath:AT2G03160 ASK19; ASK19 (ARABIDOPSIS SKP1-LIKE 19); protein... 51.6 6e-07
ath:AT3G21860 ASK10; ASK10 (ARABIDOPSIS SKP1-LIKE 10); protein... 51.6 6e-07
pfa:MAL13P1.337 Skp1 family protein, putative; K03094 S-phase ... 50.4 1e-06
cel:F46A9.5 skr-1; SKp1 Related (ubiquitin ligase complex comp... 50.1 2e-06
ath:AT3G60010 ASK13; ASK13 (ARABIDOPSIS SKP1-LIKE 13); protein... 49.7 2e-06
ath:AT3G21850 ASK9; ASK9 (ARABIDOPSIS SKP1-LIKE 9); protein bi... 48.1 6e-06
ath:AT1G10230 ASK18; ASK18 (ARABIDOPSIS SKP1-LIKE 18); protein... 47.8 9e-06
cel:F44G3.6 skr-3; SKp1 Related (ubiquitin ligase complex comp... 47.0 1e-05
sce:YDR328C SKP1, MGO1; Evolutionarily conserved kinetochore p... 46.2 3e-05
bbo:BBOV_IV006860 23.m05921; cytosolic glycoprotein FP21; K030... 46.2 3e-05
ath:AT3G25650 ASK15; ASK15 (ARABIDOPSIS SKP1-LIKE 15); protein... 45.4 5e-05
ath:AT3G21840 ASK7; ASK7 (ARABIDOPSIS SKP1-LIKE 7); protein bi... 45.4 5e-05
ath:AT2G03170 ASK14; ASK14 (ARABIDOPSIS SKP1-LIKE 14); protein... 44.7 7e-05
ath:AT3G21830 ASK8; ASK8 (ARABIDOPSIS SKP1-LIKE 8); protein bi... 44.3 9e-05
cel:F46A9.4 skr-2; SKp1 Related (ubiquitin ligase complex comp... 42.0 5e-04
ath:AT2G03190 ASK16; ASK16 (ARABIDOPSIS SKP1-LIKE 16); protein... 40.8 0.001
cel:F47H4.10 skr-5; SKp1 Related (ubiquitin ligase complex com... 40.0 0.002
ath:AT2G20160 MEO; MEO (MEIDOS); protein binding / ubiquitin-p... 38.5 0.005
ath:AT3G53060 ASK6; ASK6 (ARABIDOPSIS SKP1-LIKE 6); protein bi... 37.4 0.011
ath:AT5G59140 SKP1 family protein; K03872 transcription elonga... 37.0 0.016
dre:447935 tceb1a, tceb1, zgc:101879; transcription elongation... 33.9 0.12
xla:379820 tceb1, MGC64412; transcription elongation factor B ... 33.9 0.12
mmu:67923 Tceb1, 2610043E24Rik, 2610301I15Rik, AA407206, AI987... 33.9 0.12
mmu:100045866 transcription elongation factor B polypeptide 1-... 33.9 0.12
hsa:6921 TCEB1, SIII; transcription elongation factor B (SIII)... 33.9 0.12
dre:436713 tceb1b, TCEB1, fb09f09, wu:fb09f09, zgc:111990, zgc... 33.9 0.14
cel:Y37H2C.2 skr-6; SKp1 Related (ubiquitin ligase complex com... 33.5 0.18
cel:Y82E9BR.15 elc-1; ELongin C family member (elc-1); K03872 ... 33.1 0.23
cel:C52D10.9 skr-8; SKp1 Related (ubiquitin ligase complex com... 32.7 0.26
cel:C42D4.6 skr-16; SKp1 Related (ubiquitin ligase complex com... 32.7 0.33
cel:F54D10.1 skr-15; SKp1 Related (ubiquitin ligase complex co... 32.3 0.38
sce:YPL046C ELC1; Elc1p; K03872 transcription elongation facto... 31.6 0.62
cel:C06A8.4 skr-17; SKp1 Related (ubiquitin ligase complex com... 31.6 0.68
cel:C52D10.7 skr-9; SKp1 Related (ubiquitin ligase complex com... 31.6 0.72
> tgo:TGME49_007680 S-phase kinase-associated protein, putative
; K03094 S-phase kinase-associated protein 1
Length=170
Score = 78.6 bits (192), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 38/68 (55%), Positives = 48/68 (70%), Gaps = 1/68 (1%)
Query 17 AEPRCVVLVSSEGEEFKVPREVASASLLVKSMTE-DGDDTDVVPLPKVSSYILKKVVEYC 75
+ R V LVS EG+EF V EVAS S L+K+M E D D + +PLP V + ILKK++EYC
Sbjct 7 GDARKVTLVSQEGDEFDVDIEVASMSALIKTMVEEDSDCQESIPLPNVDTCILKKIIEYC 66
Query 76 THHHDNPP 83
HHH+NPP
Sbjct 67 EHHHNNPP 74
> tpv:TP03_0225 hypothetical protein; K03094 S-phase kinase-associated
protein 1
Length=182
Score = 62.8 bits (151), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 28/70 (40%), Positives = 45/70 (64%), Gaps = 1/70 (1%)
Query 15 MEAEPRCVVLVSSEGEEFKVPREVASASLLVKSMTEDGDD-TDVVPLPKVSSYILKKVVE 73
M + + LVS+EG V R+V S ++K++ D DD T+ +PLP + + +L K++E
Sbjct 1 MTLSKKIITLVSAEGVSCTVNRDVICMSNVIKNILNDIDDETEPIPLPNIKTNVLNKIIE 60
Query 74 YCTHHHDNPP 83
YC HH++NPP
Sbjct 61 YCKHHYNNPP 70
> dre:393716 skp1, MGC73186, skp1a, zgc:73186; S-phase kinase-associated
protein 1 (EC:3.1.1.3); K03094 S-phase kinase-associated
protein 1
Length=163
Score = 61.2 bits (147), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 32/63 (50%), Positives = 44/63 (69%), Gaps = 7/63 (11%)
Query 22 VVLVSSEGEEFKVPREVASASLLVKSMTED------GDDTDVVPLPKVSSYILKKVVEYC 75
+ L SS+GE F+V E+A S+ +K+M ED GDD D VPLP V++ ILKKV+++C
Sbjct 4 IKLQSSDGEMFEVDVEIAKQSVTIKTMLEDLGMDDEGDD-DPVPLPNVNAAILKKVIQWC 62
Query 76 THH 78
THH
Sbjct 63 THH 65
> mmu:21402 Skp1a, 15kDa, 2610043E24Rik, 2610206H23Rik, EMC19,
OCP-II, OCP2, SKP1, Tceb1l, p19A, p19Skp1; S-phase kinase-associated
protein 1A (EC:3.1.1.3); K03094 S-phase kinase-associated
protein 1
Length=163
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 32/63 (50%), Positives = 44/63 (69%), Gaps = 7/63 (11%)
Query 22 VVLVSSEGEEFKVPREVASASLLVKSMTED------GDDTDVVPLPKVSSYILKKVVEYC 75
+ L SS+GE F+V E+A S+ +K+M ED GDD D VPLP V++ ILKKV+++C
Sbjct 4 IKLQSSDGEIFEVDVEIAKQSVTIKTMLEDLGMDDEGDD-DPVPLPNVNAAILKKVIQWC 62
Query 76 THH 78
THH
Sbjct 63 THH 65
> xla:380538 skp1, MGC64317, skp1a; S-phase kinase-associated
protein 1 (EC:3.1.1.3); K03094 S-phase kinase-associated protein
1
Length=163
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 32/63 (50%), Positives = 44/63 (69%), Gaps = 7/63 (11%)
Query 22 VVLVSSEGEEFKVPREVASASLLVKSMTED------GDDTDVVPLPKVSSYILKKVVEYC 75
+ L SS+GE F+V E+A S+ +K+M ED GDD D VPLP V++ ILKKV+++C
Sbjct 4 IKLQSSDGEIFEVDVEIAKQSVTIKTMLEDLGMDDEGDD-DPVPLPNVNAAILKKVIQWC 62
Query 76 THH 78
THH
Sbjct 63 THH 65
> cpv:cgd7_2500 Skp1 family protein ; K03094 S-phase kinase-associated
protein 1
Length=162
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 33/62 (53%), Positives = 43/62 (69%), Gaps = 1/62 (1%)
Query 22 VVLVSSEGEEFKVPREVASASLLVKSMTE-DGDDTDVVPLPKVSSYILKKVVEYCTHHHD 80
V L+SSEGEEF V VA+AS LV+++ E D D VPLP V +L+KV++YC +H D
Sbjct 6 VRLLSSEGEEFAVDVRVATASTLVRNIIEADVGIDDPVPLPNVRGDVLRKVLDYCEYHVD 65
Query 81 NP 82
NP
Sbjct 66 NP 67
> hsa:6500 SKP1, EMC19, MGC34403, OCP-II, OCP2, SKP1A, TCEB1L,
p19A; S-phase kinase-associated protein 1 (EC:3.1.1.3); K03094
S-phase kinase-associated protein 1
Length=160
Score = 60.1 bits (144), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 32/63 (50%), Positives = 44/63 (69%), Gaps = 7/63 (11%)
Query 22 VVLVSSEGEEFKVPREVASASLLVKSMTED------GDDTDVVPLPKVSSYILKKVVEYC 75
+ L SS+GE F+V E+A S+ +K+M ED GDD D VPLP V++ ILKKV+++C
Sbjct 4 IKLQSSDGEIFEVDVEIAKQSVTIKTMLEDLGMDDEGDD-DPVPLPNVNAAILKKVIQWC 62
Query 76 THH 78
THH
Sbjct 63 THH 65
> hsa:100506332 s-phase kinase-associated protein 1-like; K03094
S-phase kinase-associated protein 1
Length=163
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 31/67 (46%), Positives = 43/67 (64%), Gaps = 5/67 (7%)
Query 22 VVLVSSEGEEFKVPREVASASLLVKSMTED-----GDDTDVVPLPKVSSYILKKVVEYCT 76
+ L SS GE F+V E+ S+ +K+M ED D D VPLP V++ ILKKV+++CT
Sbjct 4 IKLQSSGGEIFEVDVEIVKQSVTIKTMLEDLGMNDEGDHDPVPLPNVNAAILKKVIQWCT 63
Query 77 HHHDNPP 83
HH D+ P
Sbjct 64 HHEDDSP 70
> ath:AT3G60020 ASK5; ASK5 (ARABIDOPSIS SKP1-LIKE 5); protein
binding / ubiquitin-protein ligase; K03094 S-phase kinase-associated
protein 1
Length=153
Score = 57.8 bits (138), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 26/57 (45%), Positives = 39/57 (68%), Gaps = 0/57 (0%)
Query 22 VVLVSSEGEEFKVPREVASASLLVKSMTEDGDDTDVVPLPKVSSYILKKVVEYCTHH 78
++L SS+G+ F++ +VA S+ + M EDG TDV+PL V+S ILK V++YC H
Sbjct 5 IMLKSSDGKSFEIDEDVARKSIAINHMVEDGCATDVIPLRNVTSKILKIVIDYCEKH 61
> ath:AT5G42190 ASK2; ASK2 (ARABIDOPSIS SKP1-LIKE 2); protein
binding / ubiquitin-protein ligase; K03094 S-phase kinase-associated
protein 1
Length=171
Score = 55.5 bits (132), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 27/59 (45%), Positives = 36/59 (61%), Gaps = 0/59 (0%)
Query 20 RCVVLVSSEGEEFKVPREVASASLLVKSMTEDGDDTDVVPLPKVSSYILKKVVEYCTHH 78
R + L SS+GE F++ VA S +K M ED + +PLP V+S IL KV+EYC H
Sbjct 5 RKITLKSSDGENFEIDEAVALESQTIKHMIEDDCTDNGIPLPNVTSKILSKVIEYCKRH 63
> cel:Y60A3A.18 skr-4; SKp1 Related (ubiquitin ligase complex
component) family member (skr-4); K03094 S-phase kinase-associated
protein 1
Length=159
Score = 54.7 bits (130), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 25/64 (39%), Positives = 40/64 (62%), Gaps = 4/64 (6%)
Query 20 RCVVLVSSEGEEFKVPREVASASLLVKSMTEDGDDTDVVPLPKVSSYILKKVVEYCTHHH 79
+ + L+SS+ + F V R+V S S + T + D +PLPKV+S IL+K++ +C HH
Sbjct 8 KQIKLISSDDKTFTVSRKVISQSKTISGFTSE----DTIPLPKVTSAILEKIITWCEHHA 63
Query 80 DNPP 83
D+ P
Sbjct 64 DDEP 67
> ath:AT1G20140 ASK4; ASK4 (ARABIDOPSIS SKP1-LIKE 4); protein
binding / ubiquitin-protein ligase; K03094 S-phase kinase-associated
protein 1
Length=163
Score = 54.7 bits (130), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 26/64 (40%), Positives = 37/64 (57%), Gaps = 0/64 (0%)
Query 15 MEAEPRCVVLVSSEGEEFKVPREVASASLLVKSMTEDGDDTDVVPLPKVSSYILKKVVEY 74
M + ++L SS+GE F++ VA S +K M ED + +PLP V+ IL KV+EY
Sbjct 1 MAETKKMIILKSSDGESFEIEEAVAVKSQTIKHMIEDDCADNGIPLPNVTGAILAKVIEY 60
Query 75 CTHH 78
C H
Sbjct 61 CKKH 64
> ath:AT4G34210 ASK11; ASK11 (ARABIDOPSIS SKP1-LIKE 11); protein
binding / ubiquitin-protein ligase; K03094 S-phase kinase-associated
protein 1
Length=152
Score = 54.3 bits (129), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 27/60 (45%), Positives = 36/60 (60%), Gaps = 0/60 (0%)
Query 20 RCVVLVSSEGEEFKVPREVASASLLVKSMTEDGDDTDVVPLPKVSSYILKKVVEYCTHHH 79
+ +VL+SS+G+ F+V VA S + M ED D +PL V S IL KV+EYC HH
Sbjct 4 KMIVLMSSDGQSFEVEEAVAIQSQTIAHMVEDDCVADGIPLANVESKILVKVIEYCKKHH 63
> ath:AT2G25700 ASK3; ASK3 (ARABIDOPSIS SKP1-LIKE 3); protein
binding / ubiquitin-protein ligase; K03094 S-phase kinase-associated
protein 1
Length=163
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 27/64 (42%), Positives = 37/64 (57%), Gaps = 0/64 (0%)
Query 15 MEAEPRCVVLVSSEGEEFKVPREVASASLLVKSMTEDGDDTDVVPLPKVSSYILKKVVEY 74
M + ++L SS+GE F+V VA S +K M ED + +PLP V+ IL KV+EY
Sbjct 1 MAETKKMIILKSSDGESFEVEEAVAVESQTIKHMIEDDCVDNGIPLPNVTGAILAKVIEY 60
Query 75 CTHH 78
C H
Sbjct 61 CKKH 64
> ath:AT1G75950 SKP1; SKP1 (S PHASE KINASE-ASSOCIATED PROTEIN
1); protein binding / ubiquitin-protein ligase; K03094 S-phase
kinase-associated protein 1
Length=160
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 28/57 (49%), Positives = 35/57 (61%), Gaps = 0/57 (0%)
Query 22 VVLVSSEGEEFKVPREVASASLLVKSMTEDGDDTDVVPLPKVSSYILKKVVEYCTHH 78
+VL SS+GE F+V VA S + M ED + VPLP V+S IL KV+EYC H
Sbjct 6 IVLKSSDGESFEVEEAVALESQTIAHMVEDDCVDNGVPLPNVTSKILAKVIEYCKRH 62
> ath:AT4G34470 ASK12; ASK12 (ARABIDOPSIS SKP1-LIKE 12); protein
binding / ubiquitin-protein ligase; K03094 S-phase kinase-associated
protein 1
Length=152
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 26/60 (43%), Positives = 36/60 (60%), Gaps = 0/60 (0%)
Query 20 RCVVLVSSEGEEFKVPREVASASLLVKSMTEDGDDTDVVPLPKVSSYILKKVVEYCTHHH 79
+ +VL+SS+G+ F+V VA S + M ED D +PL V S IL KV+EYC +H
Sbjct 4 KMIVLMSSDGQSFEVEEAVAIQSQTIAHMVEDDCVADGIPLANVESKILVKVIEYCKKYH 63
> ath:AT2G03160 ASK19; ASK19 (ARABIDOPSIS SKP1-LIKE 19); protein
binding / ubiquitin-protein ligase; K03094 S-phase kinase-associated
protein 1
Length=200
Score = 51.6 bits (122), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 26/59 (44%), Positives = 36/59 (61%), Gaps = 0/59 (0%)
Query 20 RCVVLVSSEGEEFKVPREVASASLLVKSMTEDGDDTDVVPLPKVSSYILKKVVEYCTHH 78
+ +VL SS+GE FKV VA +V + ED T+ +P+P V+ IL KV+EYC H
Sbjct 4 KKIVLTSSDGESFKVEEVVARKLQIVGHIIEDDCATNKIPIPNVTGEILAKVIEYCKKH 62
> ath:AT3G21860 ASK10; ASK10 (ARABIDOPSIS SKP1-LIKE 10); protein
binding / ubiquitin-protein ligase; K03094 S-phase kinase-associated
protein 1
Length=152
Score = 51.6 bits (122), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 23/58 (39%), Positives = 35/58 (60%), Gaps = 0/58 (0%)
Query 22 VVLVSSEGEEFKVPREVASASLLVKSMTEDGDDTDVVPLPKVSSYILKKVVEYCTHHH 79
++L SS+G F+V E A + M+ED + +PLP+V+ IL+ V+EYC HH
Sbjct 6 IILKSSDGHSFEVEEEAACQCQTIAHMSEDDCTDNGIPLPEVTGKILEMVIEYCNKHH 63
> pfa:MAL13P1.337 Skp1 family protein, putative; K03094 S-phase
kinase-associated protein 1
Length=162
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 40/62 (64%), Gaps = 1/62 (1%)
Query 22 VVLVSSEGEEFKVPREVASASLLVKSMTE-DGDDTDVVPLPKVSSYILKKVVEYCTHHHD 80
+ LVS EG+EF V + AS S ++ ++ E + D +PLP + + ILKK++EY +H +
Sbjct 6 IKLVSFEGDEFIVDKNTASMSTVIMNILEVMTAEEDTIPLPNIKTPILKKIIEYMEYHIN 65
Query 81 NP 82
NP
Sbjct 66 NP 67
> cel:F46A9.5 skr-1; SKp1 Related (ubiquitin ligase complex component)
family member (skr-1); K03094 S-phase kinase-associated
protein 1
Length=176
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 42/73 (57%), Gaps = 7/73 (9%)
Query 17 AEPRCVVLVSSEGEEFKVPREVASASLLVKSM-------TEDGDDTDVVPLPKVSSYILK 69
A+ R + + SS+ E F VPR V S + ++ E+G + + +P+ V++ ILK
Sbjct 11 AKEREIKISSSDNEIFLVPRNVIRLSNTINTLLMDLGLDDEEGTNAEPIPVQNVTASILK 70
Query 70 KVVEYCTHHHDNP 82
KV+ +C HHH +P
Sbjct 71 KVISWCNHHHSDP 83
> ath:AT3G60010 ASK13; ASK13 (ARABIDOPSIS SKP1-LIKE 13); protein
binding / ubiquitin-protein ligase; K03094 S-phase kinase-associated
protein 1
Length=154
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 25/59 (42%), Positives = 35/59 (59%), Gaps = 0/59 (0%)
Query 20 RCVVLVSSEGEEFKVPREVASASLLVKSMTEDGDDTDVVPLPKVSSYILKKVVEYCTHH 78
+ V+L+SS+GE F+V VA S + M ED + VP+ V+ IL KV+EYC H
Sbjct 3 KMVMLLSSDGESFQVEEAVAVQSQTIAHMIEDDCVANGVPIANVTGVILSKVIEYCKKH 61
> ath:AT3G21850 ASK9; ASK9 (ARABIDOPSIS SKP1-LIKE 9); protein
binding / ubiquitin-protein ligase; K03094 S-phase kinase-associated
protein 1
Length=153
Score = 48.1 bits (113), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 22/59 (37%), Positives = 35/59 (59%), Gaps = 1/59 (1%)
Query 22 VVLVSSEGEEFKVPREVA-SASLLVKSMTEDGDDTDVVPLPKVSSYILKKVVEYCTHHH 79
++L SS+G F+V E A +++ M+E+ + +PLP V+ IL V+EYC HH
Sbjct 6 IILKSSDGHSFEVEEEAARQCQIIIAHMSENDCTDNGIPLPNVTGKILAMVIEYCNKHH 64
> ath:AT1G10230 ASK18; ASK18 (ARABIDOPSIS SKP1-LIKE 18); protein
binding / ubiquitin-protein ligase; K03094 S-phase kinase-associated
protein 1
Length=183
Score = 47.8 bits (112), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 22/61 (36%), Positives = 35/61 (57%), Gaps = 0/61 (0%)
Query 22 VVLVSSEGEEFKVPREVASASLLVKSMTEDGDDTDVVPLPKVSSYILKKVVEYCTHHHDN 81
++L SS+GE F++ VA L++ M ED + +PL V+ IL K++EY H +
Sbjct 31 ILLTSSDGESFEIDEAVARKFLIIVHMMEDNCAGEAIPLENVTGDILSKIIEYAKMHVNE 90
Query 82 P 82
P
Sbjct 91 P 91
> cel:F44G3.6 skr-3; SKp1 Related (ubiquitin ligase complex component)
family member (skr-3); K03094 S-phase kinase-associated
protein 1
Length=167
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 39/71 (54%), Gaps = 7/71 (9%)
Query 20 RCVVLVSSEGEEFKVPREVASASLLVKSMTED-------GDDTDVVPLPKVSSYILKKVV 72
+ + L SS+ + F V R+V S S + + ++ D +PL KV+S IL+K++
Sbjct 8 KQIKLTSSDDKTFTVSRKVISQSKTITDIIQNLGIEESGSTSEDTIPLQKVTSTILEKII 67
Query 73 EYCTHHHDNPP 83
+C HH D+ P
Sbjct 68 TWCEHHADDEP 78
> sce:YDR328C SKP1, MGO1; Evolutionarily conserved kinetochore
protein that is part of multiple protein complexes, including
the SCF ubiquitin ligase complex, the CBF3 complex that binds
centromeric DNA, and the RAVE complex that regulates assembly
of the V-ATPase; K03094 S-phase kinase-associated protein
1
Length=194
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 29/94 (30%), Positives = 42/94 (44%), Gaps = 35/94 (37%)
Query 22 VVLVSSEGEEFKVPREVASASLLVKSMTEDGDDTD------------------------- 56
VVLVS EGE F V +++A SLL+K+ D D++
Sbjct 6 VVLVSGEGERFTVDKKIAERSLLLKNYLNDMHDSNLQNNSDSESDSDSETNHKSKDNNNG 65
Query 57 ----------VVPLPKVSSYILKKVVEYCTHHHD 80
V+P+P V S +L+KV+E+ HH D
Sbjct 66 DDDDEDDDEIVMPVPNVRSSVLQKVIEWAEHHRD 99
> bbo:BBOV_IV006860 23.m05921; cytosolic glycoprotein FP21; K03094
S-phase kinase-associated protein 1
Length=161
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 24/64 (37%), Positives = 42/64 (65%), Gaps = 3/64 (4%)
Query 22 VVLVSSEGEEFKVPREVASASLLVKSMTEDGD-DTDVVP--LPKVSSYILKKVVEYCTHH 78
V LVS+EG+ F V EV + S+L+ +M + D +T++ P L + + L K+++YC +H
Sbjct 3 VKLVSAEGDTFTVNSEVLTPSVLLTNMLQGYDEETELAPIELKNIPTRTLGKILDYCKYH 62
Query 79 HDNP 82
++NP
Sbjct 63 YNNP 66
> ath:AT3G25650 ASK15; ASK15 (ARABIDOPSIS SKP1-LIKE 15); protein
binding / ubiquitin-protein ligase; K03094 S-phase kinase-associated
protein 1
Length=177
Score = 45.4 bits (106), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 24/57 (42%), Positives = 33/57 (57%), Gaps = 0/57 (0%)
Query 22 VVLVSSEGEEFKVPREVASASLLVKSMTEDGDDTDVVPLPKVSSYILKKVVEYCTHH 78
+VL SS+GE F+V VA +VK + ED + +PL V+ IL V+EYC H
Sbjct 6 IVLTSSDGESFQVEEVVARKLQIVKHLLEDDCVINEIPLQNVTGNILSIVLEYCKKH 62
> ath:AT3G21840 ASK7; ASK7 (ARABIDOPSIS SKP1-LIKE 7); protein
binding / ubiquitin-protein ligase; K03094 S-phase kinase-associated
protein 1
Length=125
Score = 45.4 bits (106), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 21/58 (36%), Positives = 34/58 (58%), Gaps = 0/58 (0%)
Query 22 VVLVSSEGEEFKVPREVASASLLVKSMTEDGDDTDVVPLPKVSSYILKKVVEYCTHHH 79
++L SS+G+ F++ E A + M E +V+P+ V+S IL+ V+EYC HH
Sbjct 6 IMLKSSDGKMFEIEEETARQCQTIAHMIEAECTDNVIPVSNVTSEILEMVIEYCNKHH 63
> ath:AT2G03170 ASK14; ASK14 (ARABIDOPSIS SKP1-LIKE 14); protein
binding / ubiquitin-protein ligase; K03094 S-phase kinase-associated
protein 1
Length=149
Score = 44.7 bits (104), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 26/57 (45%), Positives = 32/57 (56%), Gaps = 0/57 (0%)
Query 22 VVLVSSEGEEFKVPREVASASLLVKSMTEDGDDTDVVPLPKVSSYILKKVVEYCTHH 78
+VL SS+GE F+V VA +V+ M ED VPL V+ IL VVEYC H
Sbjct 6 IVLSSSDGESFEVEEAVARKLKIVEHMIEDDCVVTEVPLQNVTGKILSIVVEYCKKH 62
> ath:AT3G21830 ASK8; ASK8 (ARABIDOPSIS SKP1-LIKE 8); protein
binding / ubiquitin-protein ligase; K03094 S-phase kinase-associated
protein 1
Length=152
Score = 44.3 bits (103), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 21/58 (36%), Positives = 34/58 (58%), Gaps = 0/58 (0%)
Query 22 VVLVSSEGEEFKVPREVASASLLVKSMTEDGDDTDVVPLPKVSSYILKKVVEYCTHHH 79
++L SSEG+ F++ E A + M E +V+ + K++S IL+ V+EYC HH
Sbjct 6 IMLKSSEGKTFEIEEETARQCQTIAHMIEAECTDNVILVLKMTSEILEMVIEYCNKHH 63
> cel:F46A9.4 skr-2; SKp1 Related (ubiquitin ligase complex component)
family member (skr-2); K03094 S-phase kinase-associated
protein 1
Length=174
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 23/73 (31%), Positives = 40/73 (54%), Gaps = 7/73 (9%)
Query 17 AEPRCVVLVSSEGEEFKVPREVASASLLVKSMTE-------DGDDTDVVPLPKVSSYILK 69
A+ R + + SS+ E F VPR V S + ++ +G + + +P+ V++ ILK
Sbjct 11 AKNREIKISSSDDEIFLVPRNVIRLSNTLNTLLVDLGLDDDEGTNAEPIPVQNVTASILK 70
Query 70 KVVEYCTHHHDNP 82
KV+ +CT H +P
Sbjct 71 KVINWCTKHQSDP 83
> ath:AT2G03190 ASK16; ASK16 (ARABIDOPSIS SKP1-LIKE 16); protein
binding / ubiquitin-protein ligase; K03094 S-phase kinase-associated
protein 1
Length=170
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 21/57 (36%), Positives = 30/57 (52%), Gaps = 0/57 (0%)
Query 22 VVLVSSEGEEFKVPREVASASLLVKSMTEDGDDTDVVPLPKVSSYILKKVVEYCTHH 78
+VL SS+ E F+V VA ++ M +D +PL V+ IL V+EYC H
Sbjct 6 IVLTSSDDESFEVEEAVARKLKVIAHMIDDDCADKAIPLENVTGNILALVIEYCKKH 62
> cel:F47H4.10 skr-5; SKp1 Related (ubiquitin ligase complex component)
family member (skr-5); K03094 S-phase kinase-associated
protein 1
Length=145
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/69 (33%), Positives = 41/69 (59%), Gaps = 1/69 (1%)
Query 15 MEAEPRCVVLVSSEGEEFKVPREVASASLLVKSMTEDGDDTDVVPLPKVSSYILKKVVEY 74
M +E + V +V+S+ EF V ++A+ S L+ + + +PL V+S I KKV+E+
Sbjct 1 MSSEEQDVKIVTSDDVEFIVSPKIANQSKLLADFVV-LNQREPIPLKNVTSEIFKKVIEW 59
Query 75 CTHHHDNPP 83
C +H ++ P
Sbjct 60 CEYHAEDIP 68
> ath:AT2G20160 MEO; MEO (MEIDOS); protein binding / ubiquitin-protein
ligase; K03094 S-phase kinase-associated protein 1
Length=150
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 31/60 (51%), Gaps = 0/60 (0%)
Query 22 VVLVSSEGEEFKVPREVASASLLVKSMTEDGDDTDVVPLPKVSSYILKKVVEYCTHHHDN 81
+VL SS+ E F++ VA +V M +D + L V+ IL ++EYC H D+
Sbjct 6 IVLTSSDDECFEIDEAVARKMQMVAHMIDDDCADKAIRLQNVTGKILAIIIEYCKKHVDD 65
> ath:AT3G53060 ASK6; ASK6 (ARABIDOPSIS SKP1-LIKE 6); protein
binding / ubiquitin-protein ligase; K03094 S-phase kinase-associated
protein 1
Length=85
Score = 37.4 bits (85), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 16/36 (44%), Positives = 23/36 (63%), Gaps = 0/36 (0%)
Query 43 LLVKSMTEDGDDTDVVPLPKVSSYILKKVVEYCTHH 78
+++K M ED + +PLP V+S IL V+EYC H
Sbjct 1 MMIKGMAEDDCADNGIPLPNVTSKILLLVIEYCKKH 36
> ath:AT5G59140 SKP1 family protein; K03872 transcription elongation
factor B, polypeptide 1
Length=96
Score = 37.0 bits (84), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 23/58 (39%), Positives = 34/58 (58%), Gaps = 5/58 (8%)
Query 22 VVLVSSEGEEFKVPREVASASLLVKSM-TEDGDDTD----VVPLPKVSSYILKKVVEY 74
V L+S EG EF + RE A S ++SM T G ++ VV P +S+ IL+K+ +Y
Sbjct 5 VKLISMEGFEFVIDREAAMVSQTIRSMLTSPGGFSESKDGVVTFPDISTTILEKICQY 62
> dre:447935 tceb1a, tceb1, zgc:101879; transcription elongation
factor B (SIII), polypeptide 1a
Length=113
Score = 33.9 bits (76), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 22/61 (36%), Positives = 34/61 (55%), Gaps = 5/61 (8%)
Query 22 VVLVSSEGEEFKVPREVASASLLVKSMTED-----GDDTDVVPLPKVSSYILKKVVEYCT 76
V L+SS+G EF V RE A S +K+M ++T+ V ++ S++L KV Y T
Sbjct 20 VKLISSDGHEFIVKREHALTSGTIKAMLSGPGQFAENETNEVNFREIPSHVLSKVCMYFT 79
Query 77 H 77
+
Sbjct 80 Y 80
> xla:379820 tceb1, MGC64412; transcription elongation factor
B (SIII), polypeptide 1 (15kDa, elongin C); K03872 transcription
elongation factor B, polypeptide 1
Length=112
Score = 33.9 bits (76), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 22/61 (36%), Positives = 34/61 (55%), Gaps = 5/61 (8%)
Query 22 VVLVSSEGEEFKVPREVASASLLVKSMTED-----GDDTDVVPLPKVSSYILKKVVEYCT 76
V L+SS+G EF V RE A S +K+M ++T+ V ++ S++L KV Y T
Sbjct 19 VKLISSDGHEFIVKREHALTSGTIKAMLSGPGQFAENETNEVNFREIPSHVLSKVCMYFT 78
Query 77 H 77
+
Sbjct 79 Y 79
> mmu:67923 Tceb1, 2610043E24Rik, 2610301I15Rik, AA407206, AI987979,
AW049146; transcription elongation factor B (SIII), polypeptide
1; K03872 transcription elongation factor B, polypeptide
1
Length=112
Score = 33.9 bits (76), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 22/61 (36%), Positives = 34/61 (55%), Gaps = 5/61 (8%)
Query 22 VVLVSSEGEEFKVPREVASASLLVKSMTED-----GDDTDVVPLPKVSSYILKKVVEYCT 76
V L+SS+G EF V RE A S +K+M ++T+ V ++ S++L KV Y T
Sbjct 19 VKLISSDGHEFIVKREHALTSGTIKAMLSGPGQFAENETNEVNFREIPSHVLSKVCMYFT 78
Query 77 H 77
+
Sbjct 79 Y 79
> mmu:100045866 transcription elongation factor B polypeptide
1-like; K03872 transcription elongation factor B, polypeptide
1
Length=112
Score = 33.9 bits (76), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 22/61 (36%), Positives = 34/61 (55%), Gaps = 5/61 (8%)
Query 22 VVLVSSEGEEFKVPREVASASLLVKSMTED-----GDDTDVVPLPKVSSYILKKVVEYCT 76
V L+SS+G EF V RE A S +K+M ++T+ V ++ S++L KV Y T
Sbjct 19 VKLISSDGHEFIVKREHALTSGTIKAMLSGPGQFAENETNEVNFREIPSHVLSKVCMYFT 78
Query 77 H 77
+
Sbjct 79 Y 79
> hsa:6921 TCEB1, SIII; transcription elongation factor B (SIII),
polypeptide 1 (15kDa, elongin C); K03872 transcription elongation
factor B, polypeptide 1
Length=112
Score = 33.9 bits (76), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 22/61 (36%), Positives = 34/61 (55%), Gaps = 5/61 (8%)
Query 22 VVLVSSEGEEFKVPREVASASLLVKSMTED-----GDDTDVVPLPKVSSYILKKVVEYCT 76
V L+SS+G EF V RE A S +K+M ++T+ V ++ S++L KV Y T
Sbjct 19 VKLISSDGHEFIVKREHALTSGTIKAMLSGPGQFAENETNEVNFREIPSHVLSKVCMYFT 78
Query 77 H 77
+
Sbjct 79 Y 79
> dre:436713 tceb1b, TCEB1, fb09f09, wu:fb09f09, zgc:111990, zgc:92635;
transcription elongation factor B (SIII), polypeptide
1b; K03872 transcription elongation factor B, polypeptide
1
Length=112
Score = 33.9 bits (76), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 22/61 (36%), Positives = 34/61 (55%), Gaps = 5/61 (8%)
Query 22 VVLVSSEGEEFKVPREVASASLLVKSMTED-----GDDTDVVPLPKVSSYILKKVVEYCT 76
V L+SS+G EF V RE A S +K+M ++T+ V ++ S++L KV Y T
Sbjct 19 VKLISSDGHEFIVKREHALTSGTIKAMLSGPGQFAENETNEVNFREIPSHVLSKVCMYFT 78
Query 77 H 77
+
Sbjct 79 Y 79
> cel:Y37H2C.2 skr-6; SKp1 Related (ubiquitin ligase complex component)
family member (skr-6); K03094 S-phase kinase-associated
protein 1
Length=106
Score = 33.5 bits (75), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 11/23 (47%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 56 DVVPLPKVSSYILKKVVEYCTHH 78
D +PL KV + I +K++EYC H
Sbjct 2 DAIPLTKVDAKIFEKIIEYCEHQ 24
> cel:Y82E9BR.15 elc-1; ELongin C family member (elc-1); K03872
transcription elongation factor B, polypeptide 1
Length=124
Score = 33.1 bits (74), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 36/61 (59%), Gaps = 5/61 (8%)
Query 22 VVLVSSEGEEFKVPREVASASLLVKSMTED-----GDDTDVVPLPKVSSYILKKVVEYCT 76
V LVSS+ EF + RE+A S +++M ++++VV ++ S++L+KV +Y
Sbjct 31 VKLVSSDDHEFIIKRELALTSGTIRAMLSGPGVYAENESNVVYFREIPSHVLQKVCQYFA 90
Query 77 H 77
+
Sbjct 91 Y 91
> cel:C52D10.9 skr-8; SKp1 Related (ubiquitin ligase complex component)
family member (skr-8); K03094 S-phase kinase-associated
protein 1
Length=194
Score = 32.7 bits (73), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 17/59 (28%), Positives = 31/59 (52%), Gaps = 5/59 (8%)
Query 26 SSEGEEFKVPREVASASLLVKSMT-----EDGDDTDVVPLPKVSSYILKKVVEYCTHHH 79
S++G+ F++ E S ++ ++ ED D +P+ V+ ILK V+E+C H
Sbjct 27 SNDGKVFEISDEAVKQSNILSNLISTCAPEDVASMDPIPITNVTGNILKMVIEWCEKHK 85
> cel:C42D4.6 skr-16; SKp1 Related (ubiquitin ligase complex component)
family member (skr-16); K03094 S-phase kinase-associated
protein 1
Length=181
Score = 32.7 bits (73), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 16/60 (26%), Positives = 33/60 (55%), Gaps = 1/60 (1%)
Query 23 VLVSSEGEEFKVPREVASASLLVKSMTEDGDDTDV-VPLPKVSSYILKKVVEYCTHHHDN 81
+L+SS+GE+F+ S ++ ++ + D + + KV L +V+E+C +H D+
Sbjct 33 ILISSDGEKFQTDGHCIRHSKVLMQASKSLETPDTPIQVEKVQGDTLNRVLEWCNNHRDD 92
> cel:F54D10.1 skr-15; SKp1 Related (ubiquitin ligase complex
component) family member (skr-15); K03094 S-phase kinase-associated
protein 1
Length=184
Score = 32.3 bits (72), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 20/76 (26%), Positives = 38/76 (50%), Gaps = 14/76 (18%)
Query 13 FAMEAEPRCVVLVSSEGEEFKVPREVASASLLVKSMT------EDGDDTDVVPLPKVSSY 66
+++E+ R V+ +S + + ++ L S+T E+ + +P+ KV+
Sbjct 21 YSIESNDRVVLKISEQA--------IKQSATLSNSITNLGYSAENAESMVPIPIEKVNGK 72
Query 67 ILKKVVEYCTHHHDNP 82
LK VVE+C HH +P
Sbjct 73 TLKLVVEWCEHHKADP 88
> sce:YPL046C ELC1; Elc1p; K03872 transcription elongation factor
B, polypeptide 1
Length=99
Score = 31.6 bits (70), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 33/59 (55%), Gaps = 2/59 (3%)
Query 22 VVLVSSEGEEFKVPREVASASLLVKSMTED--GDDTDVVPLPKVSSYILKKVVEYCTHH 78
V LVS + +E+++ R A S +K+M E + + L + S+IL+K VEY ++
Sbjct 6 VTLVSKDDKEYEISRSAAMISPTLKAMIEGPFRESKGRIELKQFDSHILEKAVEYLNYN 64
> cel:C06A8.4 skr-17; SKp1 Related (ubiquitin ligase complex component)
family member (skr-17); K03094 S-phase kinase-associated
protein 1
Length=180
Score = 31.6 bits (70), Expect = 0.68, Method: Compositional matrix adjust.
Identities = 21/72 (29%), Positives = 36/72 (50%), Gaps = 7/72 (9%)
Query 19 PRCVVLVSSEGEEFKVP-REVASASLLVKSMTEDGDDTDV------VPLPKVSSYILKKV 71
PR + L SS+G + R + +S L ++ E G D + VP+ V + LK +
Sbjct 22 PRLLQLTSSDGHLLQGDIRALLLSSTLAATIRELGYDKEYCAELKPVPVNNVVGFTLKLL 81
Query 72 VEYCTHHHDNPP 83
+E+C H ++ P
Sbjct 82 IEWCDKHKEDDP 93
> cel:C52D10.7 skr-9; SKp1 Related (ubiquitin ligase complex component)
family member (skr-9); K03094 S-phase kinase-associated
protein 1
Length=194
Score = 31.6 bits (70), Expect = 0.72, Method: Compositional matrix adjust.
Identities = 17/58 (29%), Positives = 30/58 (51%), Gaps = 5/58 (8%)
Query 26 SSEGEEFKVPREVASASLLVKSMT-----EDGDDTDVVPLPKVSSYILKKVVEYCTHH 78
S++G+ F++ E S ++ ++ ED D +P+ V ILK V+E+C H
Sbjct 27 SNDGKVFEISDEAVKQSNILSNLISTCAPEDVASMDPIPITNVIGNILKMVIEWCEKH 84
Lambda K H
0.314 0.131 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2044675024
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40