bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0285_orf1
Length=170
Score E
Sequences producing significant alignments: (Bits) Value
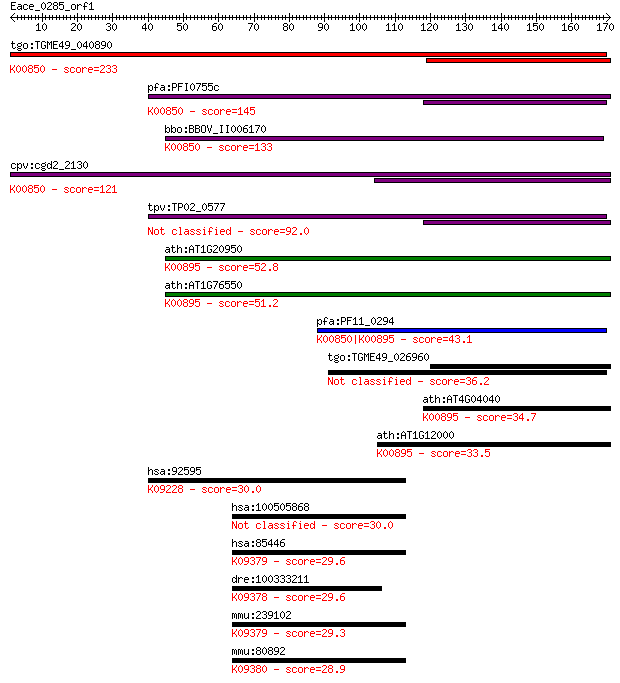
tgo:TGME49_040890 phosphofructokinase, putative (EC:2.7.1.90);... 233 2e-61
pfa:PFI0755c 6-phosphofructokinase, putative; K00850 6-phospho... 145 9e-35
bbo:BBOV_II006170 18.m06510; 6-phosphofructokinase (EC:2.7.1.9... 133 3e-31
cpv:cgd2_2130 pyrophosphate-dependent phosphofructokinase (EC:... 121 1e-27
tpv:TP02_0577 6-phosphofructokinase 92.0 8e-19
ath:AT1G20950 pyrophosphate--fructose-6-phosphate 1-phosphotra... 52.8 6e-07
ath:AT1G76550 pyrophosphate--fructose-6-phosphate 1-phosphotra... 51.2 2e-06
pfa:PF11_0294 ATP-dependent phosphofructokinase, putative; K00... 43.1 5e-04
tgo:TGME49_026960 phosphofructokinase, putative (EC:2.7.1.90) 36.2 0.055
ath:AT4G04040 MEE51; MEE51 (maternal effect embryo arrest 51);... 34.7 0.16
ath:AT1G12000 pyrophosphate--fructose-6-phosphate 1-phosphotra... 33.5 0.38
hsa:92595 ZNF764, MGC13138; zinc finger protein 764; K09228 KR... 30.0 4.3
hsa:100505868 zinc finger homeobox protein 2-like 30.0 4.4
hsa:85446 ZFHX2, FLJ17566, FLJ42478, KIAA1056, KIAA1762, MGC14... 29.6 4.9
dre:100333211 zinc finger homeobox 3-like; K09378 AT-binding t... 29.6 5.2
mmu:239102 Zfhx2, ZFH-5; zinc finger homeobox 2; K09379 zinc f... 29.3 6.9
mmu:80892 Zfhx4, A930021B15, C130041O22Rik, Zfh-4, Zfh4; zinc ... 28.9 9.9
> tgo:TGME49_040890 phosphofructokinase, putative (EC:2.7.1.90);
K00850 6-phosphofructokinase [EC:2.7.1.11]
Length=1399
Score = 233 bits (594), Expect = 2e-61, Method: Compositional matrix adjust.
Identities = 106/173 (61%), Positives = 134/173 (77%), Gaps = 7/173 (4%)
Query 1 QFEGPTADEPAYAVSIPTKEELLH----QSEAICHVGVGGRVVKSFGFLSALQRERLTHR 56
QF G TADEP YAV +P+ E+LL ++ C GV V K G S+LQ++RL +R
Sbjct 671 QFAGVTADEPCYAVMVPSAEDLLAGNDPRANLTCTTGV---VEKDLGTYSSLQQQRLLYR 727
Query 57 PPVPPLCLDPKCKARPMKQVLCKDPYTQRQVLMHYPYLANSTLFYLHEVAHDKYTPPINY 116
P VPPLC D K +ARP KQ+ C+DPYTQRQVLMHYPYL+NS+LF+LH+V HDKY PPI +
Sbjct 728 PTVPPLCQDAKARARPSKQMFCRDPYTQRQVLMHYPYLSNSSLFFLHDVCHDKYIPPIGF 787
Query 117 GLRVGFVLLSRQSPGVMNVLWGLHERLKMVQGKCLAFFGLTGLIEKNYIEITD 169
G+R+G V +SRQSPGV NVLWGLHERLK+VQGKC+AFFGL GL+++ +++I D
Sbjct 788 GVRIGLVFISRQSPGVANVLWGLHERLKLVQGKCIAFFGLNGLVDRKFLQIED 840
Score = 37.7 bits (86), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 22/55 (40%), Positives = 30/55 (54%), Gaps = 3/55 (5%)
Query 119 RVGFVLLSRQSPGVMNVLWGLHERLKMVQGKCLAF---FGLTGLIEKNYIEITDE 170
R+G VL +PG NV+ GLH+ +K + F GL G+I K Y +TDE
Sbjct 228 RIGIVLSGGPAPGGHNVIAGLHDFVKSRHPDSVVFGFMGGLDGVINKKYKVLTDE 282
> pfa:PFI0755c 6-phosphofructokinase, putative; K00850 6-phosphofructokinase
[EC:2.7.1.11]
Length=1418
Score = 145 bits (365), Expect = 9e-35, Method: Composition-based stats.
Identities = 64/131 (48%), Positives = 90/131 (68%), Gaps = 0/131 (0%)
Query 40 KSFGFLSALQRERLTHRPPVPPLCLDPKCKARPMKQVLCKDPYTQRQVLMHYPYLANSTL 99
KS G +S LQ RL ++ +P LC D K K R KQ + DPYTQ+Q+L +YP+++
Sbjct 773 KSLGCMSELQTSRLYNKLELPELCSDLKAKVRAGKQYISNDPYTQKQILSNYPHMSYENK 832
Query 100 FYLHEVAHDKYTPPINYGLRVGFVLLSRQSPGVMNVLWGLHERLKMVQGKCLAFFGLTGL 159
F + E+ HDKY PI++ +R+G V LSRQ+PG MNVL GL+ RLK+++G C+AF+GL GL
Sbjct 833 FQIQEIFHDKYASPISFEIRIGIVFLSRQAPGAMNVLCGLYRRLKLLKGVCIAFYGLYGL 892
Query 160 IEKNYIEITDE 170
+ YI I D+
Sbjct 893 LHNKYIIIDDD 903
Score = 37.4 bits (85), Expect = 0.024, Method: Composition-based stats.
Identities = 20/55 (36%), Positives = 32/55 (58%), Gaps = 3/55 (5%)
Query 118 LRVGFVLLSRQSPGVMNVLWGLHERLKMV--QGKCLAFFG-LTGLIEKNYIEITD 169
L++G +L +PG NV+ G+++ K Q + + F G + GL KNY+ ITD
Sbjct 180 LKIGIILSGGPAPGGHNVISGIYDYAKRYNEQSQVIGFLGGIDGLYSKNYVTITD 234
> bbo:BBOV_II006170 18.m06510; 6-phosphofructokinase (EC:2.7.1.90);
K00850 6-phosphofructokinase [EC:2.7.1.11]
Length=1337
Score = 133 bits (335), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 64/129 (49%), Positives = 84/129 (65%), Gaps = 5/129 (3%)
Query 45 LSALQRERLTHRPPVPPLCLDPKCKARPMKQVLCKDPYTQRQVLMHYPYLANSTLFYLHE 104
+S LQ RL RPP+P LC DP+ + R K+V D YT+ QV ++YPY+ + F LHE
Sbjct 693 MSPLQLSRLEWRPPIPMLCHDPRARMRNFKEVHSNDSYTRDQVALNYPYMTRRSHFSLHE 752
Query 105 VAHDKYTPPINY-----GLRVGFVLLSRQSPGVMNVLWGLHERLKMVQGKCLAFFGLTGL 159
V N GLRVG VLLS+Q+PGV+NV+WG+HER+ ++ G C+AF G GL
Sbjct 753 VVGHYSGDRTNLNKATQGLRVGVVLLSKQAPGVLNVIWGIHERMSIIGGSCVAFHGALGL 812
Query 160 IEKNYIEIT 168
IE NYIE+T
Sbjct 813 IEGNYIELT 821
> cpv:cgd2_2130 pyrophosphate-dependent phosphofructokinase (EC:2.7.1.11);
K00850 6-phosphofructokinase [EC:2.7.1.11]
Length=1426
Score = 121 bits (304), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 72/182 (39%), Positives = 105/182 (57%), Gaps = 12/182 (6%)
Query 1 QFEGPTADEPAYAVSIPTKEELLHQSEAIC----------HVGVGGRVV--KSFGFLSAL 48
QFEGP ++ Y V P ++LL +A + G G R V + G LS+L
Sbjct 648 QFEGPCSETIPYIVRTPELQDLLCGEDASVTDTDINIVQPNNGGGKRGVFNRIQGTLSSL 707
Query 49 QRERLTHRPPVPPLCLDPKCKARPMKQVLCKDPYTQRQVLMHYPYLANSTLFYLHEVAHD 108
Q R T+RPP+P L K + +P Q DP +RQ+ YP+L NS FY E D
Sbjct 708 QLYRTTYRPPLPLLLTHLKARCKPTTQYNLSDPLIKRQITTCYPHLCNSNHFYCQEAQLD 767
Query 109 KYTPPINYGLRVGFVLLSRQSPGVMNVLWGLHERLKMVQGKCLAFFGLTGLIEKNYIEIT 168
+ N GLR+G +L+SRQ+PG+ NV+WGL+ RLKMV+G+CL F+G+ G ++ +I +T
Sbjct 768 ISSEEPNVGLRIGVLLMSRQAPGIANVVWGLYHRLKMVRGRCLGFYGVKGFLQGKHITLT 827
Query 169 DE 170
++
Sbjct 828 EK 829
Score = 33.9 bits (76), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 24/72 (33%), Positives = 37/72 (51%), Gaps = 5/72 (6%)
Query 104 EVAHD--KYTPPINYGLRVGFVLLSRQSPGVMNVLWGLHERLKMV--QGKCLAFF-GLTG 158
E+ D K + ++ LRVG VL +PG NV+ G+++ +K + F GL G
Sbjct 140 EITRDESKNSENSDHALRVGIVLSGGPAPGGHNVICGVYDFIKNHHPDSQLFGFLGGLDG 199
Query 159 LIEKNYIEITDE 170
L + Y I+DE
Sbjct 200 LFKSTYKVISDE 211
> tpv:TP02_0577 6-phosphofructokinase
Length=1692
Score = 92.0 bits (227), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 51/146 (34%), Positives = 78/146 (53%), Gaps = 18/146 (12%)
Query 40 KSFGFLSALQRERLTHRPPVPPLCLDPKCKARPMKQVLCKDPYTQRQVLMHYPYLANSTL 99
K F+S L+ RL P +P +C D K + P KQ + D Y + Q++++YPY
Sbjct 990 KDKSFMSELELRRLEVIPDIPIVCSDVKSRLVPFKQHVEFDQYIKNQIMLYYPYQHKLNH 1049
Query 100 FYLHEVAHDKYTPPINYG----------------LRVGFVLLSRQSPGVMNVLWGLHERL 143
F +E+ + T P G LRVG V + +Q+PGV+NV WG+ R+
Sbjct 1050 FNQYELVPN--TIPTGMGRREIGSVSSESLKSGSLRVGVVPVGKQAPGVLNVFWGIFTRV 1107
Query 144 KMVQGKCLAFFGLTGLIEKNYIEITD 169
+ GKCLAF G+ GL++ +YIE+ +
Sbjct 1108 SKMGGKCLAFHGIKGLLQNSYIELKE 1133
Score = 29.3 bits (64), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 20/56 (35%), Positives = 31/56 (55%), Gaps = 3/56 (5%)
Query 118 LRVGFVLLSRQSPGVMNVLWGLHERLKMV--QGKCLAFF-GLTGLIEKNYIEITDE 170
LR+G +L +PG NV+ G+ + LK + + + F GL GL K Y IT++
Sbjct 302 LRIGLILSGGPAPGGHNVIAGVFDYLKHRNPESQLIGFMGGLDGLKYKRYQIITED 357
> ath:AT1G20950 pyrophosphate--fructose-6-phosphate 1-phosphotransferase-related
/ pyrophosphate-dependent 6-phosphofructose-1-kinase-related;
K00895 pyrophosphate--fructose-6-phosphate
1-phosphotransferase [EC:2.7.1.90]
Length=614
Score = 52.8 bits (125), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 40/132 (30%), Positives = 59/132 (44%), Gaps = 6/132 (4%)
Query 45 LSALQRERLTHRPPVPPLCLDPKCKARPMKQVLCKDPYTQRQVLMHYPY-LANSTLFYLH 103
LS LQ+ R ++P +PP + + + +P+ L +L
Sbjct 11 LSPLQQLRSQYKPELPPCLQGTTVRVELGDGTTVAEAADSHTMARAFPHTLGQPLAHFLR 70
Query 104 EVAH--DKYTPPINYGLRVGFVLLSRQSPGVMNVLWGLHERLKMVQGKC--LAFFGLT-G 158
E A D + +RVG V RQ+PG NV+WGL E LK+ K L F G + G
Sbjct 71 ETAQVPDAHIITELPSVRVGIVFCGRQAPGGHNVIWGLFEALKVHNAKSTLLGFLGGSEG 130
Query 159 LIEKNYIEITDE 170
L + +EITD+
Sbjct 131 LFAQKTLEITDD 142
> ath:AT1G76550 pyrophosphate--fructose-6-phosphate 1-phosphotransferase
alpha subunit, putative / pyrophosphate-dependent
6-phosphofructose-1-kinase, putative; K00895 pyrophosphate--fructose-6-phosphate
1-phosphotransferase [EC:2.7.1.90]
Length=617
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 47/145 (32%), Positives = 64/145 (44%), Gaps = 32/145 (22%)
Query 45 LSALQRERLTHRPPVPPLCLDPK------------CKARPMKQVLCKDPYTQRQVLMHY- 91
LS LQ+ R + P +PP CL KA + P+T Q L H+
Sbjct 11 LSPLQQLRSQYHPELPP-CLQGTTVRVELGDGTTVAKAGDAHIIARAFPHTLGQPLAHFL 69
Query 92 ---PYLANSTLFYLHEVAHDKYTPPINYGLRVGFVLLSRQSPGVMNVLWGLHERLKMVQG 148
+A++ + H V RVG V RQ+PG NV+WGL+E LK+
Sbjct 70 RATAKVADAQIITEHPVK------------RVGIVFCGRQAPGGHNVVWGLYEALKVHNA 117
Query 149 K--CLAFFGLT-GLIEKNYIEITDE 170
K L F G + GL + +EITDE
Sbjct 118 KNTLLGFLGGSEGLFAQKTLEITDE 142
> pfa:PF11_0294 ATP-dependent phosphofructokinase, putative; K00850
6-phosphofructokinase [EC:2.7.1.11]; K00895 pyrophosphate--fructose-6-phosphate
1-phosphotransferase [EC:2.7.1.90]
Length=1570
Score = 43.1 bits (100), Expect = 5e-04, Method: Composition-based stats.
Identities = 31/88 (35%), Positives = 43/88 (48%), Gaps = 11/88 (12%)
Query 88 LMHYP-----YLANSTLFYLHEVAHDKYTPPINYGLRVGFVLLSRQSPGVMNVLWGLHER 142
+H P Y+ F+L + + T P N VG VLLS +PG N+L GLH+R
Sbjct 1024 FIHIPKIKDFYILQDNQFHLKQ-NYRNVTCPKN----VGVVLLSHCTPGTNNILVGLHQR 1078
Query 143 LKMVQGKCLAFF-GLTGLIEKNYIEITD 169
L + K + F GL GL+ + I D
Sbjct 1079 LSINNFKLIGFIKGLKGLLNNDICLIND 1106
> tgo:TGME49_026960 phosphofructokinase, putative (EC:2.7.1.90)
Length=1225
Score = 36.2 bits (82), Expect = 0.055, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 29/51 (56%), Gaps = 0/51 (0%)
Query 120 VGFVLLSRQSPGVMNVLWGLHERLKMVQGKCLAFFGLTGLIEKNYIEITDE 170
VG VL+ + PG NVL GL +R+ ++ G F G GL+ + + I ++
Sbjct 677 VGVVLVGPERPGYANVLCGLVQRVALLGGTVKGFKGARGLLTNDCVVIGEK 727
Score = 32.0 bits (71), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 23/82 (28%), Positives = 37/82 (45%), Gaps = 5/82 (6%)
Query 91 YPYLANSTLFYLHEVAHDKYTPPINYGLRVGFVLLSRQSPGVMNVLWGLHERLKMVQGKC 150
+P N+ L + D+ + LRVG VL Q+ G NV+ G+ + +K V
Sbjct 106 FPNTINTNFCLLAPASGDRQAS--SEPLRVGVVLSGGQAAGGHNVICGIFDYVKRVNPAS 163
Query 151 LAF---FGLTGLIEKNYIEITD 169
F G G+ Y+E+T+
Sbjct 164 TVFGFLGGPHGVFSHEYVELTE 185
> ath:AT4G04040 MEE51; MEE51 (maternal effect embryo arrest 51);
diphosphate-fructose-6-phosphate 1-phosphotransferase (EC:2.7.1.90);
K00895 pyrophosphate--fructose-6-phosphate 1-phosphotransferase
[EC:2.7.1.90]
Length=569
Score = 34.7 bits (78), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 33/56 (58%), Gaps = 3/56 (5%)
Query 118 LRVGFVLLSRQSPGVMNVLWGLHERLK-MVQGKCLAFF--GLTGLIEKNYIEITDE 170
L++G VL Q+PG NV+ G+ + L+ +G L F G G+++ YIE+T E
Sbjct 98 LKIGVVLSGGQAPGGHNVICGIFDYLQEYARGSSLFGFRGGPAGIMKGKYIELTSE 153
> ath:AT1G12000 pyrophosphate--fructose-6-phosphate 1-phosphotransferase
beta subunit, putative / pyrophosphate-dependent
6-phosphofructose-1-kinase, putative (EC:2.7.1.90); K00895 pyrophosphate--fructose-6-phosphate
1-phosphotransferase [EC:2.7.1.90]
Length=566
Score = 33.5 bits (75), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 22/70 (31%), Positives = 36/70 (51%), Gaps = 5/70 (7%)
Query 105 VAHDKYTPPINYGLRVGFVLLSRQSPGVMNVLWGLHERLKMVQGKCLAFFGL----TGLI 160
V D+ P L++G VL Q+PG NV+ GL + L+ + K F+G G++
Sbjct 83 VVPDQDAPSSAPKLKIGVVLSGGQAPGGHNVISGLFDYLQE-RAKGSTFYGFKGGPAGIM 141
Query 161 EKNYIEITDE 170
+ Y+E+ E
Sbjct 142 KCKYVELNAE 151
> hsa:92595 ZNF764, MGC13138; zinc finger protein 764; K09228
KRAB domain-containing zinc finger protein
Length=407
Score = 30.0 bits (66), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 24/89 (26%), Positives = 31/89 (34%), Gaps = 28/89 (31%)
Query 40 KSFGFLSALQRERLTHRPPVPPLCLDPKCKARPMKQVLCKDPYTQRQVLMHY-------- 91
K FG S+L + R HR P CL+ C +TQR L +
Sbjct 209 KGFGHASSLSKHRAIHRGERPHRCLE------------CGRAFTQRSALTSHLRVHTGEK 256
Query 92 PY--------LANSTLFYLHEVAHDKYTP 112
PY + S+ Y H H TP
Sbjct 257 PYGCADCGRRFSQSSALYQHRRVHSGETP 285
> hsa:100505868 zinc finger homeobox protein 2-like
Length=2706
Score = 30.0 bits (66), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 28/49 (57%), Gaps = 4/49 (8%)
Query 64 LDPKCKARPMKQVLCKDPYTQRQVLMHYPYLANSTLFYLHEVAHDKYTP 112
LDP ARP K +CK+ +TQ+ +L+ + Y + S L + + A D P
Sbjct 1319 LDP---ARPYKCTVCKESFTQKNILLVH-YNSVSHLHKMKKAAIDPSAP 1363
> hsa:85446 ZFHX2, FLJ17566, FLJ42478, KIAA1056, KIAA1762, MGC141820,
ZFH-5, ZNF409; zinc finger homeobox 2; K09379 zinc finger
homeobox protein 2
Length=2572
Score = 29.6 bits (65), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 28/49 (57%), Gaps = 4/49 (8%)
Query 64 LDPKCKARPMKQVLCKDPYTQRQVLMHYPYLANSTLFYLHEVAHDKYTP 112
LDP ARP K +CK+ +TQ+ +L+ + Y + S L + + A D P
Sbjct 1185 LDP---ARPYKCTVCKESFTQKNILLVH-YNSVSHLHKMKKAAIDPSAP 1229
> dre:100333211 zinc finger homeobox 3-like; K09378 AT-binding
transcription factor 1
Length=3809
Score = 29.6 bits (65), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 28/43 (65%), Gaps = 9/43 (20%)
Query 64 LDPKCKARPMKQVLCKDPYTQRQVLM-HYPYLANSTLFYLHEV 105
LDP +RP K +CK+ +TQ+ +L+ HY +++ +LH+V
Sbjct 1664 LDP---SRPFKCTVCKESFTQKNILLVHY-----NSVSHLHKV 1698
> mmu:239102 Zfhx2, ZFH-5; zinc finger homeobox 2; K09379 zinc
finger homeobox protein 2
Length=2562
Score = 29.3 bits (64), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 28/49 (57%), Gaps = 4/49 (8%)
Query 64 LDPKCKARPMKQVLCKDPYTQRQVLMHYPYLANSTLFYLHEVAHDKYTP 112
LDP ARP K +CK+ +TQ+ +L+ + Y + S L + + A D P
Sbjct 1179 LDP---ARPYKCTVCKESFTQKNILLVH-YNSVSHLHKMKKAAIDPSGP 1223
> mmu:80892 Zfhx4, A930021B15, C130041O22Rik, Zfh-4, Zfh4; zinc
finger homeodomain 4; K09380 zinc finger homeobox protein
4
Length=3581
Score = 28.9 bits (63), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 29/49 (59%), Gaps = 4/49 (8%)
Query 64 LDPKCKARPMKQVLCKDPYTQRQVLMHYPYLANSTLFYLHEVAHDKYTP 112
LDP +RP K +CK+ +TQ+ +L+ + Y + S L L +V + +P
Sbjct 1506 LDP---SRPYKCTVCKESFTQKNILLVH-YNSVSHLHKLKKVLQEASSP 1550
Lambda K H
0.323 0.140 0.440
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4287611064
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40