bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0289_orf1
Length=143
Score E
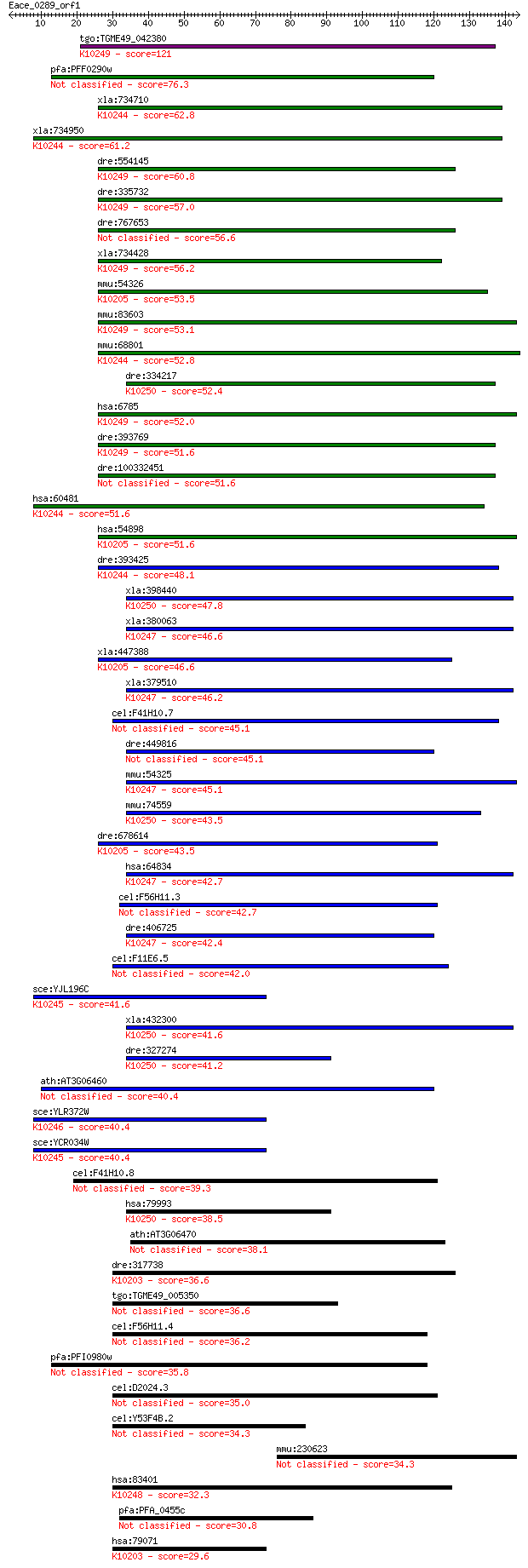
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_042380 elongation of very long chain fatty acids 4 ... 121 8e-28
pfa:PFF0290w long chain polyunsaturated fatty acid elongation ... 76.3 3e-14
xla:734710 hypothetical protein MGC114802; K10244 elongation o... 62.8 4e-10
xla:734950 elovl5, MGC131143; ELOVL family member 5, elongatio... 61.2 1e-09
dre:554145 im:7139490; zgc:112263; K10249 elongation of very l... 60.8 1e-09
dre:335732 elovl4b, elovl4, wu:fk62b04, zgc:73054; elongation ... 57.0 2e-08
dre:767653 MGC153394; zgc:153394 56.6 3e-08
xla:734428 hypothetical protein MGC115163; K10249 elongation o... 56.2 3e-08
mmu:54326 Elovl2, AI317360, Ssc2; elongation of very long chai... 53.5 2e-07
mmu:83603 Elovl4; elongation of very long chain fatty acids (F... 53.1 3e-07
mmu:68801 Elovl5, 1110059L23Rik, AI747313, AU043003, HELO1; EL... 52.8 3e-07
dre:334217 elovl7a, MGC174838, elovl7, fi36f02, wu:fi36f02, zg... 52.4 5e-07
hsa:6785 ELOVL4, ADMD, CT118, FLJ17667, FLJ92876, STGD2, STGD3... 52.0 7e-07
dre:393769 elovl4a, MGC73341, zgc:73341; elongation of very lo... 51.6 7e-07
dre:100332451 elongation of very long chain fatty acids-like 4... 51.6 8e-07
hsa:60481 ELOVL5, HELO1, dJ483K16.1; ELOVL family member 5, el... 51.6 8e-07
hsa:54898 ELOVL2, FLJ20334, SSC2; elongation of very long chai... 51.6 9e-07
dre:393425 elovl5, MGC63549, zgc:63549; ELOVL family member 5,... 48.1 1e-05
xla:398440 elovl7; ELOVL family member 7, elongation of long c... 47.8 1e-05
xla:380063 elovl1, MGC52731; elongation of very long chain fat... 46.6 3e-05
xla:447388 elovl2, MGC84669; elongation of very long chain fat... 46.6 3e-05
xla:379510 hypothetical protein MGC64517; K10247 elongation of... 46.2 3e-05
cel:F41H10.7 elo-5; fatty acid ELOngation family member (elo-5) 45.1 8e-05
dre:449816 elovl1a, zgc:103538; elongation of very long chain ... 45.1 8e-05
mmu:54325 Elovl1, AA407424, BB151133, Ssc1; elongation of very... 45.1 8e-05
mmu:74559 Elovl7, 9130013K24Rik, AI840082; ELOVL family member... 43.5 2e-04
dre:678614 elovl2, MGC136352, MGC158526, zgc:136352; elongatio... 43.5 2e-04
hsa:64834 ELOVL1, Ssc1; elongation of very long chain fatty ac... 42.7 4e-04
cel:F56H11.3 elo-7; fatty acid ELOngation family member (elo-7) 42.7 4e-04
dre:406725 elovl1b, wu:fj32h05, zgc:56567; elongation of very ... 42.4 4e-04
cel:F11E6.5 elo-2; fatty acid ELOngation family member (elo-2) 42.0 6e-04
sce:YJL196C ELO1; Elo1p; K10245 fatty acid elongase 2 [EC:2.3.... 41.6 8e-04
xla:432300 hypothetical protein MGC80262; K10250 elongation of... 41.6 8e-04
dre:327274 elovl7b, elovl1, wu:fd20a06, zgc:56422; ELOVL famil... 41.2 0.001
ath:AT3G06460 GNS1/SUR4 membrane family protein 40.4 0.002
sce:YLR372W SUR4, APA1, ELO3, SRE1, VBM1; Elongase, involved i... 40.4 0.002
sce:YCR034W FEN1, ELO2, GNS1, VBM2; Fatty acid elongase, invol... 40.4 0.002
cel:F41H10.8 elo-6; fatty acid ELOngation family member (elo-6) 39.3 0.004
hsa:79993 ELOVL7, FLJ23563; ELOVL family member 7, elongation ... 38.5 0.006
ath:AT3G06470 GNS1/SUR4 membrane family protein 38.1 0.009
dre:317738 elovl6, cb618, lce, zgc:73089; ELOVL family member ... 36.6 0.025
tgo:TGME49_005350 fatty acid elongation protein, putative 36.6 0.028
cel:F56H11.4 elo-1; fatty acid ELOngation family member (elo-1) 36.2 0.038
pfa:PFI0980w long chain fatty acid elongation enzyme, putative 35.8 0.050
cel:D2024.3 elo-3; fatty acid ELOngation family member (elo-3) 35.0 0.075
cel:Y53F4B.2 elo-9; fatty acid ELOngation family member (elo-9) 34.3 0.14
mmu:230623 Skint11, A630098G03Rik, Gm569; selection and upkeep... 34.3 0.15
hsa:83401 ELOVL3, CIG-30, CIG30, MGC21435; elongation of very ... 32.3 0.56
pfa:PFA_0455c fatty acid elongation protein, GNS1/SUR4 family,... 30.8 1.4
hsa:79071 ELOVL6, FACE, FAE, FLJ23378, LCE, MGC5487; ELOVL fam... 29.6 3.2
> tgo:TGME49_042380 elongation of very long chain fatty acids
4 protein, putative ; K10249 elongation of very long chain fatty
acids protein 4
Length=350
Score = 121 bits (303), Expect = 8e-28, Method: Compositional matrix adjust.
Identities = 58/116 (50%), Positives = 77/116 (66%), Gaps = 2/116 (1%)
Query 21 ECGYDGEIYYVVALNALVHVVMYAYYFMASLNSPLARTIKIFVTQLQMAQFLSMTAHACY 80
GYDG+IY+ VA N +HVVMYAYY MASLN AR IK F+T++QM QFL M Y
Sbjct 224 SVGYDGDIYFAVAANGAIHVVMYAYYLMASLNIGAARYIKPFITRMQMTQFLGMLGQGFY 283
Query 81 HVYFYKSCRYPVRVTIGYFFYVLSLFLLFRNFSKKTYGKKPATAAVGAASSKDGRS 136
H+ FY+ C+YP+R+ + Y Y++S++ LF F+K+TYG K A G A K R+
Sbjct 284 HICFYQDCQYPIRIGVVYLGYIVSMYALFDRFAKRTYGTK--NAGKGNACQKPVRA 337
> pfa:PFF0290w long chain polyunsaturated fatty acid elongation
enzyme, putative
Length=293
Score = 76.3 bits (186), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 42/107 (39%), Positives = 63/107 (58%), Gaps = 0/107 (0%)
Query 13 YLHLLGERECGYDGEIYYVVALNALVHVVMYAYYFMASLNSPLARTIKIFVTQLQMAQFL 72
+L + GYDG+IYY++ +N+ VH VMY YY+++S+ + K VT LQM QFL
Sbjct 181 FLIMWVNTSVGYDGDIYYIIVVNSFVHFVMYLYYYLSSVKFKVPIFAKACVTYLQMLQFL 240
Query 73 SMTAHACYHVYFYKSCRYPVRVTIGYFFYVLSLFLLFRNFSKKTYGK 119
S+ Y ++ C YP ++ F+Y +SL +LF NF+ TY K
Sbjct 241 SIILPGFYVLFVRHYCPYPRKLVGLSFYYCISLLILFGNFALHTYIK 287
> xla:734710 hypothetical protein MGC114802; K10244 elongation
of very long chain fatty acids protein 5 [EC:2.3.1.-]
Length=295
Score = 62.8 bits (151), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 42/125 (33%), Positives = 72/125 (57%), Gaps = 21/125 (16%)
Query 26 GEIYYVVALNALVHVVMYAYYFMASLNSPLART---IKIFVTQLQMAQF-LSMTAHACYH 81
G Y+ LN+ +HV+MY+YY ++++ P R K ++TQ Q+ QF L+MT C
Sbjct 164 GHSYFGATLNSFIHVLMYSYYGLSAI--PAIRPYLWWKKYITQCQLTQFVLTMTQTTCAM 221
Query 82 VYFYKSCRYPVRVTIGYFF----YVLSLFLLFRNFSKKTYGKKPATA----AVGAASSKD 133
++ C++P +G+ + Y++SL +LF NF KTY KK ++ G+AS+ +
Sbjct 222 IW---PCKFP----MGWLYFQNSYMISLIILFTNFYIKTYKKKTSSRRKEYQNGSASAVN 274
Query 134 GRSNT 138
G +N+
Sbjct 275 GHTNS 279
> xla:734950 elovl5, MGC131143; ELOVL family member 5, elongation
of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like); K10244
elongation of very long chain fatty acids protein 5 [EC:2.3.1.-]
Length=295
Score = 61.2 bits (147), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 44/143 (30%), Positives = 79/143 (55%), Gaps = 24/143 (16%)
Query 8 SSLFCYLHLLGERECGYDGEIYYVVALNALVHVVMYAYYFMASLNSPLART---IKIFVT 64
S L + ++ CG+ ++ LN+ +HV+MY+YY ++++ P R K ++T
Sbjct 149 SMLNIWWFVMNWVPCGHS---FFGATLNSFIHVLMYSYYGLSAI--PAIRPYLWWKKYIT 203
Query 65 QLQMAQF-LSMTAHACYHVYFYKSCRYPVRVTIGYFF----YVLSLFLLFRNFSKKTYGK 119
Q Q+ QF L+MT C ++ C++P +G+ + Y++SL +LF NF KTY K
Sbjct 204 QCQLTQFVLTMTQTTCAMIW---PCKFP----MGWLYFQNSYMISLIILFTNFYLKTYNK 256
Query 120 KPATA----AVGAASSKDGRSNT 138
K ++ G+AS+ +G +N+
Sbjct 257 KTSSRRKEYQNGSASAVNGYTNS 279
> dre:554145 im:7139490; zgc:112263; K10249 elongation of very
long chain fatty acids protein 4
Length=264
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 33/102 (32%), Positives = 60/102 (58%), Gaps = 4/102 (3%)
Query 26 GEIYYVVALNALVHVVMYAYYFMASLNSPLARTI--KIFVTQLQMAQFLSMTAHACYHVY 83
G+ +++ LN VH+ MY+YY +A+L L + + K ++T LQ+ QF+ +T H Y++
Sbjct 165 GQSFFIGLLNTFVHIWMYSYYGLAALGPHLQKYLWWKRYLTSLQLVQFILLTVHTGYNL- 223
Query 84 FYKSCRYPVRVTIGYFFYVLSLFLLFRNFSKKTYGKKPATAA 125
+ C +P + F Y +SL +LF NF ++Y K+ + +
Sbjct 224 -FTECEFPDSMNAVVFAYCVSLIILFSNFYYQSYIKRKSKKS 264
> dre:335732 elovl4b, elovl4, wu:fk62b04, zgc:73054; elongation
of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like
4b; K10249 elongation of very long chain fatty acids
protein 4
Length=303
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 34/120 (28%), Positives = 62/120 (51%), Gaps = 9/120 (7%)
Query 26 GEIYYVVALNALVHVVMYAYYFMASLNSPLARTI--KIFVTQLQMAQFLSMTAHACYHVY 83
G+ ++ +N+ +HV+MY YY +A+ + + + K ++T +QM QF HA + +Y
Sbjct 168 GQSFFGATINSGIHVLMYGYYGLAAFGPKIQKYLWWKKYLTIIQMIQFHVTIGHAAHSLY 227
Query 84 FYKSCRYPVRVTIGYFFYVLSLFLLFRNFSKKTYGKKP-----ATAAVGAASSKDGRSNT 138
C +P + Y ++ +LF NF +TY ++P +A G + S +G S T
Sbjct 228 --TGCPFPAWMQWALIGYAVTFIILFANFYYQTYRRQPRLKTAKSAVNGVSMSTNGTSKT 285
> dre:767653 MGC153394; zgc:153394
Length=268
Score = 56.6 bits (135), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 34/102 (33%), Positives = 56/102 (54%), Gaps = 4/102 (3%)
Query 26 GEIYYVVALNALVHVVMYAYYFMASLNSPLARTI--KIFVTQLQMAQFLSMTAHACYHVY 83
G+ + + +N+ VHVVMY YY +A+L + + + K ++T LQ+ QF +T H +++
Sbjct 169 GQSFLIGLINSFVHVVMYMYYGLAALGPQMQKYLWWKRYLTSLQLLQFFIVTIHTAFNL- 227
Query 84 FYKSCRYPVRVTIGYFFYVLSLFLLFRNFSKKTYGKKPATAA 125
Y C +P + + Y LSL LF NF ++Y K A
Sbjct 228 -YADCDFPDSMNMVVLGYALSLIALFSNFYYQSYLSKKTKLA 268
> xla:734428 hypothetical protein MGC115163; K10249 elongation
of very long chain fatty acids protein 4
Length=265
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 32/98 (32%), Positives = 57/98 (58%), Gaps = 4/98 (4%)
Query 26 GEIYYVVALNALVHVVMYAYYFMASLNSPLARTI--KIFVTQLQMAQFLSMTAHACYHVY 83
G+ +++ LN+ VH+ MY YY +A L + + + K ++T LQ+ QF ++ H+ Y++
Sbjct 165 GQAFFIGMLNSFVHIFMYLYYGLAVLGPKMQKYLWWKRYLTLLQLTQFGAIALHSSYNL- 223
Query 84 FYKSCRYPVRVTIGYFFYVLSLFLLFRNFSKKTYGKKP 121
C +P F Y++SL +LF NF +TY ++P
Sbjct 224 -VTDCPFPDGFNGVVFAYIVSLIILFLNFYYQTYLRRP 260
> mmu:54326 Elovl2, AI317360, Ssc2; elongation of very long chain
fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2; K10205
elongation of very long chain fatty acids protein 2 [EC:2.3.1.-]
Length=292
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 35/112 (31%), Positives = 59/112 (52%), Gaps = 7/112 (6%)
Query 26 GEIYYVVALNALVHVVMYAYYFMA---SLNSPLARTIKIFVTQLQMAQFLSMTAHACYHV 82
G+ ++ LN+ +H++MY+YY ++ S++ L K ++TQ Q+ QF+ H V
Sbjct 167 GQSFFGPTLNSFIHILMYSYYGLSVFPSMHKYLWW--KKYLTQAQLVQFVLTITHTLSAV 224
Query 83 YFYKSCRYPVRVTIGYFFYVLSLFLLFRNFSKKTYGKKPATAAVGAASSKDG 134
K C +P I Y+++L +LF NF +TY KKP + K+G
Sbjct 225 V--KPCGFPFGCLIFQSSYMMTLVILFLNFYIQTYRKKPVKKELQEKEVKNG 274
> mmu:83603 Elovl4; elongation of very long chain fatty acids
(FEN1/Elo2, SUR4/Elo3, yeast)-like 4; K10249 elongation of very
long chain fatty acids protein 4
Length=312
Score = 53.1 bits (126), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 31/119 (26%), Positives = 57/119 (47%), Gaps = 5/119 (4%)
Query 26 GEIYYVVALNALVHVVMYAYYFMASLNSPLARTI--KIFVTQLQMAQFLSMTAHACYHVY 83
G+ ++ +N+ +HV+MY+YY + + + + + K ++T LQ+ QF H +
Sbjct 179 GQAFFGAQMNSFIHVIMYSYYGLTAFGPWIQKYLWWKRYLTMLQLVQFHVTIGHTA--LS 236
Query 84 FYKSCRYPVRVTIGYFFYVLSLFLLFRNFSKKTYGKKPATAAVGAASSKDGRSNTKAKA 142
Y C +P + Y +S LF NF +TY +P + G ++ SN K+
Sbjct 237 LYTDCPFPKWMHWALIAYAISFIFLFLNFYTRTY-NEPKQSKTGKTATNGISSNGVNKS 294
> mmu:68801 Elovl5, 1110059L23Rik, AI747313, AU043003, HELO1;
ELOVL family member 5, elongation of long chain fatty acids
(yeast); K10244 elongation of very long chain fatty acids protein
5 [EC:2.3.1.-]
Length=299
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 42/128 (32%), Positives = 63/128 (49%), Gaps = 24/128 (18%)
Query 26 GEIYYVVALNALVHVVMYAYYFMASLNSPLART---IKIFVTQLQMAQFLSMTAHACYHV 82
G Y+ LN+ +HV+MY+YY ++S+ P R K ++TQ Q+ QF+ V
Sbjct 164 GHSYFGATLNSFIHVLMYSYYGLSSI--PSMRPYLWWKKYITQGQLVQFVLTIIQTTCGV 221
Query 83 YFYKSCRYPVRVTIGYFF----YVLSLFLLFRNFSKKTYGKKPATAAVGAASSKD---GR 135
++ C +P +G+ F Y++SL LF NF +TY KK GA+ KD G
Sbjct 222 FW--PCSFP----LGWLFFQIGYMISLIALFTNFYIQTYNKK------GASRRKDHLKGH 269
Query 136 SNTKAKAA 143
N A
Sbjct 270 QNGSVAAV 277
> dre:334217 elovl7a, MGC174838, elovl7, fi36f02, wu:fi36f02,
zgc:55879; ELOVL family member 7, elongation of long chain fatty
acids (yeast) a; K10250 elongation of very long chain fatty
acids protein 7
Length=288
Score = 52.4 bits (124), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 37/112 (33%), Positives = 56/112 (50%), Gaps = 14/112 (12%)
Query 34 LNALVHVVMYAYYFMASLNSPLARTI--KIFVTQLQMAQFLSMTAHACYHVYFYKSCRYP 91
LN +VHV+MY YY +++L R + K +T LQ+ QF+ +T H + +F K C YP
Sbjct 176 LNCIVHVIMYTYYLLSALGPSFQRFLWWKKHLTSLQLIQFVLVTVHISQY-FFMKDCPYP 234
Query 92 VRVTIGYFFYVLSLF-----LLFRNFSKKTY--GKKPATAAVGAASSKDGRS 136
+ F Y+++L+ LLF NF Y GK+ S K+
Sbjct 235 YPL----FMYIIALYGIIFLLLFLNFWHHAYTKGKRLPKILQKTISPKNNND 282
> hsa:6785 ELOVL4, ADMD, CT118, FLJ17667, FLJ92876, STGD2, STGD3;
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3,
yeast)-like 4; K10249 elongation of very long chain
fatty acids protein 4
Length=314
Score = 52.0 bits (123), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 32/119 (26%), Positives = 57/119 (47%), Gaps = 5/119 (4%)
Query 26 GEIYYVVALNALVHVVMYAYYFMASLNSPLARTI--KIFVTQLQMAQFLSMTAHACYHVY 83
G+ ++ LN+ +HV+MY+YY + + + + + K ++T LQ+ QF H +
Sbjct 179 GQAFFGAQLNSFIHVIMYSYYGLTAFGPWIQKYLWWKRYLTMLQLIQFHVTIGHTA--LS 236
Query 84 FYKSCRYPVRVTIGYFFYVLSLFLLFRNFSKKTYGKKPATAAVGAASSKDGRSNTKAKA 142
Y C +P + Y +S LF NF +TY K+P G + +N +K+
Sbjct 237 LYTDCPFPKWMHWALIAYAISFIFLFLNFYIRTY-KEPKKPKAGKTAMNGISANGVSKS 294
> dre:393769 elovl4a, MGC73341, zgc:73341; elongation of very
long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4a;
K10249 elongation of very long chain fatty acids protein 4
Length=309
Score = 51.6 bits (122), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 34/122 (27%), Positives = 60/122 (49%), Gaps = 13/122 (10%)
Query 26 GEIYYVVALNALVHVVMYAYYFMASLNSPLARTI--KIFVTQLQMAQFLSMTAHACYHVY 83
G+ ++ +NA +HV+MY YY +A+ + + + K ++T +QM QF H +
Sbjct 168 GQSFFGAHMNAAIHVLMYLYYGLAAFGPKIQKFLWWKKYLTIIQMVQFHVTIGHTA--LS 225
Query 84 FYKSCRYPVRVTIGYFFYVLSLFLLFRNFSKKTYGKKPAT---------AAVGAASSKDG 134
Y C +P + Y L+ +LF NF +TY ++P A+ GA +S +G
Sbjct 226 LYSDCPFPKWMHWCLIGYALTFIILFGNFYYQTYRRQPRRDKPRALHNGASNGALTSSNG 285
Query 135 RS 136
+
Sbjct 286 NT 287
> dre:100332451 elongation of very long chain fatty acids-like
4-like
Length=298
Score = 51.6 bits (122), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 34/122 (27%), Positives = 60/122 (49%), Gaps = 13/122 (10%)
Query 26 GEIYYVVALNALVHVVMYAYYFMASLNSPLARTI--KIFVTQLQMAQFLSMTAHACYHVY 83
G+ ++ +NA +HV+MY YY +A+ + + + K ++T +QM QF H +
Sbjct 157 GQSFFGAHMNAAIHVLMYLYYGLAAFGPKIQKFLWWKKYLTIIQMVQFHVTIGHTA--LS 214
Query 84 FYKSCRYPVRVTIGYFFYVLSLFLLFRNFSKKTYGKKPAT---------AAVGAASSKDG 134
Y C +P + Y L+ +LF NF +TY ++P A+ GA +S +G
Sbjct 215 LYSDCPFPKWMHWCLIGYALTFIILFGNFYYQTYRRQPRRDKPRALHNGASNGALTSSNG 274
Query 135 RS 136
+
Sbjct 275 NT 276
> hsa:60481 ELOVL5, HELO1, dJ483K16.1; ELOVL family member 5,
elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like,
yeast); K10244 elongation of very long chain fatty acids
protein 5 [EC:2.3.1.-]
Length=299
Score = 51.6 bits (122), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 41/130 (31%), Positives = 67/130 (51%), Gaps = 18/130 (13%)
Query 8 SSLFCYLHLLGERECGYDGEIYYVVALNALVHVVMYAYYFMASLNSPLART---IKIFVT 64
S L + ++ CG+ Y+ LN+ +HV+MY+YY ++S+ P R K ++T
Sbjct 149 SMLNIWWFVMNWVPCGHS---YFGATLNSFIHVLMYSYYGLSSV--PSMRPYLWWKKYIT 203
Query 65 QLQMAQF-LSMTAHACYHVYFYKSCRYPVRVTIGYFFYVLSLFLLFRNFSKKTYGKKPAT 123
Q Q+ QF L++ +C ++ C +P+ Y++SL LF NF +TY KK
Sbjct 204 QGQLLQFVLTIIQTSCGVIW---PCTFPLGWLYFQIGYMISLIALFTNFYIQTYNKK--- 257
Query 124 AAVGAASSKD 133
GA+ KD
Sbjct 258 ---GASRRKD 264
> hsa:54898 ELOVL2, FLJ20334, SSC2; elongation of very long chain
fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2; K10205
elongation of very long chain fatty acids protein 2 [EC:2.3.1.-]
Length=296
Score = 51.6 bits (122), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 36/119 (30%), Positives = 61/119 (51%), Gaps = 5/119 (4%)
Query 26 GEIYYVVALNALVHVVMYAYYFMASLNSPLARTI--KIFVTQLQMAQFLSMTAHACYHVY 83
G+ ++ LN+ +H++MY+YY ++ S + + + K ++TQ Q+ QF+ H V
Sbjct 167 GQSFFGPTLNSFIHILMYSYYGLSVFPS-MHKYLWWKKYLTQAQLVQFVLTITHTMSAVV 225
Query 84 FYKSCRYPVRVTIGYFFYVLSLFLLFRNFSKKTYGKKPATAAVGAASSKDGRSNTKAKA 142
K C +P I Y+L+L +LF NF +TY KKP + + N +KA
Sbjct 226 --KPCGFPFGCLIFQSSYMLTLVILFLNFYVQTYRKKPMKKDMQEPPAGKEVKNGFSKA 282
> dre:393425 elovl5, MGC63549, zgc:63549; ELOVL family member
5, elongation of long chain fatty acids (yeast); K10244 elongation
of very long chain fatty acids protein 5 [EC:2.3.1.-]
Length=291
Score = 48.1 bits (113), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 36/117 (30%), Positives = 61/117 (52%), Gaps = 10/117 (8%)
Query 26 GEIYYVVALNALVHVVMYAYYFMASLNSPLART---IKIFVTQLQMAQF-LSMTAHACYH 81
G Y+ N+ +HV+MY+YY ++++ P R K ++TQ Q+ QF L+M +C
Sbjct 164 GHSYFGATFNSFIHVLMYSYYGLSAV--PALRPYLWWKKYITQGQLVQFVLTMFQTSCAV 221
Query 82 VYFYKSCRYPVRVTIGYFFYVLSLFLLFRNFSKKTYGKKPATAAVG-AASSKDGRSN 137
V+ C +P+ Y+++L LLF NF +TY K+ + S +G +N
Sbjct 222 VW---PCGFPMGWLYFQISYMVTLILLFSNFYIQTYKKRSGSRKSDYPNGSVNGHTN 275
> xla:398440 elovl7; ELOVL family member 7, elongation of long
chain fatty acids; K10250 elongation of very long chain fatty
acids protein 7
Length=302
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 37/120 (30%), Positives = 61/120 (50%), Gaps = 19/120 (15%)
Query 34 LNALVHVVMYAYYFMASLNSPLARTI--KIFVTQLQMAQFLSMTAHACYHVYFYKSC--R 89
+N +VHV+MY+YY +++L + + K ++T +Q+ QFL +T H +F ++C +
Sbjct 176 VNCVVHVIMYSYYGLSALGPAYQKYLWWKKYMTSIQLTQFLMVTFHIG-QFFFMENCPYQ 234
Query 90 YPVRVTIGYFFYVLSLF-----LLFRNFSKKTYGKK---PATAAVGAASSKDGRSNTKAK 141
YPV F YV+ L+ +LF NF Y K P G + + + NT K
Sbjct 235 YPV------FLYVIWLYGFVFLILFLNFWFHAYIKGQRLPKAVQNGHCKNNNNQENTWCK 288
> xla:380063 elovl1, MGC52731; elongation of very long chain fatty
acids (FEN1/Elo2, SUR4/Elo3)-like 1; K10247 elongation
of very long chain fatty acids protein 1
Length=290
Score = 46.6 bits (109), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 37/116 (31%), Positives = 57/116 (49%), Gaps = 9/116 (7%)
Query 34 LNALVHVVMYAYYFMASLNSPLARTI--KIFVTQLQMAQFLSMTAHACYHVYFYKSCRYP 91
+N+LVHV+MY YY +++ + + K +T +Q+ QF+ ++ H + YF SC Y
Sbjct 174 INSLVHVIMYFYYGLSAAGPRFQKYLWWKKHMTAIQLIQFVLVSIHITQY-YFMPSCDYQ 232
Query 92 VRVTIGY-FFYVLSLFLLFRNFSKKTYGK-----KPATAAVGAASSKDGRSNTKAK 141
+ I + Y F+LF NF + Y K K +T A GA N K K
Sbjct 233 FPIFIHLIWIYGTVFFILFSNFWYQAYTKGKRLPKGSTVANGALHQNGKAKNGKYK 288
> xla:447388 elovl2, MGC84669; elongation of very long chain fatty
acids (FEN1/Elo2, SUR4/Elo3)-like 2; K10205 elongation
of very long chain fatty acids protein 2 [EC:2.3.1.-]
Length=296
Score = 46.6 bits (109), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 31/100 (31%), Positives = 52/100 (52%), Gaps = 3/100 (3%)
Query 26 GEIYYVVALNALVHVVMYAYYFMASLNSPLA-RTIKIFVTQLQMAQFLSMTAHACYHVYF 84
G+ ++ LN+ +HV+MY+YY ++ + S K ++TQ Q+ QFL H
Sbjct 167 GQSFFGPTLNSFIHVLMYSYYGLSVIPSMHKYLWWKKYLTQAQLVQFLLTITHTLSAA-- 224
Query 85 YKSCRYPVRVTIGYFFYVLSLFLLFRNFSKKTYGKKPATA 124
K C +P + Y+ +L +LF NF K Y K+P+ +
Sbjct 225 VKPCGFPFGCLMFQSSYMTTLVMLFVNFYLKAYKKRPSKS 264
> xla:379510 hypothetical protein MGC64517; K10247 elongation
of very long chain fatty acids protein 1
Length=290
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 37/116 (31%), Positives = 57/116 (49%), Gaps = 9/116 (7%)
Query 34 LNALVHVVMYAYYFMASLNSPLARTI--KIFVTQLQMAQFLSMTAHACYHVYFYKSCRYP 91
+N+LVHV+MY YY +++ + + K +T +Q+ QF+ ++ H + YF SC Y
Sbjct 174 INSLVHVIMYFYYGLSAAGPRFQKYLWWKKHMTAIQLIQFVLVSIHISQY-YFMPSCDYQ 232
Query 92 VRVTIGY-FFYVLSLFLLFRNFSKKTYGK-----KPATAAVGAASSKDGRSNTKAK 141
+ I + Y F+LF NF + Y K K +T A GA N K K
Sbjct 233 FPIFIHLIWIYGTVFFILFSNFWYQAYTKGRRLPKGSTVANGALHRNGKAKNGKYK 288
> cel:F41H10.7 elo-5; fatty acid ELOngation family member (elo-5)
Length=286
Score = 45.1 bits (105), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 34/116 (29%), Positives = 53/116 (45%), Gaps = 8/116 (6%)
Query 30 YVVALNALVHVVMYAYYFMASLNSPLARTIKIFVTQLQMAQF---LSMTAHACYHVYF-- 84
+VV +N ++H MY YY + SL P+ ++ +T QM QF + H Y Y
Sbjct 171 WVVWMNYIIHAFMYGYYLLKSLKVPIPPSVAQAITTSQMVQFAVAIFAQVHVSYKHYVEG 230
Query 85 YKSCRYPVRVTIGYFFYVLSLFLLFRNFSKKTY---GKKPATAAVGAASSKDGRSN 137
+ Y R T FF + + F L+ F K+ Y G K A A ++ ++N
Sbjct 231 VEGLAYSFRGTAIGFFMLTTYFYLWIQFYKEHYLKNGGKKYNLAKDQAKTQTKKAN 286
> dre:449816 elovl1a, zgc:103538; elongation of very long chain
fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1a
Length=315
Score = 45.1 bits (105), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 33/94 (35%), Positives = 50/94 (53%), Gaps = 14/94 (14%)
Query 34 LNALVHVVMYAYYFMASLNSPLARTI--KIFVTQLQMAQFLSMTAHACYHVYFYKSCRYP 91
+NA VHV+MY YY +A+ + + K ++T +Q+ QF+ +T H + YF + C Y
Sbjct 176 VNACVHVIMYTYYGLAAAGPRFQKYLWWKKYMTAIQLIQFVLVTGHISQY-YFMEKCDYQ 234
Query 92 VRVTI------GYFFYVLSLFLLFRNFSKKTYGK 119
V + I G FF F+LF NF + Y K
Sbjct 235 VPIFIHLILIYGTFF-----FILFSNFWIQAYIK 263
> mmu:54325 Elovl1, AA407424, BB151133, Ssc1; elongation of very
long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like
1; K10247 elongation of very long chain fatty acids protein
1
Length=202
Score = 45.1 bits (105), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 36/114 (31%), Positives = 60/114 (52%), Gaps = 10/114 (8%)
Query 34 LNALVHVVMYAYYFMASLNSPLART---IKIFVTQLQMAQFLSMTAHACYHVYFYKSC-- 88
+N+ VHVVMY YY +++L P+A+ K +T +Q+ QF+ ++ H + YF SC
Sbjct 93 INSSVHVVMYLYYGLSALG-PVAQPYLWWKKHMTAIQLIQFVLVSLHISQY-YFMPSCNY 150
Query 89 RYPVRVTIGYFFYVLSLFLLFRNFSKKTYGKKPATAAVGAASSKDGRSNTKAKA 142
+YP+ + + + + + F+LF NF +Y K A + TK KA
Sbjct 151 QYPIIIHLIWMYGTI-FFILFSNFWYHSYTKGKRLPR--AVQQNGAPATTKVKA 201
> mmu:74559 Elovl7, 9130013K24Rik, AI840082; ELOVL family member
7, elongation of long chain fatty acids (yeast); K10250 elongation
of very long chain fatty acids protein 7
Length=281
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 35/107 (32%), Positives = 55/107 (51%), Gaps = 11/107 (10%)
Query 34 LNALVHVVMYAYYFMASLNSPLARTI--KIFVTQLQMAQFLSMTAHACYHVYFYKSC--R 89
LN VHVVMY+YY + ++ + + K +T LQ+ QF+ +T H ++F + C +
Sbjct 176 LNTAVHVVMYSYYGLCAMGPAYQKYLWWKKHLTSLQLVQFVLVTIHIG-QIFFMEDCNYQ 234
Query 90 YPVRVTI----GYFFYVLSLFLLFRNFSKKTYGKKPATAAVGAASSK 132
YPV + I G F +L L +R ++K + P T G SK
Sbjct 235 YPVFLYIIMSYGCIFLLLFLHFWYRAYTKGQ--RLPKTLENGNCKSK 279
> dre:678614 elovl2, MGC136352, MGC158526, zgc:136352; elongation
of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like
2; K10205 elongation of very long chain fatty acids
protein 2 [EC:2.3.1.-]
Length=260
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 30/96 (31%), Positives = 52/96 (54%), Gaps = 3/96 (3%)
Query 26 GEIYYVVALNALVHVVMYAYYFMASLNSPLA-RTIKIFVTQLQMAQFLSMTAHACYHVYF 84
G+ ++ LN+ +HV+MY+YY +A++ S K ++TQ Q+ QF+ H +
Sbjct 167 GQSFFGPTLNSFIHVLMYSYYGLATIPSMHKYLWWKRYLTQAQLVQFVLTITHTVSA--W 224
Query 85 YKSCRYPVRVTIGYFFYVLSLFLLFRNFSKKTYGKK 120
C +P+ FY+ +L +LF NF +TY K+
Sbjct 225 VVPCGFPLGCLKFQTFYMCTLVVLFVNFYIQTYKKR 260
> hsa:64834 ELOVL1, Ssc1; elongation of very long chain fatty
acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1; K10247 elongation
of very long chain fatty acids protein 1
Length=279
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 33/113 (29%), Positives = 58/113 (51%), Gaps = 8/113 (7%)
Query 34 LNALVHVVMYAYYFMASLNSPLART---IKIFVTQLQMAQFLSMTAHACYHVYFYKSC-- 88
+N+ VHV+MY YY +++ P+A+ K +T +Q+ QF+ ++ H + YF SC
Sbjct 170 INSSVHVIMYLYYGLSAFG-PVAQPYLWWKKHMTAIQLIQFVLVSLHISQY-YFMSSCNY 227
Query 89 RYPVRVTIGYFFYVLSLFLLFRNFSKKTYGKKPATAAVGAASSKDGRSNTKAK 141
+YPV + + + + + F+LF NF +Y K + G + KA
Sbjct 228 QYPVIIHLIWMYGTI-FFMLFSNFWYHSYTKGKRLPRALQQNGAPGIAKVKAN 279
> cel:F56H11.3 elo-7; fatty acid ELOngation family member (elo-7)
Length=309
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 32/94 (34%), Positives = 46/94 (48%), Gaps = 6/94 (6%)
Query 32 VALNALVHVVMYAYYFMASLNSPLARTIKIFVTQLQMAQFLSMTAHACYHVYF-----YK 86
V +N VH MY YYF S+N + I + VT LQ+ QF+ + C +Y+
Sbjct 203 VIVNLFVHAFMYPYYFTRSMNIKVPAKISMAVTVLQLTQFMCFI-YGCTLMYYSLATNQY 261
Query 87 SCRYPVRVTIGYFFYVLSLFLLFRNFSKKTYGKK 120
+C P+ V F S F+LF NF K Y ++
Sbjct 262 TCDTPMFVLHSTFALSSSYFVLFANFFHKAYLQR 295
> dre:406725 elovl1b, wu:fj32h05, zgc:56567; elongation of very
long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like
1b; K10247 elongation of very long chain fatty acids protein
1
Length=320
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 29/89 (32%), Positives = 49/89 (55%), Gaps = 4/89 (4%)
Query 34 LNALVHVVMYAYYFMASLNSPLARTI--KIFVTQLQMAQFLSMTAHACYHVYFYKSCRYP 91
+N+ VHV+MY YY +++ + + K ++T +Q+ QF+ ++ H YF +SC +
Sbjct 175 VNSCVHVIMYFYYGLSAAGPRFQKFLWWKKYMTAIQLTQFVLVSLHVS-QWYFMESCDFQ 233
Query 92 VRVTIGY-FFYVLSLFLLFRNFSKKTYGK 119
V V I + Y F+LF NF + Y K
Sbjct 234 VPVIIHLIWLYGTFFFVLFSNFWYQAYIK 262
> cel:F11E6.5 elo-2; fatty acid ELOngation family member (elo-2)
Length=274
Score = 42.0 bits (97), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 35/104 (33%), Positives = 51/104 (49%), Gaps = 14/104 (13%)
Query 30 YVVALNALVHVVMYAYYFMASLNSPLARTIKIFVTQLQMAQFLSMTAHACY---HVYFYK 86
+ +ALN VH VMY Y+ + +LN R + F+T +Q+ QF+ +CY H+ F K
Sbjct 169 WSLALNLAVHTVMYFYFAVRALNIQTPRPVAKFITTIQIVQFVI----SCYIFGHLVFIK 224
Query 87 S------CRYPVRV-TIGYFFYVLSLFLLFRNFSKKTYGKKPAT 123
S C V +IG Y+ LFL + F K K+ T
Sbjct 225 SADSVPGCAVSWNVLSIGGLMYISYLFLFAKFFYKAYIQKRSPT 268
> sce:YJL196C ELO1; Elo1p; K10245 fatty acid elongase 2 [EC:2.3.1.-]
Length=310
Score = 41.6 bits (96), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 38/65 (58%), Gaps = 7/65 (10%)
Query 8 SSLFCYLHLLGERECGYDGEIYYVVALNALVHVVMYAYYFMASLNSPLARTIKIFVTQLQ 67
++L CY L+G Y + V LN VHV+MY YYF+++ S + K +VT+LQ
Sbjct 179 TALLCYNQLVG-----YTAVTWVPVTLNLAVHVLMYWYYFLSA--SGIRVWWKAWVTRLQ 231
Query 68 MAQFL 72
+ QF+
Sbjct 232 IVQFM 236
> xla:432300 hypothetical protein MGC80262; K10250 elongation
of very long chain fatty acids protein 7
Length=301
Score = 41.6 bits (96), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 34/114 (29%), Positives = 56/114 (49%), Gaps = 8/114 (7%)
Query 34 LNALVHVVMYAYYFMASLNSPLARTI--KIFVTQLQMAQFLSMTAHACYHVYFYKSC--R 89
+N +VHV+MY+YY +++L + + K ++T +Q+ QFL +T H +F ++C +
Sbjct 176 VNCVVHVIMYSYYGLSALGPAYQKYLWWKKYMTSIQLTQFLMVTFHIG-QFFFMENCPYQ 234
Query 90 YPV--RVTIGYFFYVLSLFLLFRNFSKKTYGKKPATAAVGAASSKDGRSNTKAK 141
YPV V Y F L LFL F F G++ + + N K
Sbjct 235 YPVFLYVIWSYGFVFLILFLNFW-FHAYIKGQRLPKFIQNGDCKNNNQENAHCK 287
> dre:327274 elovl7b, elovl1, wu:fd20a06, zgc:56422; ELOVL family
member 7, elongation of long chain fatty acids (yeast) b;
K10250 elongation of very long chain fatty acids protein 7
Length=282
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 21/59 (35%), Positives = 37/59 (62%), Gaps = 3/59 (5%)
Query 34 LNALVHVVMYAYYFMASLNSPLARTI--KIFVTQLQMAQFLSMTAHACYHVYFYKSCRY 90
LN +VHV+MY+YY +++L + + K ++T +Q+ QF+ +TAH +F + C Y
Sbjct 176 LNCIVHVIMYSYYLLSALGPKYQKYLWWKKYMTTIQLVQFVLVTAHIG-QFFFMQDCPY 233
> ath:AT3G06460 GNS1/SUR4 membrane family protein
Length=298
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 39/116 (33%), Positives = 51/116 (43%), Gaps = 16/116 (13%)
Query 10 LFCYLHLLGERECGYDGEIYYVVALNALVHVVMYAYYFMASLNSPLARTIKIFVTQLQMA 69
+ CYL L + G + LN+ VHV+MY YYF+ ++ S K VT QM
Sbjct 160 ILCYLWLRTRQSMFPVG-----LVLNSTVHVIMYGYYFLCAIGS--RPKWKKLVTNFQMV 212
Query 70 QF-LSMTAHACYHV---YFYKSCRYPVRVTIGYF--FYVLSLFLLFRNFSKKTYGK 119
QF M A + + YF C + YF + SL LF NF K Y K
Sbjct 213 QFAFGMGLGAAWMLPEHYFGSGC---AGIWTVYFNGVFTASLLALFYNFHSKNYEK 265
> sce:YLR372W SUR4, APA1, ELO3, SRE1, VBM1; Elongase, involved
in fatty acid and sphingolipid biosynthesis; synthesizes very
long chain 20-26-carbon fatty acids from C18-CoA primers;
involved in regulation of sphingolipid biosynthesis; K10246
fatty acid elongase 3 [EC:2.3.1.-]
Length=345
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/65 (36%), Positives = 36/65 (55%), Gaps = 7/65 (10%)
Query 8 SSLFCYLHLLGERECGYDGEIYYVVALNALVHVVMYAYYFMASLNSPLARTIKIFVTQLQ 67
++L CY L+G + V+ LN VHV+MY YYF++S + K +VT+ Q
Sbjct 192 TALLCYTQLIGRTSVEW-----VVILLNLGVHVIMYWYYFLSSCG--IRVWWKQWVTRFQ 244
Query 68 MAQFL 72
+ QFL
Sbjct 245 IIQFL 249
> sce:YCR034W FEN1, ELO2, GNS1, VBM2; Fatty acid elongase, involved
in sphingolipid biosynthesis; acts on fatty acids of up
to 24 carbons in length; mutations have regulatory effects
on 1,3-beta-glucan synthase, vacuolar ATPase, and the secretory
pathway; K10245 fatty acid elongase 2 [EC:2.3.1.-]
Length=347
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 25/69 (36%), Positives = 38/69 (55%), Gaps = 15/69 (21%)
Query 8 SSLFCYLHLLGERECGYDGEIYYVVALNALVHVVMYAYYFMASLNSPLARTIKI----FV 63
++L CY L+G + ++LN VHVVMY YYF+A AR I++ +V
Sbjct 185 TALLCYTQLMGTTSISWVP-----ISLNLGVHVVMYWYYFLA------ARGIRVWWKEWV 233
Query 64 TQLQMAQFL 72
T+ Q+ QF+
Sbjct 234 TRFQIIQFV 242
> cel:F41H10.8 elo-6; fatty acid ELOngation family member (elo-6)
Length=274
Score = 39.3 bits (90), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 32/110 (29%), Positives = 53/110 (48%), Gaps = 12/110 (10%)
Query 19 ERECGYDGEIYYVVALNALVHVVMYAYYFMASLNSPLARTIKIFVTQLQMAQFLSMTAHA 78
E G++ ++ +N VH +MY YY + S + I +T +Q+ QF+ +T
Sbjct 157 EANLGFNT---WITWMNFSVHSIMYGYYMLRSFGVKVPAWIAKNITTMQILQFV-ITHFI 212
Query 79 CYHVYFYKSCRYPVRVTIGYFFYVL----SLFLLFRNFSKKTY----GKK 120
+HV + V T GY+++ L S +LF NF ++Y GKK
Sbjct 213 LFHVGYLAVTGQSVDSTPGYYWFCLLMEISYVVLFGNFYYQSYIKGGGKK 262
> hsa:79993 ELOVL7, FLJ23563; ELOVL family member 7, elongation
of long chain fatty acids (yeast); K10250 elongation of very
long chain fatty acids protein 7
Length=281
Score = 38.5 bits (88), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 21/59 (35%), Positives = 35/59 (59%), Gaps = 3/59 (5%)
Query 34 LNALVHVVMYAYYFMASLNSPLARTI--KIFVTQLQMAQFLSMTAHACYHVYFYKSCRY 90
LN VHVVMY+YY +++L + + K ++T LQ+ QF+ + H +F + C+Y
Sbjct 176 LNTAVHVVMYSYYGLSALGPAYQKYLWWKKYLTSLQLVQFVIVAIHIS-QFFFMEDCKY 233
> ath:AT3G06470 GNS1/SUR4 membrane family protein
Length=278
Score = 38.1 bits (87), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 31/94 (32%), Positives = 42/94 (44%), Gaps = 12/94 (12%)
Query 35 NALVHVVMYAYYFMASLNSPLARTIKIFVTQLQMAQFL---SMTAHACYHVYFYKSCRYP 91
N+ VHV+MY YYF+ ++ S K VT Q+ QF+ ++ F C
Sbjct 183 NSTVHVIMYGYYFLCAVGS--RPKWKRLVTDCQIVQFVFSFGLSGWMLREHLFGSGCTG- 239
Query 92 VRVTIGYFF---YVLSLFLLFRNFSKKTYGKKPA 122
G+ F + SL LF NF K Y KKP
Sbjct 240 ---IWGWCFNAAFNASLLALFSNFHSKNYVKKPT 270
> dre:317738 elovl6, cb618, lce, zgc:73089; ELOVL family member
6, elongation of long chain fatty acids (yeast); K10203 elongation
of very long chain fatty acids protein 6 [EC:2.3.1.-]
Length=266
Score = 36.6 bits (83), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 30/101 (29%), Positives = 50/101 (49%), Gaps = 6/101 (5%)
Query 30 YVVALNALVHVVMYAYYFMASLNSPLARTIKIFVTQLQMAQFLSMTAHACYHVYFY---- 85
+ + +N LVH VMY+YY + + ++R +F+T Q+ Q + M Y VY +
Sbjct 163 WFMTMNYLVHAVMYSYYALRAAGFKISRKFAMFITLTQITQ-MVMGCVVNYLVYLWMQQG 221
Query 86 KSCRYPVRVTIGYFFYVLSLFLLFRNFSKKTY-GKKPATAA 125
+ C V+ + LS F+LF F + Y K+ + AA
Sbjct 222 QECPSHVQNIVWSSLMYLSYFVLFCQFFFEAYITKRKSNAA 262
> tgo:TGME49_005350 fatty acid elongation protein, putative
Length=318
Score = 36.6 bits (83), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 26/64 (40%), Positives = 35/64 (54%), Gaps = 5/64 (7%)
Query 30 YVVALNALVHVVMYAYYFMAS-LNSPLARTIKIFVTQLQMAQFLSMTAHACYHVYFYKSC 88
Y VA+N VH +MY YYF+A+ L PL IFVT Q++Q C +Y+ S
Sbjct 198 YFVAMNYSVHAIMYFYYFLAAQLQRPLPW--GIFVTIAQISQMFVGMGVTCVSLYY--SF 253
Query 89 RYPV 92
YP+
Sbjct 254 AYPL 257
> cel:F56H11.4 elo-1; fatty acid ELOngation family member (elo-1)
Length=288
Score = 36.2 bits (82), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 29/92 (31%), Positives = 47/92 (51%), Gaps = 4/92 (4%)
Query 30 YVVALNALVHVVMYAYYFMASLNSPLARTIKIFVTQLQMAQFL---SMTAHACYHVYFYK 86
Y + LN +VH MY+YYF+ S+ + I +T LQ+ QF+ ++ AH Y ++F
Sbjct 179 YGIYLNFVVHAFMYSYYFLRSMKIRVPGFIAQAITSLQIVQFIISCAVLAHLGYLMHFTN 238
Query 87 -SCRYPVRVTIGYFFYVLSLFLLFRNFSKKTY 117
+C + V F + LF NF ++Y
Sbjct 239 ANCDFEPSVFKLAVFMDTTYLALFVNFFLQSY 270
> pfa:PFI0980w long chain fatty acid elongation enzyme, putative
Length=642
Score = 35.8 bits (81), Expect = 0.050, Method: Composition-based stats.
Identities = 34/112 (30%), Positives = 50/112 (44%), Gaps = 10/112 (8%)
Query 13 YLHLLGERECGYDGEI-----YYVVALNALVHVVMYAYYFMASLNSPLARTIKIFVTQLQ 67
Y HL C Y +I +Y V LN +VH +MY Y+ + + I+ F+T LQ
Sbjct 129 YHHLSVVIYCLYSQKILVSHAHYFVLLNLIVHSIMYFYFGFIYIFPRILTKIRKFITCLQ 188
Query 68 MAQFLSMTAHACYHVYFYKSCRYPVRVT--IGYFFYVLSLFLLFRNFSKKTY 117
+ Q M A Y K+ P+ V+ + F L+ +LF NF Y
Sbjct 189 IFQ---MFAGIYISYYAIKNVDNPLYVSNAMASFILYLTYAILFLNFYFSNY 237
> cel:D2024.3 elo-3; fatty acid ELOngation family member (elo-3)
Length=320
Score = 35.0 bits (79), Expect = 0.075, Method: Compositional matrix adjust.
Identities = 34/96 (35%), Positives = 48/96 (50%), Gaps = 7/96 (7%)
Query 30 YVVALNALVHVVMYAYYFMASLNSPLARTIKIFVTQLQMAQFLSMTAHACYHVYFYKS-- 87
+ + +N VH +MY+YY + SL L + + + VT LQ+AQ + M VY KS
Sbjct 174 WFIWMNYGVHALMYSYYALRSLKFRLPKQMAMVVTTLQLAQ-MVMGVIIGVTVYRIKSSG 232
Query 88 --CRYPVRVTIGYFFYV-LSLFLLFRNFSKKTYGKK 120
C+ +G F V + FLLF NF Y KK
Sbjct 233 EYCQ-QTWDNLGLCFGVYFTYFLLFANFFYHAYVKK 267
> cel:Y53F4B.2 elo-9; fatty acid ELOngation family member (elo-9)
Length=286
Score = 34.3 bits (77), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 30/54 (55%), Gaps = 0/54 (0%)
Query 30 YVVALNALVHVVMYAYYFMASLNSPLARTIKIFVTQLQMAQFLSMTAHACYHVY 83
+ + +N LVH +MY YY + S+ L + + + VT LQ Q L + +C +Y
Sbjct 183 WFIFMNYLVHSIMYTYYAITSIGYRLPKIVSMTVTFLQTLQMLIGVSISCIVLY 236
> mmu:230623 Skint11, A630098G03Rik, Gm569; selection and upkeep
of intraepithelial T cells 11
Length=364
Score = 34.3 bits (77), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 21/68 (30%), Positives = 34/68 (50%), Gaps = 6/68 (8%)
Query 76 AHACYHVYFYKSCRYPVRVTIGYFFYVLSLFLLFRNFSKKTYGKKPATAAVG-AASSKDG 134
AH Y+ + +K C+ + + I F VL LFLL+ + YGK P +++ S+ D
Sbjct 217 AHILYNNWLWKVCKTLIAMMI--LFTVLILFLLW---TLNRYGKMPCLSSMNIDVSTHDA 271
Query 135 RSNTKAKA 142
N+ A
Sbjct 272 EQNSSKSA 279
> hsa:83401 ELOVL3, CIG-30, CIG30, MGC21435; elongation of very
long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like
3; K10248 elongation of very long chain fatty acids protein
3
Length=270
Score = 32.3 bits (72), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 24/98 (24%), Positives = 45/98 (45%), Gaps = 4/98 (4%)
Query 30 YVVALNALVHVVMYAYYFMASLNSPLARTIKIFVTQLQMAQFLSMTAHACYHVYFYKS-- 87
+ V +N VH +MY YY + + N + + + +T LQ+ Q + + A Y ++
Sbjct 169 WFVTMNFGVHAIMYTYYTLKAANVKPPKMLPMLITSLQILQ-MFVGAIVSILTYIWRQDQ 227
Query 88 -CRYPVRVTIGYFFYVLSLFLLFRNFSKKTYGKKPATA 124
C + F ++ F+LF +F +TY + A
Sbjct 228 GCHTTMEHLFWSFILYMTYFILFAHFFCQTYIRPKVKA 265
> pfa:PFA_0455c fatty acid elongation protein, GNS1/SUR4 family,
putative
Length=322
Score = 30.8 bits (68), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 20/56 (35%), Positives = 32/56 (57%), Gaps = 2/56 (3%)
Query 32 VALNALVHVVMYAYYFMAS-LNSPLARTIKIFVTQL-QMAQFLSMTAHACYHVYFY 85
+ +NA VH +MY YYF+AS N +I + + Q+ QM + +T++ Y Y Y
Sbjct 202 ICINAFVHSIMYFYYFLASCYNKKFKWSIIVTLIQICQMFLGVLLTSYCLYISYIY 257
> hsa:79071 ELOVL6, FACE, FAE, FLJ23378, LCE, MGC5487; ELOVL family
member 6, elongation of long chain fatty acids (FEN1/Elo2,
SUR4/Elo3-like, yeast); K10203 elongation of very long
chain fatty acids protein 6 [EC:2.3.1.-]
Length=265
Score = 29.6 bits (65), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 25/43 (58%), Gaps = 0/43 (0%)
Query 30 YVVALNALVHVVMYAYYFMASLNSPLARTIKIFVTQLQMAQFL 72
+ + +N VH VMY+YY + + ++R +F+T Q+ Q L
Sbjct 165 WFMTMNYGVHAVMYSYYALRAAGFRVSRKFAMFITLSQITQML 207
Lambda K H
0.329 0.139 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2749206264
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40