bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0310_orf1
Length=164
Score E
Sequences producing significant alignments: (Bits) Value
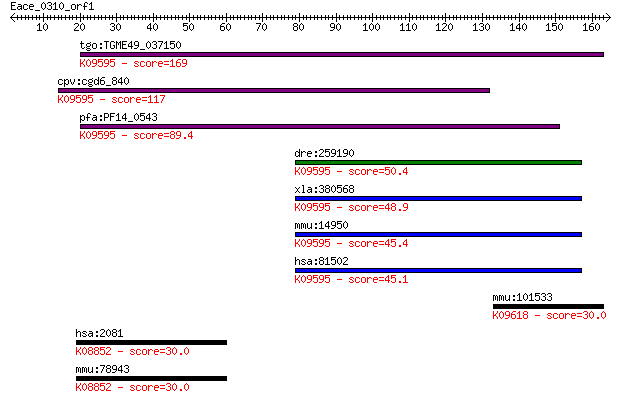
tgo:TGME49_037150 signal peptide peptidase domain-containing p... 169 4e-42
cpv:cgd6_840 shanti/Ykl100cp/Minor histocompatibility antigen ... 117 1e-26
pfa:PF14_0543 SPP; signal peptide peptidase; K09595 minor hist... 89.4 5e-18
dre:259190 hm13, H13, SPP, cb228, wu:fc11a06, zgc:56660, zgc:8... 50.4 3e-06
xla:380568 hm13, MGC68919, h13; histocompatibility (minor) 13 ... 48.9 7e-06
mmu:14950 H13, 1200006O09Rik, 4930443L17Rik, 5031424B04Rik, AV... 45.4 7e-05
hsa:81502 HM13, H13, IMP1, IMPAS, IMPAS-1, MSTP086, PSENL3, PS... 45.1 1e-04
mmu:101533 Klk9, 1200016C12Rik, AI324041, MGC130095, MGC130096... 30.0 3.3
hsa:2081 ERN1, FLJ30999, IRE1, IRE1P, IRE1a, MGC163277, MGC163... 30.0 3.7
mmu:78943 Ern1, 9030414B18Rik, AI225830, C85377, Ire1a, Ire1al... 30.0 3.8
> tgo:TGME49_037150 signal peptide peptidase domain-containing
protein ; K09595 minor histocompatibility antigen H13 [EC:3.4.23.-]
Length=417
Score = 169 bits (427), Expect = 4e-42, Method: Compositional matrix adjust.
Identities = 89/146 (60%), Positives = 98/146 (67%), Gaps = 3/146 (2%)
Query 20 LFYSCFVCLAVVMVTPWLYPLPVLLQMGLYTAPILYVGSHLSLKQNEVDATTGEKLNKGE 79
LFY CF L M LPVLLQM +YT PI+Y+GSH SL+QNEVD TGEK NKGE
Sbjct 22 LFYGCFALLGATMAVSHFLALPVLLQMVVYTFPIVYIGSHASLRQNEVDEVTGEKSNKGE 81
Query 80 AMNRTDAMLFPVFGSIALFSLYLAYKFLGTVWVNMLLTLYLTGVGLLALGETTFAVAKPL 139
AMN TDAMLFPVFGS+ALFSLY+AYKFL WVN LLTLYLT +GLLALGET PL
Sbjct 82 AMNHTDAMLFPVFGSVALFSLYVAYKFLDASWVNFLLTLYLTAIGLLALGETLHVALFPL 141
Query 140 CPPHLVDDRWLVLH---PRGPFLWLR 162
P D+ L P P L+ R
Sbjct 142 FPDWAKDESRLAFKLCLPHIPVLYPR 167
> cpv:cgd6_840 shanti/Ykl100cp/Minor histocompatibility antigen
H13-like; presenilin, signal peptide peptidase family, with
10 transmembrane domains and a signal peptide ; K09595 minor
histocompatibility antigen H13 [EC:3.4.23.-]
Length=408
Score = 117 bits (294), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 60/118 (50%), Positives = 83/118 (70%), Gaps = 0/118 (0%)
Query 14 CLVSLTLFYSCFVCLAVVMVTPWLYPLPVLLQMGLYTAPILYVGSHLSLKQNEVDATTGE 73
C SL + ++ LA+ ++ PLP+++QM +YT+ I+Y+GS+LSL Q +D TGE
Sbjct 6 CNKSLMAYCLSYLVLALAILITNFKPLPIIVQMLMYTSSIIYIGSYLSLSQTIIDPKTGE 65
Query 74 KLNKGEAMNRTDAMLFPVFGSIALFSLYLAYKFLGTVWVNMLLTLYLTGVGLLALGET 131
K E+++R DAM+FPV GS+ALFSLYLAYKFL WVN+LLT YL +G +AL ET
Sbjct 66 KDRSTESLSRKDAMMFPVIGSVALFSLYLAYKFLPVYWVNLLLTSYLFIIGAVALMET 123
> pfa:PF14_0543 SPP; signal peptide peptidase; K09595 minor histocompatibility
antigen H13 [EC:3.4.23.-]
Length=412
Score = 89.4 bits (220), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 53/131 (40%), Positives = 79/131 (60%), Gaps = 4/131 (3%)
Query 20 LFYSCFVCLAVVMVTPWLYPLPVLLQMGLYTAPILYVGSHLSLKQNEVDATTGEKLNKGE 79
+YSC+V + + ++ +P++ QM LYT +Y+GSH SLKQ E+D +K K +
Sbjct 23 FYYSCYVIIVLTIILSKFVVIPLMAQMFLYTFITIYIGSHDSLKQLEID----DKTKKSD 78
Query 80 AMNRTDAMLFPVFGSIALFSLYLAYKFLGTVWVNMLLTLYLTGVGLLALGETTFAVAKPL 139
+ DAM+FPV GS AL +LY AYKFL +VN+LLTLYLT G+ +L + +P+
Sbjct 79 NITAYDAMMFPVIGSAALLTLYFAYKFLDPFYVNLLLTLYLTLAGVFSLQGVFTTILEPV 138
Query 140 CPPHLVDDRWL 150
P D ++
Sbjct 139 FPNFFKKDEYV 149
> dre:259190 hm13, H13, SPP, cb228, wu:fc11a06, zgc:56660, zgc:86862;
histocompatibility (minor) 13 (EC:3.4.99.-); K09595
minor histocompatibility antigen H13 [EC:3.4.23.-]
Length=366
Score = 50.4 bits (119), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 27/78 (34%), Positives = 42/78 (53%), Gaps = 0/78 (0%)
Query 79 EAMNRTDAMLFPVFGSIALFSLYLAYKFLGTVWVNMLLTLYLTGVGLLALGETTFAVAKP 138
E + DA FP+ S LF LYL +K +VNMLL++Y +G+LAL T
Sbjct 71 ETITSRDAARFPIIASCTLFGLYLFFKVFSQEYVNMLLSMYFFVLGILALSHTMSPFMCR 130
Query 139 LCPPHLVDDRWLVLHPRG 156
+ P +L + ++ +L +G
Sbjct 131 VFPANLSNKQYQLLFTQG 148
> xla:380568 hm13, MGC68919, h13; histocompatibility (minor) 13
(EC:3.4.99.-); K09595 minor histocompatibility antigen H13
[EC:3.4.23.-]
Length=392
Score = 48.9 bits (115), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 24/78 (30%), Positives = 41/78 (52%), Gaps = 0/78 (0%)
Query 79 EAMNRTDAMLFPVFGSIALFSLYLAYKFLGTVWVNMLLTLYLTGVGLLALGETTFAVAKP 138
E + DA FP+ S LF LY+ +K ++N+LL++Y +G+LAL T
Sbjct 66 ETITSRDAARFPIIASCTLFGLYIFFKIFSQEYINLLLSMYFFILGVLALAHTISPAMNR 125
Query 139 LCPPHLVDDRWLVLHPRG 156
L P + + ++ +L +G
Sbjct 126 LFPENFPNRQYQMLFTQG 143
> mmu:14950 H13, 1200006O09Rik, 4930443L17Rik, 5031424B04Rik,
AV020344, H-13, Hm13, PSL3, Spp; histocompatibility 13 (EC:3.4.99.-);
K09595 minor histocompatibility antigen H13 [EC:3.4.23.-]
Length=394
Score = 45.4 bits (106), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 23/78 (29%), Positives = 39/78 (50%), Gaps = 0/78 (0%)
Query 79 EAMNRTDAMLFPVFGSIALFSLYLAYKFLGTVWVNMLLTLYLTGVGLLALGETTFAVAKP 138
E + DA FP+ S L LYL +K ++N+LL++Y +G+LAL T
Sbjct 68 ETITSRDAARFPIIASCTLLGLYLFFKIFSQEYINLLLSMYFFVLGILALSHTISPFMNK 127
Query 139 LCPPHLVDDRWLVLHPRG 156
P + + ++ +L +G
Sbjct 128 FFPANFPNRQYQLLFTQG 145
> hsa:81502 HM13, H13, IMP1, IMPAS, IMPAS-1, MSTP086, PSENL3,
PSL3, SPP, dJ324O17.1; histocompatibility (minor) 13; K09595
minor histocompatibility antigen H13 [EC:3.4.23.-]
Length=377
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 23/78 (29%), Positives = 38/78 (48%), Gaps = 0/78 (0%)
Query 79 EAMNRTDAMLFPVFGSIALFSLYLAYKFLGTVWVNMLLTLYLTGVGLLALGETTFAVAKP 138
E + DA FP+ S L LYL +K ++N+LL++Y +G+LAL T
Sbjct 68 ETITSRDAARFPIIASCTLLGLYLFFKIFSQEYINLLLSMYFFVLGILALSHTISPFMNK 127
Query 139 LCPPHLVDDRWLVLHPRG 156
P + ++ +L +G
Sbjct 128 FFPASFPNRQYQLLFTQG 145
> mmu:101533 Klk9, 1200016C12Rik, AI324041, MGC130095, MGC130096;
kallikrein related-peptidase 9; K09618 kallikrein 9 [EC:3.4.21.-]
Length=251
Score = 30.0 bits (66), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 13/32 (40%), Positives = 21/32 (65%), Gaps = 2/32 (6%)
Query 133 FAVAKPLCPPHLVDDRWLVL--HPRGPFLWLR 162
F + + LC L++D+WL+ H R P+LW+R
Sbjct 41 FYLTRQLCGATLINDQWLLTAAHCRKPYLWVR 72
> hsa:2081 ERN1, FLJ30999, IRE1, IRE1P, IRE1a, MGC163277, MGC163279,
hIRE1p; endoplasmic reticulum to nucleus signaling 1
(EC:2.7.11.1); K08852 serine/threonine-protein kinase/endoribonuclease
IRE1 [EC:2.7.11.1 3.1.26.-]
Length=977
Score = 30.0 bits (66), Expect = 3.7, Method: Composition-based stats.
Identities = 15/41 (36%), Positives = 19/41 (46%), Gaps = 0/41 (0%)
Query 19 TLFYSCFVCLAVVMVTPWLYPLPVLLQMGLYTAPILYVGSH 59
TL Y F+ V +T W YP P + P LYVG +
Sbjct 249 TLRYLTFMSGEVGRITKWKYPFPKETEAKSKLTPTLYVGKY 289
> mmu:78943 Ern1, 9030414B18Rik, AI225830, C85377, Ire1a, Ire1alpha,
Ire1p; endoplasmic reticulum (ER) to nucleus signalling
1 (EC:2.7.11.1); K08852 serine/threonine-protein kinase/endoribonuclease
IRE1 [EC:2.7.11.1 3.1.26.-]
Length=977
Score = 30.0 bits (66), Expect = 3.8, Method: Composition-based stats.
Identities = 15/41 (36%), Positives = 19/41 (46%), Gaps = 0/41 (0%)
Query 19 TLFYSCFVCLAVVMVTPWLYPLPVLLQMGLYTAPILYVGSH 59
TL Y F+ V +T W YP P + P LYVG +
Sbjct 251 TLRYLTFMSGEVGRITKWKYPFPKETEAKSKLTPTLYVGKY 291
Lambda K H
0.325 0.138 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3897828240
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40