bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0315_orf3
Length=501
Score E
Sequences producing significant alignments: (Bits) Value
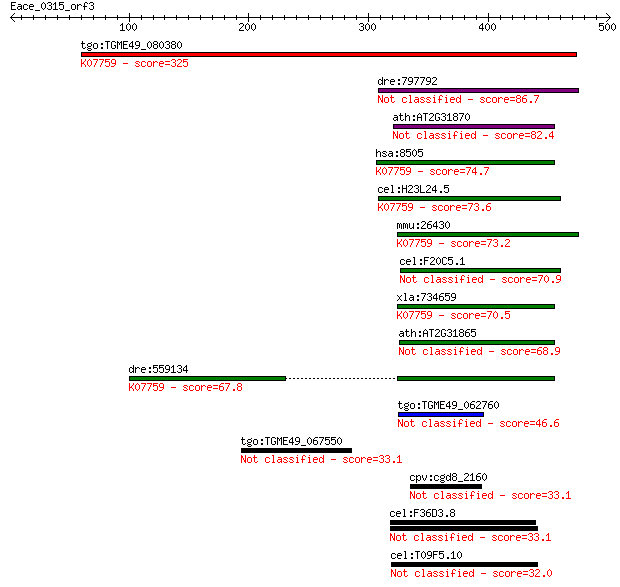
tgo:TGME49_080380 non-transmembrane antigen (EC:3.2.1.143); K0... 325 3e-88
dre:797792 si:dkey-259k14.2 86.7 2e-16
ath:AT2G31870 TEJ; TEJ (Sanskrit for 'bright'); poly(ADP-ribos... 82.4 4e-15
hsa:8505 PARG, FLJ54459, FLJ60257, FLJ60456, PARG99; poly (ADP... 74.7 8e-13
cel:H23L24.5 pme-4; Poly(ADP-ribose) Metabolism Enzyme family ... 73.6 2e-12
mmu:26430 Parg, AI413217; poly (ADP-ribose) glycohydrolase (EC... 73.2 2e-12
cel:F20C5.1 pme-3; Poly(ADP-ribose) Metabolism Enzyme family m... 70.9 1e-11
xla:734659 parg, MGC115697; poly (ADP-ribose) glycohydrolase; ... 70.5 2e-11
ath:AT2G31865 poly (ADP-ribose) glycohydrolase (PARG) family p... 68.9 5e-11
dre:559134 poly(ADP-ribose) glycohydrolase 63 kDa-like; K07759... 67.8 1e-10
tgo:TGME49_062760 poly(ADP-ribose) glycohydrolase, putative 46.6 2e-04
tgo:TGME49_067550 hypothetical protein 33.1 2.6
cpv:cgd8_2160 shares a domain with poly(ADP) ribose glycohydro... 33.1 2.6
cel:F36D3.8 hypothetical protein 33.1 2.9
cel:T09F5.10 hypothetical protein 32.0 6.6
> tgo:TGME49_080380 non-transmembrane antigen (EC:3.2.1.143);
K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=553
Score = 325 bits (832), Expect = 3e-88, Method: Compositional matrix adjust.
Identities = 178/426 (41%), Positives = 249/426 (58%), Gaps = 15/426 (3%)
Query 60 PSAVLWPAGESHQRRIQKILSGIEGGDIPSSASEFEDFFVDLLRAAGNYRGGGQLRI-QR 118
P A+L E + + IL+ + G P+ + LL+ +GN R RI QR
Sbjct 45 PCAILRTPVEEDWGKTKYILNFVASGFGPADPQQLASLQKRLLKLSGNIRTEKNKRILQR 104
Query 119 -LTTFLAHDRA-FSTTTLPFLASVAMRIQVLFPQGLKHMTRLRQSTHLRRIQVLCLMAAS 176
L +L + F + LPF+A++ MRI LFP GL+++T HLR+IQV L+AA+
Sbjct 105 GLLNYLEENPGKFFSHDLPFMATLVMRIDELFPSGLQYITPENPQVHLRKIQVFTLIAAA 164
Query 177 FFGIIPQNQRKLLRAWGR---LRLNALDMHHRGFLEREPKLKAALIYFNSMRKTMGTCWQ 233
F G+IP NQR LL + + N LDM +RGF+ER+PK ++ LIYF SMR+ +G CW
Sbjct 165 FLGVIPHNQRALLAHHQKKFVMNQNKLDMFYRGFMERKPKFQSVLIYFASMRERLGNCWN 224
Query 234 QMLKDSDFTQARCTATCLCQEITFDGKTRNVSPADEIISFYLHTPAATTFEGPEGE-SVS 292
+M+K A CT TC+C + ++ +++P DE+I FY + F +G +
Sbjct 225 EMVKKGFVDMAPCTGTCICSTLE---ESVSIAPTDELIGFYRQSDKVADFVWSDGSVKGT 281
Query 293 RDEVDVNTIITQPLPLPDATVDIYGDITKSDGDLQVDFADMYVGGLSMWEGHVAQEELIF 352
V ++ I L V GDIT SDGDLQVDFAD Y+GGLSM+EG++AQEELIF
Sbjct 282 SASVSIDDIAKSTKALGGFEVFEEGDITTSDGDLQVDFADKYLGGLSMFEGYIAQEELIF 341
Query 353 ALRPELHVIMLFMSAMKDDEAVVVKGAESFVLYSGYEGTFQVFPPV-----VPVGSHWSI 407
+ PE+ +MLF MK +EAVVVKG E FV GYE TFQ+ +P H+S+
Sbjct 342 IIYPEMLCVMLFSDIMKPNEAVVVKGVERFVFSQGYEWTFQITAAAGDWTELPSNKHFSV 401
Query 408 HGYTPLDSMGRRETTVVAIDAIIVKNEREQYSTPSMKREILKAALGFQGDPFEDAIHATR 467
G PLDS+GRR +V IDA+ +QYS + RE++KAA+GF+GDP+E + +R
Sbjct 402 QGVVPLDSLGRRNVAIVGIDAVQFHEPNKQYSPVMVNRELMKAAVGFKGDPYELIVSGSR 461
Query 468 AGVATG 473
+ATG
Sbjct 462 QPIATG 467
> dre:797792 si:dkey-259k14.2
Length=609
Score = 86.7 bits (213), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 61/168 (36%), Positives = 83/168 (49%), Gaps = 19/168 (11%)
Query 308 LPDATVDIYGDITKS-DGDLQVDFADMYVGGLSMWEGHVAQEELIFALRPELHVIMLFMS 366
L + V G I K G LQVDFA ++GG + G V QEE++F + PEL + LF
Sbjct 366 LKNLHVSADGSIEKEGTGMLQVDFASKFIGGGVLKSGLV-QEEILFLMSPELILARLFTE 424
Query 367 AMKDDEAVVVKGAESFVLYSGYEGTFQVFPPVVPVGSHWSIHGYTPLDSMGRRETTVVAI 426
+ D E V + G + + L SGY +F P + T D RR +VAI
Sbjct 425 KLDDHECVRITGPQMYSLTSGYSRSFSWTGPYM---------DRTKRDVWKRRFRQIVAI 475
Query 427 DAIIVKNEREQYSTPSMKREILKAALGFQGDPFEDAIHATRAGVATGK 474
DA+ KN EQYS ++ RE+ KA +GF G P + +ATG
Sbjct 476 DALDFKNPLEQYSRENITRELNKAFVGFCGQP--------KTAIATGN 515
> ath:AT2G31870 TEJ; TEJ (Sanskrit for 'bright'); poly(ADP-ribose)
glycohydrolase
Length=547
Score = 82.4 bits (202), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 51/134 (38%), Positives = 72/134 (53%), Gaps = 11/134 (8%)
Query 321 KSDGDLQVDFADMYVGGLSMWEGHVAQEELIFALRPELHVIMLFMSAMKDDEAVVVKGAE 380
+ D L+VDFA+ Y+GG S+ G V QEE+ F + PEL MLF+ M D+EA+ + GAE
Sbjct 247 QPDNALEVDFANKYLGGGSLSRGCV-QEEIRFMINPELIAGMLFLPRMDDNEAIEIVGAE 305
Query 381 SFVLYSGYEGTFQVFPPVVPVGSHWSIHGYTPLDSMGRRETTVVAIDAIIVKNEREQYST 440
F Y+GY +F+ + + +D RR T +VAIDA+ R +
Sbjct 306 RFSCYTGYASSFRFAGEYIDKKA---------MDPFKRRRTRIVAIDALCTPKMR-HFKD 355
Query 441 PSMKREILKAALGF 454
+ REI KA GF
Sbjct 356 ICLLREINKALCGF 369
> hsa:8505 PARG, FLJ54459, FLJ60257, FLJ60456, PARG99; poly (ADP-ribose)
glycohydrolase (EC:3.2.1.143); K07759 poly(ADP-ribose)
glycohydrolase [EC:3.2.1.143]
Length=976
Score = 74.7 bits (182), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 52/151 (34%), Positives = 76/151 (50%), Gaps = 15/151 (9%)
Query 307 PLPDATVDIYGDITKS-DGDLQVDFADMYVGGLSMWEGHVAQEELIFALRPELHVIMLFM 365
PL V G I ++ G LQVDFA+ +VGG G V QEE+ F + PEL + LF
Sbjct 714 PLTRLHVTYEGTIEENGQGMLQVDFANRFVGGGVTSAGLV-QEEIRFLINPELIISRLFT 772
Query 366 SAMKDDEAVVVKGAESFVLYSGYEGTFQVFPPVVPVGSHWSI--HGYTPLDSMGRRETTV 423
+ +E +++ G E + Y+GY T++ WS + D RR T +
Sbjct 773 EVLDHNECLIITGTEQYSEYTGYAETYR-----------WSRSHEDGSERDDWQRRCTEI 821
Query 424 VAIDAIIVKNEREQYSTPSMKREILKAALGF 454
VAIDA+ + +Q+ M+RE+ KA GF
Sbjct 822 VAIDALHFRRYLDQFVPEKMRRELNKAYCGF 852
> cel:H23L24.5 pme-4; Poly(ADP-ribose) Metabolism Enzyme family
member (pme-4); K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=485
Score = 73.6 bits (179), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 55/161 (34%), Positives = 82/161 (50%), Gaps = 15/161 (9%)
Query 308 LPDATVDIYGDITKSDGDLQVDFADMYVGGLSMWEGHVAQEELIFALRPELHVIMLFMSA 367
LPD V I ++ Q+DFA+ +GG + +G QEE+ F + PE+ V +L
Sbjct 232 LPDVQVFDKMSIEETALCTQIDFANKRLGG-GVLKGGAVQEEIRFMMCPEMMVAILLNDV 290
Query 368 MKDDEAVVVKGAESFVLYSGYEGTFQVFPPVVPVGSHWSIHGYTPLDSMGRRETTVVAID 427
+D EA+ + GA F Y+GY T + + + P H + + + D GR +T VAID
Sbjct 291 TQDLEAISIVGAYVFSSYTGYSNTLK-WAKITP--KHSAQNNNSFRDQFGRLQTETVAID 347
Query 428 AIIVKNE-------REQYSTPSMKREILKAALGF--QGDPF 459
A V+N Q +T + RE+ KAA+GF GD F
Sbjct 348 A--VRNAGTPLECLLNQLTTEKLTREVRKAAIGFLSAGDGF 386
> mmu:26430 Parg, AI413217; poly (ADP-ribose) glycohydrolase (EC:3.2.1.143);
K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=961
Score = 73.2 bits (178), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 54/155 (34%), Positives = 78/155 (50%), Gaps = 21/155 (13%)
Query 324 GDLQVDFADMYVGGLSMWEGHVAQEELIFALRPELHVIMLFMSAMKDDEAVVVKGAESFV 383
G LQVDFA+ +VGG G V QEE+ F + PEL V LF + +E +++ G E +
Sbjct 725 GMLQVDFANRFVGGGVTGAGLV-QEEIRFLINPELIVSRLFTEVLDHNECLIITGTEQYS 783
Query 384 LYSGYEGTFQVFPPVVPVGSHW--SIHGYTPLDSMGRRETTVVAIDAIIVKNEREQYSTP 441
Y+GY T++ W S + D RR T +VAIDA+ + +Q+
Sbjct 784 EYTGYAETYR-----------WARSHEDGSEKDDWQRRCTEIVAIDALHFRRYLDQFVPE 832
Query 442 SMKREILKAALGF--QGDPFEDAIHATRAGVATGK 474
++RE+ KA GF G P E+ + VATG
Sbjct 833 KVRRELNKAYCGFLRPGVPSENL-----SAVATGN 862
> cel:F20C5.1 pme-3; Poly(ADP-ribose) Metabolism Enzyme family
member (pme-3)
Length=764
Score = 70.9 bits (172), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 51/140 (36%), Positives = 76/140 (54%), Gaps = 11/140 (7%)
Query 327 QVDFADMYVGGLSMWEGHVAQEELIFALRPELHVIMLFMSAMKDDEAVVVKGAESFVLYS 386
QVDFA+ ++GG + G V QEE+ F + PE+ V ML MK EA+ + GA F Y+
Sbjct 517 QVDFANEHLGGGVLNHGSV-QEEIRFLMCPEMMVGMLLCEKMKQLEAISIVGAYVFSSYT 575
Query 387 GYEGTFQVFPPVVPVGSHWSIHGYTPLDSMGRRETTVVAIDAIIVKNER-----EQYSTP 441
GY T + + + P S + + + D GR +AIDAI+ K + EQ +
Sbjct 576 GYGHTLK-WAELQPNHSRQNTNEFR--DRFGRLRVETIAIDAILFKGSKLDCQTEQLNKA 632
Query 442 SMKREILKAALGF--QGDPF 459
++ RE+ KA++GF QG F
Sbjct 633 NIIREMKKASIGFMSQGPKF 652
> xla:734659 parg, MGC115697; poly (ADP-ribose) glycohydrolase;
K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=759
Score = 70.5 bits (171), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 49/133 (36%), Positives = 72/133 (54%), Gaps = 14/133 (10%)
Query 324 GDLQVDFADMYVGGLSMWEGHVAQEELIFALRPELHVIMLFMSAMKDDEAVVVKGAESFV 383
G LQVDFA+ +VGG G + QEE+ F + PEL V LF + +E +++ GAE +
Sbjct 529 GMLQVDFANRFVGGGVT-GGGLVQEEIRFLINPELIVSRLFTEVLDSNECLIITGAEQYS 587
Query 384 LYSGYEGTFQVFPPVVPVGSHWS-IH-GYTPLDSMGRRETTVVAIDAIIVKNEREQYSTP 441
Y+GY T++ W+ +H +P D RR T +VAIDA + +Q+
Sbjct 588 EYTGYSETYK-----------WACVHEDESPRDEWQRRTTEIVAIDAFHFRRPIDQFVPE 636
Query 442 SMKREILKAALGF 454
+KRE+ KA GF
Sbjct 637 KIKRELNKAFCGF 649
> ath:AT2G31865 poly (ADP-ribose) glycohydrolase (PARG) family
protein
Length=522
Score = 68.9 bits (167), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 48/129 (37%), Positives = 68/129 (52%), Gaps = 11/129 (8%)
Query 326 LQVDFADMYVGGLSMWEGHVAQEELIFALRPELHVIMLFMSAMKDDEAVVVKGAESFVLY 385
L+VDFAD Y GGL++ QEE+ F + PEL M+F+ M +EA+ + G E F Y
Sbjct 263 LEVDFADEYFGGLTL-SYDTLQEEIRFVINPELIAGMIFLPRMDANEAIEIVGVERFSGY 321
Query 386 SGYEGTFQVFPPVVPVGSHWSIHGYTPLDSMGRRETTVVAIDAIIVKNEREQYSTPSMKR 445
+GY +FQ G + LD RR+T V+AIDA+ QY ++ R
Sbjct 322 TGYGPSFQY------AGDY---TDNKDLDIFRRRKTRVIAIDAMPDPG-MGQYKLDALIR 371
Query 446 EILKAALGF 454
E+ KA G+
Sbjct 372 EVNKAFSGY 380
> dre:559134 poly(ADP-ribose) glycohydrolase 63 kDa-like; K07759
poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=777
Score = 67.8 bits (164), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 45/133 (33%), Positives = 67/133 (50%), Gaps = 14/133 (10%)
Query 324 GDLQVDFADMYVGGLSMWEGHVAQEELIFALRPELHVIMLFMSAMKDDEAVVVKGAESFV 383
G LQVDFA+ + G + + QEE+ F + PEL V LF + +E +++ G E +
Sbjct 526 GMLQVDFANR-MVGGGVTGLGLVQEEIRFLINPELIVSRLFTEVLDHNECLIITGTEQYS 584
Query 384 LYSGYEGTFQVFPPVVPVGSHWSIH--GYTPLDSMGRRETTVVAIDAIIVKNEREQYSTP 441
YSGY +F+ W + P D RR T +VA+DA+ +N +Q+
Sbjct 585 KYSGYAESFK-----------WEDNHKDKIPRDGWQRRCTEIVAMDALHYRNFMDQFQPE 633
Query 442 SMKREILKAALGF 454
M RE+ KA GF
Sbjct 634 KMTRELNKAYCGF 646
Score = 37.7 bits (86), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 36/155 (23%), Positives = 61/155 (39%), Gaps = 24/155 (15%)
Query 100 DLLRAA--GNYRGGGQLRIQRLTTFLAHDRAFSTTTLPFLASVAMR---IQVLFPQGLKH 154
+L+R+A G + +R L+ AH + + T L L + M +Q LF L
Sbjct 332 ELIRSALEGECKSSLDIRDAILSYNTAHAKRWDFTALNVLCTEGMENDEVQHLFNTILPK 391
Query 155 M-------------------TRLRQSTHLRRIQVLCLMAASFFGIIPQNQRKLLRAWGRL 195
M T++ QS + + Q+ CL+A +FF P+ +
Sbjct 392 MVKLVLNTPKICTQPIPLLKTKMNQSLTMSQEQIACLLANAFFCTFPRRNSRKSEYANYP 451
Query 196 RLNALDMHHRGFLEREPKLKAALIYFNSMRKTMGT 230
+N + + KLK L YF + ++M T
Sbjct 452 EINFYRLFEGSSQRKIEKLKTLLCYFRRVTESMPT 486
> tgo:TGME49_062760 poly(ADP-ribose) glycohydrolase, putative
Length=952
Score = 46.6 bits (109), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 26/71 (36%), Positives = 42/71 (59%), Gaps = 1/71 (1%)
Query 325 DLQVDFADMYVGGLSMWEGHVAQEELIFALRPELHVIMLFMSAMKDDEAVVVKGAESFVL 384
++ DFA+ ++GG +++ G V QEE+ FA PEL ++ LF + +E+ + GA F
Sbjct 479 EIMADFANQWIGGGALYRGCV-QEEIFFATHPELLLLRLFQQRLAINESCAMSGAMQFSR 537
Query 385 YSGYEGTFQVF 395
YSGY +F
Sbjct 538 YSGYADSFTCL 548
> tgo:TGME49_067550 hypothetical protein
Length=369
Score = 33.1 bits (74), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 26/94 (27%), Positives = 38/94 (40%), Gaps = 16/94 (17%)
Query 194 RLRLNALDMHHRGFLEREPKLKAALIYFNSMRKTMGTCWQQMLK--DSDFTQARCTATCL 251
RL LNA D+ LE P+L+ +Y N +RK G L+ D F + R
Sbjct 80 RLALNANDIEKIENLESTPELEELELYQNRVRKIEGLSTLSHLRVLDLSFNKVR------ 133
Query 252 CQEITFDGKTRNVSPADEIISFYLHTPAATTFEG 285
K N+ A +++ YL + EG
Sbjct 134 --------KIENLETAVKLVKLYLSSNKIQVIEG 159
> cpv:cgd8_2160 shares a domain with poly(ADP) ribose glycohydrolases,
some protein kinase A anchoring proteins and baculovirus
HzNV Orf103, possible transmembrane domain within N-terminus
Length=441
Score = 33.1 bits (74), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 16/62 (25%), Positives = 33/62 (53%), Gaps = 3/62 (4%)
Query 335 VGGLSMWEGHVAQEELIFALRPELHVIMLFMSAMKDDEAVVVKGAESFVLYSGY---EGT 391
V G + + + QEE++ + PE + F+ + D+++V ++G + Y G+ + T
Sbjct 235 VYGSATLQNCMNQEEIVMTVVPETLIGRFFLDDLHDEDSVTIRGVMRYSNYKGFGTDKFT 294
Query 392 FQ 393
FQ
Sbjct 295 FQ 296
> cel:F36D3.8 hypothetical protein
Length=722
Score = 33.1 bits (74), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 30/124 (24%), Positives = 54/124 (43%), Gaps = 16/124 (12%)
Query 318 DITKSDGDLQVDFADMYVGGLSMWEGH---VAQEELIFALRPELHVIMLFMSAMKDDEAV 374
++TK+D DL + + Y+ GL W G+ + ++E I+ + L + +KD V
Sbjct 28 EMTKNDVDLLTNPGEEYLDGLMKWHGNERPLFKKEDIYRWQDSFPEYRLKLICLKDTTRV 87
Query 375 VVKGAESFVLYSGYEGTFQVFPPVVPVGSHWSIHGYTPLDSMGRRETTVVAIDAIIVKNE 434
+ GA Y Y + P+ +G W+ Y R V+ + A + E
Sbjct 88 IAVGA-----YCYYNALKEGDKPLFFLGFGWTDPEY--------RTRAVMELQASMAIEE 134
Query 435 REQY 438
R++Y
Sbjct 135 RDRY 138
Score = 32.7 bits (73), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 31/126 (24%), Positives = 52/126 (41%), Gaps = 16/126 (12%)
Query 318 DITKSDGDLQVDFADMYVGGLSMWEGH---VAQEELIFALRPELHVIMLFMSAMKDDEAV 374
++TK+D DL + + Y+ GL W G V + E I+ L M +KD V
Sbjct 398 ELTKNDVDLLTNPGEEYLDGLMKWHGDERPVFKREDIYRWSDSFPEYRLRMICLKDTTRV 457
Query 375 VVKGAESFVLYSGYEGTFQVFPPVVPVGSHWSIHGYTPLDSMGRRETTVVAIDAIIVKNE 434
+ G Y ++ + P+ +G W+ Y R V+ + A + E
Sbjct 458 IAVGQ-----YCYFDALKEGEQPLFFLGLGWTDPEY--------RNRAVMELQASMALEE 504
Query 435 REQYST 440
R++Y T
Sbjct 505 RDRYPT 510
> cel:T09F5.10 hypothetical protein
Length=324
Score = 32.0 bits (71), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 32/125 (25%), Positives = 50/125 (40%), Gaps = 16/125 (12%)
Query 319 ITKSDGDLQVDFADMYVGGLSMWEGH---VAQEELIFALRPELHVIMLFMSAMKDDEAVV 375
+TK D DL + + YV L W G V ++E I+ + L M +KD V+
Sbjct 1 MTKDDVDLLTNPGEDYVDALMKWHGDERPVFKKEDIYRWKDSFPEYRLKMVCLKDTTRVI 60
Query 376 VKGAESFVLYSGYEGTFQVFPPVVPVGSHWSIHGYTPLDSMGRRETTVVAIDAIIVKNER 435
G Y + + P+ +G W+ Y R V+ + A + ER
Sbjct 61 AVGH-----YCYFNALKEGQKPLFFLGLGWTGPEY--------RTRAVMELQASMAMEER 107
Query 436 EQYST 440
E+Y T
Sbjct 108 ERYPT 112
Lambda K H
0.323 0.135 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 24124936704
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40