bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0317_orf2
Length=223
Score E
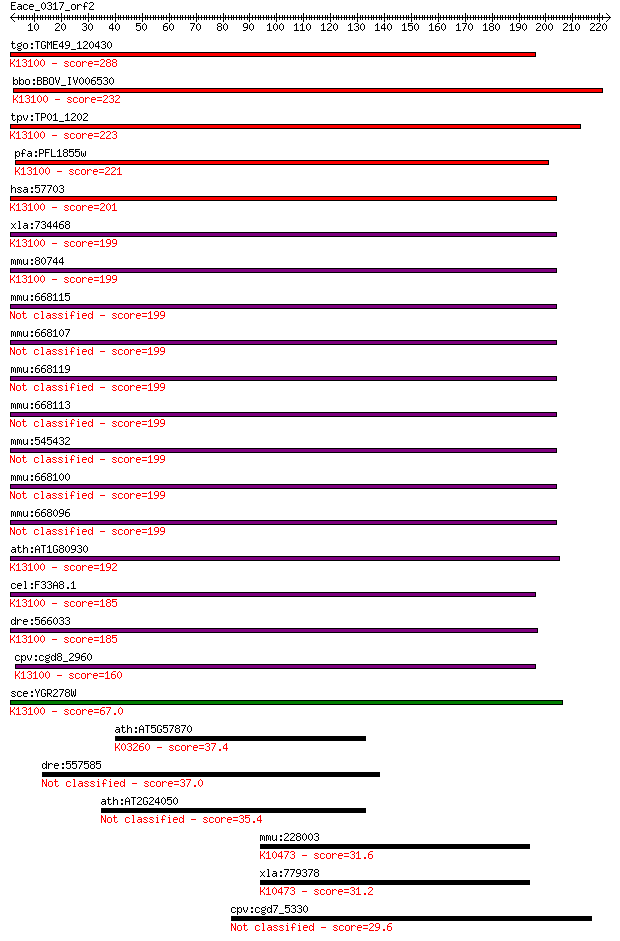
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_120430 cell cycle control protein, putative ; K1310... 288 1e-77
bbo:BBOV_IV006530 23.m06494; MIF4G/MA3 domains containing prot... 232 1e-60
tpv:TP01_1202 cell cycle control protein; K13100 pre-mRNA-spli... 223 4e-58
pfa:PFL1855w cell cycle control protein, putative; K13100 pre-... 221 1e-57
hsa:57703 CWC22, EIF4GL, KIAA1604, NCM, fSAPb; CWC22 spliceoso... 201 2e-51
xla:734468 cwc22, MGC115254; CWC22 spliceosome-associated prot... 199 6e-51
mmu:80744 Cwc22, AA684037, AI173004, AL022752, B230213M24, MGC... 199 8e-51
mmu:668115 Gm13697, OTTMUSG00000013393; predicted gene 13697 199 9e-51
mmu:668107 Gm13696, OTTMUSG00000013392; predicted gene 13696 199 9e-51
mmu:668119 Gm13691, OTTMUSG00000013386; predicted gene 13691 199 9e-51
mmu:668113 Gm13694, OTTMUSG00000013390; predicted gene 13694 199 9e-51
mmu:545432 Gm13695, Gm5838, OTTMUSG00000013391; predicted gene... 199 9e-51
mmu:668100 Gm13693, OTTMUSG00000013388; predicted gene 13693 199 9e-51
mmu:668096 Gm13698, OTTMUSG00000013395; predicted gene 13698 199 9e-51
ath:AT1G80930 MIF4G domain-containing protein / MA3 domain-con... 192 7e-49
cel:F33A8.1 let-858; LEThal family member (let-858); K13100 pr... 185 1e-46
dre:566033 MGC153452, cwc22, wu:fb13g08, wu:fi16b02; zgc:15345... 185 1e-46
cpv:cgd8_2960 NIC+MI domains containing protein. nucampholin/y... 160 3e-39
sce:YGR278W CWC22; Cwc22p; K13100 pre-mRNA-splicing factor CWC22 67.0 5e-11
ath:AT5G57870 eukaryotic translation initiation factor 4F, put... 37.4 0.047
dre:557585 nom1, si:ch211-266o5.1; nucleolar protein with MIF4... 37.0 0.057
ath:AT2G24050 MIF4G domain-containing protein / MA3 domain-con... 35.4 0.16
mmu:228003 Kbtbd10, Gm112, SARCOSIN; kelch repeat and BTB (POZ... 31.6 2.1
xla:779378 kbtbd5, MGC86259; kelch repeat and BTB (POZ) domain... 31.2 2.7
cpv:cgd7_5330 thioredoxin/PDI, cyanobacterial type, signal pep... 29.6 9.0
> tgo:TGME49_120430 cell cycle control protein, putative ; K13100
pre-mRNA-splicing factor CWC22
Length=1046
Score = 288 bits (736), Expect = 1e-77, Method: Compositional matrix adjust.
Identities = 144/195 (73%), Positives = 167/195 (85%), Gaps = 0/195 (0%)
Query 1 AALLSIVNSKLPDIGELVIKRLILQFRRAYRRDDKVVCLGCVEFFAHLVNQRIAHELLAL 60
AALL IVN+KLP+IGELV+KR+ILQFRRAYRR+DKVVCL CV+F AHLVNQR+AHELLAL
Sbjct 648 AALLCIVNAKLPEIGELVLKRVILQFRRAYRRNDKVVCLACVQFLAHLVNQRVAHELLAL 707
Query 61 QLCELLLQQPTNDSVEVCIGFLKAVGQVLEDVCRGGFDAAFERLRAILQEGETDKKTQYS 120
QLC LLL +PTNDSVEVC+GFL VGQVL +V R GFDA FERLR ILQEG TDKKTQY+
Sbjct 708 QLCALLLDEPTNDSVEVCVGFLTQVGQVLSEVSRRGFDAVFERLRGILQEGLTDKKTQYT 767
Query 121 IEALWDIRRNGFKDFPGVIEDLDLVEVEDKITHEVDIMDPSIKGEEMLNIFKPQDPDEYE 180
IE LWD+RR FKD PGV +LDLV+ +DKITHE+D++ +KGEEMLN+F QDP+E+
Sbjct 768 IEKLWDLRRQNFKDHPGVPAELDLVDEDDKITHEIDLLAEDLKGEEMLNVFHAQDPEEFA 827
Query 181 EDEKKWSALSKEILG 195
DEKKW+ LSKEILG
Sbjct 828 SDEKKWARLSKEILG 842
> bbo:BBOV_IV006530 23.m06494; MIF4G/MA3 domains containing protein;
K13100 pre-mRNA-splicing factor CWC22
Length=588
Score = 232 bits (591), Expect = 1e-60, Method: Compositional matrix adjust.
Identities = 108/224 (48%), Positives = 162/224 (72%), Gaps = 6/224 (2%)
Query 2 ALLSIVNSKLPDIGELVIKRLILQFRRAYRRDDKVVCLGCVEFFAHLVNQRIAHELLALQ 61
+ L+++NSK P+IG L+++R+ILQFRRA+R++D+++C + AHLVN R+AHE+LALQ
Sbjct 152 SFLAVINSKFPEIGNLILRRIILQFRRAFRKNDRILCQTVAKSLAHLVNYRVAHEVLALQ 211
Query 62 LCELLLQQPTNDSVEVCIGFLKAVGQVLEDVCRGGFDAAFERLRAILQEGETDKKTQYSI 121
+LL+ PT+DSV V GFL+ VG L + +A F+R + IL G+ DKKTQY+I
Sbjct 212 FLAILLENPTDDSVSVAAGFLEDVGNFLAQEAKQALEAIFDRFKQILSSGKVDKKTQYTI 271
Query 122 EALWDIRRNGFKDFPGVIEDLDLVEVEDKITHEVDIMDPSIKGEEMLNIFKPQDPDEYEE 181
EALW RN F D P V +LDLVE+ED ITH++D +D +I +EMLN+FKP +P+ Y +
Sbjct 272 EALWRSFRNKFSDHPAVKPELDLVELEDSITHDLDFLDDTITADEMLNVFKPVEPEVYIQ 331
Query 182 DEKKWSALSKEILGE-----DSTDSDASSVDSEAETSSEEETEE 220
+++KW+ + ++++G+ D+ DSD SSVDSEAE E+++E+
Sbjct 332 EQEKWTRIRRQLMGDSDDGSDTHDSD-SSVDSEAEQHDEDQSED 374
> tpv:TP01_1202 cell cycle control protein; K13100 pre-mRNA-splicing
factor CWC22
Length=596
Score = 223 bits (568), Expect = 4e-58, Method: Compositional matrix adjust.
Identities = 111/212 (52%), Positives = 161/212 (75%), Gaps = 1/212 (0%)
Query 1 AALLSIVNSKLPDIGELVIKRLILQFRRAYRRDDKVVCLGCVEFFAHLVNQRIAHELLAL 60
A+ L+++NSK P+IGEL +KR+ILQFRRAY+R+DK+VC CV+ AHLVNQ+IAHE+LAL
Sbjct 155 ASFLAVINSKFPEIGELTLKRIILQFRRAYKRNDKIVCQSCVKCVAHLVNQKIAHEILAL 214
Query 61 QLCELLLQQPTNDSVEVCIGFLKAVGQVLEDVCRGGFDAAFERLRAILQEGETDKKTQYS 120
QL +LL++PT+DSVE+ + FL+ VG L + C+ G D+ F+RL++ILQ G DK+TQYS
Sbjct 215 QLLAILLEKPTDDSVELALEFLRDVGNFLHENCKQGLDSVFDRLKSILQCGLVDKRTQYS 274
Query 121 IEALWDIRRNGFKDFPGVIEDLDLVEVEDKITHEVDIMDPSIKGEEMLNIFKPQDPDEYE 180
IEALW RNGF ++ + ++LDL+E ED+ITH++D +D +I G+EMLNIF+P +P+ Y
Sbjct 275 IEALWKHWRNGFTEY-KIPKELDLLEEEDQITHDIDFLDQTITGDEMLNIFQPVEPEVYN 333
Query 181 EDEKKWSALSKEILGEDSTDSDASSVDSEAET 212
+ KW+ + +E+ G + +S DSE +T
Sbjct 334 LENLKWNKIKQELTGAHTDSESDTSEDSEYDT 365
> pfa:PFL1855w cell cycle control protein, putative; K13100 pre-mRNA-splicing
factor CWC22
Length=967
Score = 221 bits (564), Expect = 1e-57, Method: Composition-based stats.
Identities = 110/199 (55%), Positives = 150/199 (75%), Gaps = 1/199 (0%)
Query 3 LLSIVNSKLPDIGELVIKRLILQFRRAYRRDDKVVCLGCVEFFAHLVNQRIAHELLALQL 62
LL IVNSK P+IG L I+R+IL FRRAY+R+DK++C V+F AH++NQRI HE++ LQL
Sbjct 492 LLCIVNSKFPNIGLLTIQRIILHFRRAYKRNDKILCFNTVKFIAHMINQRIVHEIVGLQL 551
Query 63 CELLLQQPTNDSVEVCIGFLKAVGQVLEDVCRGGFDAAFERLRAILQEGETDKKTQYSIE 122
C LLLQ TNDSV+VC FL VG++ ++CR G D F+RL+ I+QEG+ + KTQY IE
Sbjct 552 CSLLLQNITNDSVQVCTYFLAEVGELYTNICRKGLDIIFDRLKDIIQEGQINIKTQYDIE 611
Query 123 ALWDIRRNGFKDFPGVIEDLDLVEVEDKITHEVDIMDPSIKGEEMLNIFKPQDPDEYEED 182
LW+ R+N FKDFP V +DL+L++ EDKI HE+DI+D S +E LNIF+ ++YEE+
Sbjct 612 KLWNYRKNNFKDFPSVHDDLNLIDEEDKIVHEIDILDESFNSQEELNIFREVPYEQYEEE 671
Query 183 EKKWSALSKEIL-GEDSTD 200
E +W +S+E+L G+DS D
Sbjct 672 ENEWKHISEELLFGDDSAD 690
> hsa:57703 CWC22, EIF4GL, KIAA1604, NCM, fSAPb; CWC22 spliceosome-associated
protein homolog (S. cerevisiae); K13100 pre-mRNA-splicing
factor CWC22
Length=908
Score = 201 bits (511), Expect = 2e-51, Method: Compositional matrix adjust.
Identities = 107/203 (52%), Positives = 147/203 (72%), Gaps = 3/203 (1%)
Query 1 AALLSIVNSKLPDIGELVIKRLILQFRRAYRRDDKVVCLGCVEFFAHLVNQRIAHELLAL 60
AAL++I+NSK P IGEL++KRLIL FR+ YRR+DK +CL +F AHL+NQ +AHE+L L
Sbjct 216 AALVAIINSKFPQIGELILKRLILNFRKGYRRNDKQLCLTASKFVAHLINQNVAHEVLCL 275
Query 61 QLCELLLQQPTNDSVEVCIGFLKAVGQVLEDVCRGGFDAAFERLRAILQEGETDKKTQYS 120
++ LLL++PT+DSVEV IGFLK G L V G +A FERLR IL E E DK+ QY
Sbjct 276 EMLTLLLERPTDDSVEVAIGFLKECGLKLTQVSPRGINAIFERLRNILHESEIDKRVQYM 335
Query 121 IEALWDIRRNGFKDFPGVIEDLDLVEVEDKITHEVDIMDPSIKGEEMLNIFKPQDPDEYE 180
IE ++ +R++GFKD P ++E LDLVE +D+ TH + + D E++LN+FK DP+ +
Sbjct 336 IEVMFAVRKDGFKDHPIILEGLDLVEEDDQFTHMLPLED-DYNPEDVLNVFK-MDPN-FM 392
Query 181 EDEKKWSALSKEILGEDSTDSDA 203
E+E+K+ A+ KEIL E TDS+
Sbjct 393 ENEEKYKAIKKEILDEGDTDSNT 415
> xla:734468 cwc22, MGC115254; CWC22 spliceosome-associated protein
homolog; K13100 pre-mRNA-splicing factor CWC22
Length=803
Score = 199 bits (506), Expect = 6e-51, Method: Compositional matrix adjust.
Identities = 109/203 (53%), Positives = 146/203 (71%), Gaps = 3/203 (1%)
Query 1 AALLSIVNSKLPDIGELVIKRLILQFRRAYRRDDKVVCLGCVEFFAHLVNQRIAHELLAL 60
AAL+SI+NSK P IGEL++KRLIL FR+ YRR+DK +CL +F AHL+NQ +AHE+LAL
Sbjct 263 AALVSIINSKFPHIGELILKRLILNFRKGYRRNDKQLCLTSSKFVAHLINQNVAHEVLAL 322
Query 61 QLCELLLQQPTNDSVEVCIGFLKAVGQVLEDVCRGGFDAAFERLRAILQEGETDKKTQYS 120
++ LLL++P +DSVEV IGFLK G L V G +A FERLR IL E E DK+ QY
Sbjct 323 EMLTLLLERPNDDSVEVAIGFLKESGLKLTQVTPRGINAIFERLRNILHESEIDKRVQYM 382
Query 121 IEALWDIRRNGFKDFPGVIEDLDLVEVEDKITHEVDIMDPSIKGEEMLNIFKPQDPDEYE 180
IE ++ +R++GFKD P + E LDLVE ED+ TH + + D E++LN+FK DPD +
Sbjct 383 IEVMFAVRKDGFKDHPVIPEGLDLVEEEDQFTHMLPLED-DYNQEDVLNVFK-MDPD-FL 439
Query 181 EDEKKWSALSKEILGEDSTDSDA 203
E+E+K+ A+ KEIL E +DS+
Sbjct 440 ENEEKYKAIKKEILDEGDSDSEG 462
> mmu:80744 Cwc22, AA684037, AI173004, AL022752, B230213M24, MGC107219,
MGC107450, MGC7531, mKIAA1604; CWC22 spliceosome-associated
protein homolog (S. cerevisiae); K13100 pre-mRNA-splicing
factor CWC22
Length=908
Score = 199 bits (505), Expect = 8e-51, Method: Compositional matrix adjust.
Identities = 106/203 (52%), Positives = 147/203 (72%), Gaps = 3/203 (1%)
Query 1 AALLSIVNSKLPDIGELVIKRLILQFRRAYRRDDKVVCLGCVEFFAHLVNQRIAHELLAL 60
AAL++I+NSK P IGEL++KRLIL FR+ YRR+DK +CL +F AHL+NQ +AHE+L L
Sbjct 214 AALVAIINSKFPQIGELILKRLILNFRKGYRRNDKQLCLTASKFVAHLINQNVAHEVLCL 273
Query 61 QLCELLLQQPTNDSVEVCIGFLKAVGQVLEDVCRGGFDAAFERLRAILQEGETDKKTQYS 120
++ LLL++PT+DSVEV IGFLK G L V G +A FERLR IL E E DK+ QY
Sbjct 274 EMLTLLLERPTDDSVEVAIGFLKECGLKLTQVSPRGINAIFERLRNILHESEIDKRVQYM 333
Query 121 IEALWDIRRNGFKDFPGVIEDLDLVEVEDKITHEVDIMDPSIKGEEMLNIFKPQDPDEYE 180
IE ++ +R++GFKD P ++E LDLVE +D+ TH + + D E++LN+FK DP+ +
Sbjct 334 IEVMFAVRKDGFKDHPVILEGLDLVEEDDQFTHMLPLED-DYNPEDVLNVFK-MDPN-FM 390
Query 181 EDEKKWSALSKEILGEDSTDSDA 203
E+E+K+ A+ KEIL E +DS+
Sbjct 391 ENEEKYKAIKKEILDEGDSDSNT 413
> mmu:668115 Gm13697, OTTMUSG00000013393; predicted gene 13697
Length=830
Score = 199 bits (505), Expect = 9e-51, Method: Compositional matrix adjust.
Identities = 106/203 (52%), Positives = 147/203 (72%), Gaps = 3/203 (1%)
Query 1 AALLSIVNSKLPDIGELVIKRLILQFRRAYRRDDKVVCLGCVEFFAHLVNQRIAHELLAL 60
AAL++I+NSK P IGEL++KRLIL FR+ YRR+DK +CL +F AHL+NQ +AHE+L L
Sbjct 215 AALVAIINSKFPQIGELILKRLILNFRKGYRRNDKQLCLTASKFVAHLINQNVAHEVLCL 274
Query 61 QLCELLLQQPTNDSVEVCIGFLKAVGQVLEDVCRGGFDAAFERLRAILQEGETDKKTQYS 120
++ LLL++PT+DSVEV IGFLK G L V G +A FERLR IL E E DK+ QY
Sbjct 275 EMLTLLLERPTDDSVEVAIGFLKECGLKLAQVSPRGINAIFERLRNILHESEIDKRVQYM 334
Query 121 IEALWDIRRNGFKDFPGVIEDLDLVEVEDKITHEVDIMDPSIKGEEMLNIFKPQDPDEYE 180
IE ++ +R++GFKD P ++E LDLVE +D+ TH + + D E++LN+FK DP+ +
Sbjct 335 IEVMFAVRKDGFKDHPVILEGLDLVEEDDQFTHMLPLED-DYNPEDVLNVFK-MDPN-FM 391
Query 181 EDEKKWSALSKEILGEDSTDSDA 203
E+E+K+ A+ KEIL E +DS+
Sbjct 392 ENEEKYKAIKKEILDEGDSDSNT 414
> mmu:668107 Gm13696, OTTMUSG00000013392; predicted gene 13696
Length=830
Score = 199 bits (505), Expect = 9e-51, Method: Compositional matrix adjust.
Identities = 106/203 (52%), Positives = 147/203 (72%), Gaps = 3/203 (1%)
Query 1 AALLSIVNSKLPDIGELVIKRLILQFRRAYRRDDKVVCLGCVEFFAHLVNQRIAHELLAL 60
AAL++I+NSK P IGEL++KRLIL FR+ YRR+DK +CL +F AHL+NQ +AHE+L L
Sbjct 215 AALVAIINSKFPQIGELILKRLILNFRKGYRRNDKQLCLTASKFVAHLINQNVAHEVLCL 274
Query 61 QLCELLLQQPTNDSVEVCIGFLKAVGQVLEDVCRGGFDAAFERLRAILQEGETDKKTQYS 120
++ LLL++PT+DSVEV IGFLK G L V G +A FERLR IL E E DK+ QY
Sbjct 275 EMLTLLLERPTDDSVEVAIGFLKECGLKLAQVSPRGINAIFERLRNILHESEIDKRVQYM 334
Query 121 IEALWDIRRNGFKDFPGVIEDLDLVEVEDKITHEVDIMDPSIKGEEMLNIFKPQDPDEYE 180
IE ++ +R++GFKD P ++E LDLVE +D+ TH + + D E++LN+FK DP+ +
Sbjct 335 IEVMFAVRKDGFKDHPVILEGLDLVEEDDQFTHMLPLED-DYNPEDVLNVFK-MDPN-FM 391
Query 181 EDEKKWSALSKEILGEDSTDSDA 203
E+E+K+ A+ KEIL E +DS+
Sbjct 392 ENEEKYKAIKKEILDEGDSDSNT 414
> mmu:668119 Gm13691, OTTMUSG00000013386; predicted gene 13691
Length=830
Score = 199 bits (505), Expect = 9e-51, Method: Compositional matrix adjust.
Identities = 106/203 (52%), Positives = 147/203 (72%), Gaps = 3/203 (1%)
Query 1 AALLSIVNSKLPDIGELVIKRLILQFRRAYRRDDKVVCLGCVEFFAHLVNQRIAHELLAL 60
AAL++I+NSK P IGEL++KRLIL FR+ YRR+DK +CL +F AHL+NQ +AHE+L L
Sbjct 215 AALVAIINSKFPQIGELILKRLILNFRKGYRRNDKQLCLTASKFVAHLINQNVAHEVLCL 274
Query 61 QLCELLLQQPTNDSVEVCIGFLKAVGQVLEDVCRGGFDAAFERLRAILQEGETDKKTQYS 120
++ LLL++PT+DSVEV IGFLK G L V G +A FERLR IL E E DK+ QY
Sbjct 275 EMLTLLLERPTDDSVEVAIGFLKECGLKLAQVSPRGINAIFERLRNILHESEIDKRVQYM 334
Query 121 IEALWDIRRNGFKDFPGVIEDLDLVEVEDKITHEVDIMDPSIKGEEMLNIFKPQDPDEYE 180
IE ++ +R++GFKD P ++E LDLVE +D+ TH + + D E++LN+FK DP+ +
Sbjct 335 IEVMFAVRKDGFKDHPVILEGLDLVEEDDQFTHMLPLED-DYNPEDVLNVFK-MDPN-FM 391
Query 181 EDEKKWSALSKEILGEDSTDSDA 203
E+E+K+ A+ KEIL E +DS+
Sbjct 392 ENEEKYKAIKKEILDEGDSDSNT 414
> mmu:668113 Gm13694, OTTMUSG00000013390; predicted gene 13694
Length=830
Score = 199 bits (505), Expect = 9e-51, Method: Compositional matrix adjust.
Identities = 106/203 (52%), Positives = 147/203 (72%), Gaps = 3/203 (1%)
Query 1 AALLSIVNSKLPDIGELVIKRLILQFRRAYRRDDKVVCLGCVEFFAHLVNQRIAHELLAL 60
AAL++I+NSK P IGEL++KRLIL FR+ YRR+DK +CL +F AHL+NQ +AHE+L L
Sbjct 215 AALVAIINSKFPQIGELILKRLILNFRKGYRRNDKQLCLTASKFVAHLINQNVAHEVLCL 274
Query 61 QLCELLLQQPTNDSVEVCIGFLKAVGQVLEDVCRGGFDAAFERLRAILQEGETDKKTQYS 120
++ LLL++PT+DSVEV IGFLK G L V G +A FERLR IL E E DK+ QY
Sbjct 275 EMLTLLLERPTDDSVEVAIGFLKECGLKLAQVSPRGINAIFERLRNILHESEIDKRVQYM 334
Query 121 IEALWDIRRNGFKDFPGVIEDLDLVEVEDKITHEVDIMDPSIKGEEMLNIFKPQDPDEYE 180
IE ++ +R++GFKD P ++E LDLVE +D+ TH + + D E++LN+FK DP+ +
Sbjct 335 IEVMFAVRKDGFKDHPVILEGLDLVEEDDQFTHMLPLED-DYNPEDVLNVFK-MDPN-FM 391
Query 181 EDEKKWSALSKEILGEDSTDSDA 203
E+E+K+ A+ KEIL E +DS+
Sbjct 392 ENEEKYKAIKKEILDEGDSDSNT 414
> mmu:545432 Gm13695, Gm5838, OTTMUSG00000013391; predicted gene
13695
Length=830
Score = 199 bits (505), Expect = 9e-51, Method: Compositional matrix adjust.
Identities = 106/203 (52%), Positives = 147/203 (72%), Gaps = 3/203 (1%)
Query 1 AALLSIVNSKLPDIGELVIKRLILQFRRAYRRDDKVVCLGCVEFFAHLVNQRIAHELLAL 60
AAL++I+NSK P IGEL++KRLIL FR+ YRR+DK +CL +F AHL+NQ +AHE+L L
Sbjct 215 AALVAIINSKFPQIGELILKRLILNFRKGYRRNDKQLCLTASKFVAHLINQNVAHEVLCL 274
Query 61 QLCELLLQQPTNDSVEVCIGFLKAVGQVLEDVCRGGFDAAFERLRAILQEGETDKKTQYS 120
++ LLL++PT+DSVEV IGFLK G L V G +A FERLR IL E E DK+ QY
Sbjct 275 EMLTLLLERPTDDSVEVAIGFLKECGLKLAQVSPRGINAIFERLRNILHESEIDKRVQYM 334
Query 121 IEALWDIRRNGFKDFPGVIEDLDLVEVEDKITHEVDIMDPSIKGEEMLNIFKPQDPDEYE 180
IE ++ +R++GFKD P ++E LDLVE +D+ TH + + D E++LN+FK DP+ +
Sbjct 335 IEVMFAVRKDGFKDHPVILEGLDLVEEDDQFTHMLPLED-DYNPEDVLNVFK-MDPN-FM 391
Query 181 EDEKKWSALSKEILGEDSTDSDA 203
E+E+K+ A+ KEIL E +DS+
Sbjct 392 ENEEKYKAIKKEILDEGDSDSNT 414
> mmu:668100 Gm13693, OTTMUSG00000013388; predicted gene 13693
Length=830
Score = 199 bits (505), Expect = 9e-51, Method: Compositional matrix adjust.
Identities = 106/203 (52%), Positives = 147/203 (72%), Gaps = 3/203 (1%)
Query 1 AALLSIVNSKLPDIGELVIKRLILQFRRAYRRDDKVVCLGCVEFFAHLVNQRIAHELLAL 60
AAL++I+NSK P IGEL++KRLIL FR+ YRR+DK +CL +F AHL+NQ +AHE+L L
Sbjct 215 AALVAIINSKFPQIGELILKRLILNFRKGYRRNDKQLCLTASKFVAHLINQNVAHEVLCL 274
Query 61 QLCELLLQQPTNDSVEVCIGFLKAVGQVLEDVCRGGFDAAFERLRAILQEGETDKKTQYS 120
++ LLL++PT+DSVEV IGFLK G L V G +A FERLR IL E E DK+ QY
Sbjct 275 EMLTLLLERPTDDSVEVAIGFLKECGLKLAQVSPRGINAIFERLRNILHESEIDKRVQYM 334
Query 121 IEALWDIRRNGFKDFPGVIEDLDLVEVEDKITHEVDIMDPSIKGEEMLNIFKPQDPDEYE 180
IE ++ +R++GFKD P ++E LDLVE +D+ TH + + D E++LN+FK DP+ +
Sbjct 335 IEVMFAVRKDGFKDHPVILEGLDLVEEDDQFTHMLPLED-DYNPEDVLNVFK-MDPN-FM 391
Query 181 EDEKKWSALSKEILGEDSTDSDA 203
E+E+K+ A+ KEIL E +DS+
Sbjct 392 ENEEKYKAIKKEILDEGDSDSNT 414
> mmu:668096 Gm13698, OTTMUSG00000013395; predicted gene 13698
Length=830
Score = 199 bits (505), Expect = 9e-51, Method: Compositional matrix adjust.
Identities = 106/203 (52%), Positives = 147/203 (72%), Gaps = 3/203 (1%)
Query 1 AALLSIVNSKLPDIGELVIKRLILQFRRAYRRDDKVVCLGCVEFFAHLVNQRIAHELLAL 60
AAL++I+NSK P IGEL++KRLIL FR+ YRR+DK +CL +F AHL+NQ +AHE+L L
Sbjct 215 AALVAIINSKFPQIGELILKRLILNFRKGYRRNDKQLCLTASKFVAHLINQNVAHEVLCL 274
Query 61 QLCELLLQQPTNDSVEVCIGFLKAVGQVLEDVCRGGFDAAFERLRAILQEGETDKKTQYS 120
++ LLL++PT+DSVEV IGFLK G L V G +A FERLR IL E E DK+ QY
Sbjct 275 EMLTLLLERPTDDSVEVAIGFLKECGLKLAQVSPRGINAIFERLRNILHESEIDKRVQYM 334
Query 121 IEALWDIRRNGFKDFPGVIEDLDLVEVEDKITHEVDIMDPSIKGEEMLNIFKPQDPDEYE 180
IE ++ +R++GFKD P ++E LDLVE +D+ TH + + D E++LN+FK DP+ +
Sbjct 335 IEVMFAVRKDGFKDHPVILEGLDLVEEDDQFTHMLPLED-DYNPEDVLNVFK-MDPN-FM 391
Query 181 EDEKKWSALSKEILGEDSTDSDA 203
E+E+K+ A+ KEIL E +DS+
Sbjct 392 ENEEKYKAIKKEILDEGDSDSNT 414
> ath:AT1G80930 MIF4G domain-containing protein / MA3 domain-containing
protein; K13100 pre-mRNA-splicing factor CWC22
Length=900
Score = 192 bits (489), Expect = 7e-49, Method: Compositional matrix adjust.
Identities = 101/205 (49%), Positives = 145/205 (70%), Gaps = 6/205 (2%)
Query 1 AALLSIVNSKLPDIGELVIKRLILQFRRAYRRDDKVVCLGCVEFFAHLVNQRIAHELLAL 60
AAL++++N+K P++ EL++KR++LQ +RAY+R+DK L V+F AHLVNQ++A E++AL
Sbjct 416 AALVAVINAKFPEVAELLLKRVVLQLKRAYKRNDKPQLLAAVKFIAHLVNQQVAEEIIAL 475
Query 61 QLCELLLQQPTNDSVEVCIGFLKAVGQVLEDVCRGGFDAAFERLRAILQEGETDKKTQYS 120
+L +LL PT+DSVEV +GF+ G +L+DV G + FER R IL EGE DK+ QY
Sbjct 476 ELVTILLGDPTDDSVEVAVGFVTECGAMLQDVSPRGLNGIFERFRGILHEGEIDKRVQYL 535
Query 121 IEALWDIRRNGFKDFPGVIEDLDLVEVEDKITHEVDIMDPSIKGEEMLNIFKPQDPDEYE 180
IE+L+ R+ F+ P V +LDL VE+K +H++ +D I E L+IFKP DPD +
Sbjct 536 IESLFATRKAKFQGHPAVRPELDL--VEEKYSHDLS-LDHEIDPETALDIFKP-DPD-FV 590
Query 181 EDEKKWSALSKEILG-EDSTDSDAS 204
E+EKK+ AL KE+LG E+S D D S
Sbjct 591 ENEKKYEALKKELLGDEESEDEDGS 615
> cel:F33A8.1 let-858; LEThal family member (let-858); K13100
pre-mRNA-splicing factor CWC22
Length=897
Score = 185 bits (470), Expect = 1e-46, Method: Compositional matrix adjust.
Identities = 91/200 (45%), Positives = 143/200 (71%), Gaps = 8/200 (4%)
Query 1 AALLSIVNSKLPDIGELVIKRLILQFRRAYRRDDKVVCLGCVEFFAHLVNQRIAHELLAL 60
AAL +++NSK P +GEL+++RLI+QF+R++RR+D+ V + ++F AHL+NQ++AHE+LAL
Sbjct 247 AALAAVINSKFPHVGELLLRRLIVQFKRSFRRNDRGVTVNVIKFIAHLINQQVAHEVLAL 306
Query 61 QLCELLLQQPTNDSVEVCIGFLKAVGQVLEDVCRGGFDAAFERLRAILQEGET-----DK 115
++ L+L++PT+DSVEV I FLK G L ++ ++ ++RLRAIL E E D+
Sbjct 307 EIMILMLEEPTDDSVEVAIAFLKECGAKLLEIAPAALNSVYDRLRAILMETERSENALDR 366
Query 116 KTQYSIEALWDIRRNGFKDFPGVIEDLDLVEVEDKITHEVDIMDPSIKGEEMLNIFKPQD 175
+ QY IE IR++ F +P VIEDLDL+E ED+I H +++ D ++ E LN+FK D
Sbjct 367 RIQYMIETAMQIRKDKFAAYPAVIEDLDLIEEEDQIIHTLNLED-AVDPENGLNVFK-LD 424
Query 176 PDEYEEDEKKWSALSKEILG 195
P E+E++E+ + + KEI+G
Sbjct 425 P-EFEKNEEVYEEIRKEIIG 443
> dre:566033 MGC153452, cwc22, wu:fb13g08, wu:fi16b02; zgc:153452;
K13100 pre-mRNA-splicing factor CWC22
Length=985
Score = 185 bits (469), Expect = 1e-46, Method: Compositional matrix adjust.
Identities = 100/196 (51%), Positives = 139/196 (70%), Gaps = 3/196 (1%)
Query 1 AALLSIVNSKLPDIGELVIKRLILQFRRAYRRDDKVVCLGCVEFFAHLVNQRIAHELLAL 60
+A+++I+NSK P IGEL++KRLIL FR+ YRR+DK CL +F HL+NQ +AHE+L L
Sbjct 310 SAVVAIINSKFPQIGELILKRLILNFRKGYRRNDKQQCLTASKFVGHLINQNVAHEVLCL 369
Query 61 QLCELLLQQPTNDSVEVCIGFLKAVGQVLEDVCRGGFDAAFERLRAILQEGETDKKTQYS 120
++ LLL++PT+DSVEV I FLK G L +V G +A FERLR IL E E DK+ QY
Sbjct 370 EMLTLLLERPTDDSVEVAISFLKECGLKLTEVSPRGINAIFERLRNILHESEIDKRVQYM 429
Query 121 IEALWDIRRNGFKDFPGVIEDLDLVEVEDKITHEVDIMDPSIKGEEMLNIFKPQDPDEYE 180
IE ++ IR++GFKD P + E LDLVE ED+ TH + + D E++LN+FK DP+ +
Sbjct 430 IEVMFAIRKDGFKDHPIIPEGLDLVEEEDQFTHMLPLED-EYNTEDILNVFK-LDPN-FL 486
Query 181 EDEKKWSALSKEILGE 196
E+E+K+ + +EIL E
Sbjct 487 ENEEKYKTIKREILDE 502
> cpv:cgd8_2960 NIC+MI domains containing protein. nucampholin/yeast
Cwc22p like protein involved in mRNA splicing ; K13100
pre-mRNA-splicing factor CWC22
Length=619
Score = 160 bits (406), Expect = 3e-39, Method: Compositional matrix adjust.
Identities = 77/193 (39%), Positives = 124/193 (64%), Gaps = 0/193 (0%)
Query 3 LLSIVNSKLPDIGELVIKRLILQFRRAYRRDDKVVCLGCVEFFAHLVNQRIAHELLALQL 62
L +I+N +PD G L+++RLI QFR +Y + DK VC + F A L+NQ++ HEL+ALQ+
Sbjct 104 LSAIINCNIPDFGSLLLRRLINQFRISYSKGDKYVCKHTLLFLAQLINQKVVHELIALQI 163
Query 63 CELLLQQPTNDSVEVCIGFLKAVGQVLEDVCRGGFDAAFERLRAILQEGETDKKTQYSIE 122
C L+++ T+DS+E+CI F+ GQ L + G + + R ILQEG+ +KKT + IE
Sbjct 164 CLFLIEKLTDDSIEICIDFIFECGQFLLENTPQGLNTIMNKFRRILQEGKLNKKTNFLIE 223
Query 123 ALWDIRRNGFKDFPGVIEDLDLVEVEDKITHEVDIMDPSIKGEEMLNIFKPQDPDEYEED 182
+ RR+ F ++P + +L+++ D+ITH DI+D I ++ L+ F +P+ +EE+
Sbjct 224 RILKERRDNFMNYPINNPENELIDLNDQITHFFDILDGEIDIQDELDHFIETEPNIFEEE 283
Query 183 EKKWSALSKEILG 195
KW +SKE+L
Sbjct 284 NTKWDEISKELLS 296
> sce:YGR278W CWC22; Cwc22p; K13100 pre-mRNA-splicing factor CWC22
Length=577
Score = 67.0 bits (162), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 58/205 (28%), Positives = 106/205 (51%), Gaps = 7/205 (3%)
Query 1 AALLSIVNSKLPDIGELVIKRLILQFRRAYRRDDKVVCLGCVEFFAHLVNQRIAHELLAL 60
+AL++++NS +PDIGE + K L+L F + + R D V C ++ + L + HE++ L
Sbjct 77 SALIALLNSDIPDIGETLAKELMLMFVQQFNRKDYVSCGNILQCLSILFLYDVIHEIVIL 136
Query 61 QLCELLLQQPTNDSVEVCIGFLKAVGQVLEDVCRGGFDAAFERLRAILQEGETDKKTQYS 120
Q+ LLL++ +S+ + I +K G L V + D +E+LR ILQ E + S
Sbjct 137 QILLLLLEK---NSLRLVIAVMKICGWKLALVSKKTHDMIWEKLRYILQTQELSSTLRES 193
Query 121 IEALWDIRRNGFKDFPGVIEDLDLVEVEDKITHEVDIMDPSIKGEEMLNIFKPQDPDEYE 180
+E L++IR+ +K + LD TH + D +E+ N K ++ +
Sbjct 194 LETLFEIRQKDYKSGSQGLFILDPTSYTVH-THSYIVSDEDEANKELGNFEKCEN---FN 249
Query 181 EDEKKWSALSKEILGEDSTDSDASS 205
E + L +++L +++D++ S
Sbjct 250 ELTMAFDTLRQKLLINNTSDTNEGS 274
> ath:AT5G57870 eukaryotic translation initiation factor 4F, putative
/ eIF-4F, putative; K03260 translation initiation factor
4G
Length=776
Score = 37.4 bits (85), Expect = 0.047, Method: Compositional matrix adjust.
Identities = 22/100 (22%), Positives = 49/100 (49%), Gaps = 7/100 (7%)
Query 40 GCVEFFAHLVNQRIAHELLALQLCELLLQQ-----PTNDSVEVCIGFLKAVGQVLEDVCR 94
G + L+ Q++ E + + + LL P ++VE F K +G+ L+ +
Sbjct 339 GNIRLIGELLKQKMVPEKIVHHIVQELLGADEKVCPAEENVEAICHFFKTIGKQLDGNVK 398
Query 95 GGF--DAAFERLRAILQEGETDKKTQYSIEALWDIRRNGF 132
D F+RL+A+ + + + + ++ ++ + D+R NG+
Sbjct 399 SKRINDVYFKRLQALSKNPQLELRLRFMVQNIIDMRSNGW 438
> dre:557585 nom1, si:ch211-266o5.1; nucleolar protein with MIF4G
domain 1
Length=487
Score = 37.0 bits (84), Expect = 0.057, Method: Compositional matrix adjust.
Identities = 31/131 (23%), Positives = 57/131 (43%), Gaps = 6/131 (4%)
Query 13 DIGELVIKRLILQFRRAYRRDDKV--VCLGCVEFFAHLVNQRIAHELLALQLCELLLQQP 70
++G ++ ++ QF R Y D C V AHL N + H +L + + L+
Sbjct 60 EVGAHFLETVVRQFDRIYSELDGTDKECDNLVSIIAHLYNFHVVHAVLVFDILKKLVTSF 119
Query 71 TNDSVEVCIGFLKAVGQVL-EDVCRGGFDAAFERLRAILQEGET---DKKTQYSIEALWD 126
++ +E+ + LK VG L +D + E R EGE + ++ +E +
Sbjct 120 SSKDIELLLLVLKTVGFALRKDDPIALKELISEAQRKASAEGEQFQDQTRIRFMLETMMA 179
Query 127 IRRNGFKDFPG 137
++ N + PG
Sbjct 180 LKNNDMRKIPG 190
> ath:AT2G24050 MIF4G domain-containing protein / MA3 domain-containing
protein
Length=747
Score = 35.4 bits (80), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 24/105 (22%), Positives = 48/105 (45%), Gaps = 7/105 (6%)
Query 35 KVVCLGCVEFFAHLVNQRIAHELLALQLCELLLQQ-----PTNDSVEVCIGFLKAVGQVL 89
K+ LG + L+ Q++ E + + + LL P VE F +G+ L
Sbjct 297 KLRTLGNIRLIGELLKQKMVPEKIVHHIVQELLGDDTKACPAEGDVEALCQFFITIGKQL 356
Query 90 EDV--CRGGFDAAFERLRAILQEGETDKKTQYSIEALWDIRRNGF 132
+D RG D F RL+ + + + + + ++ ++ + D+R N +
Sbjct 357 DDSPRSRGINDTYFGRLKELARHPQLELRLRFMVQNVVDLRANKW 401
> mmu:228003 Kbtbd10, Gm112, SARCOSIN; kelch repeat and BTB (POZ)
domain containing 10; K10473 kelch repeat and BTB domain-containing
protein 5/10
Length=606
Score = 31.6 bits (70), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 27/110 (24%), Positives = 53/110 (48%), Gaps = 19/110 (17%)
Query 94 RGGFDAAFERLRAILQEGETDKKTQYSIEALWDIRRNGFK---DFPGVIEDLDLVEVEDK 150
R F A + + ++ G T+ S+EA +D++ N ++ +FP + LV +
Sbjct 485 RSMFGVAIHKGKIVIAGGVTEDGLSASVEA-FDLKTNKWEVMTEFPQERSSISLVSLAGA 543
Query 151 ITHEVDIMDPSIKGEEMLNI----FKPQDPDE---YEEDEKKWSALSKEI 193
+ +I G M+ + F P + ++ YE+D+K+W+ + KEI
Sbjct 544 LY--------AIGGFAMIQLESKEFAPTEVNDIWKYEDDKKEWAGMLKEI 585
> xla:779378 kbtbd5, MGC86259; kelch repeat and BTB (POZ) domain
containing 5; K10473 kelch repeat and BTB domain-containing
protein 5/10
Length=614
Score = 31.2 bits (69), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 28/103 (27%), Positives = 45/103 (43%), Gaps = 5/103 (4%)
Query 94 RGGFDAAFERLRAILQEGETDKKTQYSIEALWDIRRNG---FKDFPGVIEDLDLVEVEDK 150
R F A + + + G TD +IEA +DI+ N F +FP L LV +
Sbjct 493 RSLFGATVHKGKIFIAAGVTDTGLTNTIEA-YDIKTNKWEDFTEFPQERSSLSLVSMNGT 551
Query 151 ITHEVDIMDPSIKGEEMLNIFKPQDPDEYEEDEKKWSALSKEI 193
+ + EE++ + D + E+EKKW + +EI
Sbjct 552 LYAIGGFATIENESEELVPT-ELNDIWRFNEEEKKWEGILREI 593
> cpv:cgd7_5330 thioredoxin/PDI, cyanobacterial type, signal peptide
plus 4 transmembrane domains
Length=664
Score = 29.6 bits (65), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 41/134 (30%), Positives = 57/134 (42%), Gaps = 18/134 (13%)
Query 83 KAVGQVLEDVCRGGFDAAFERLRAILQEGETDKKTQYSIEALWDIRRNGFKDFPGVIEDL 142
KA QVL+D FD F++ R +K T Y W I G K GV+
Sbjct 268 KAAIQVLKD-NNSYFDCNFDQNRNSWTSCVEEKITSY---PTWLI---GDKTISGVVS-- 318
Query 143 DLVEVEDKITHEVDIMDPSIKGEEMLNIFKPQDPDEYEEDEKKWSALSKEILGEDSTDSD 202
I H V+I + EE+L + P D E E + S S+ E STD+D
Sbjct 319 -----PKTIAHMVNINLKEL-SEEVLALVHPDDKKEIE---NRISDFSEGSSTEASTDTD 369
Query 203 ASSVDSEAETSSEE 216
S +++ E S+E
Sbjct 370 TDSTEAQGEDISKE 383
Lambda K H
0.316 0.135 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7395169300
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40