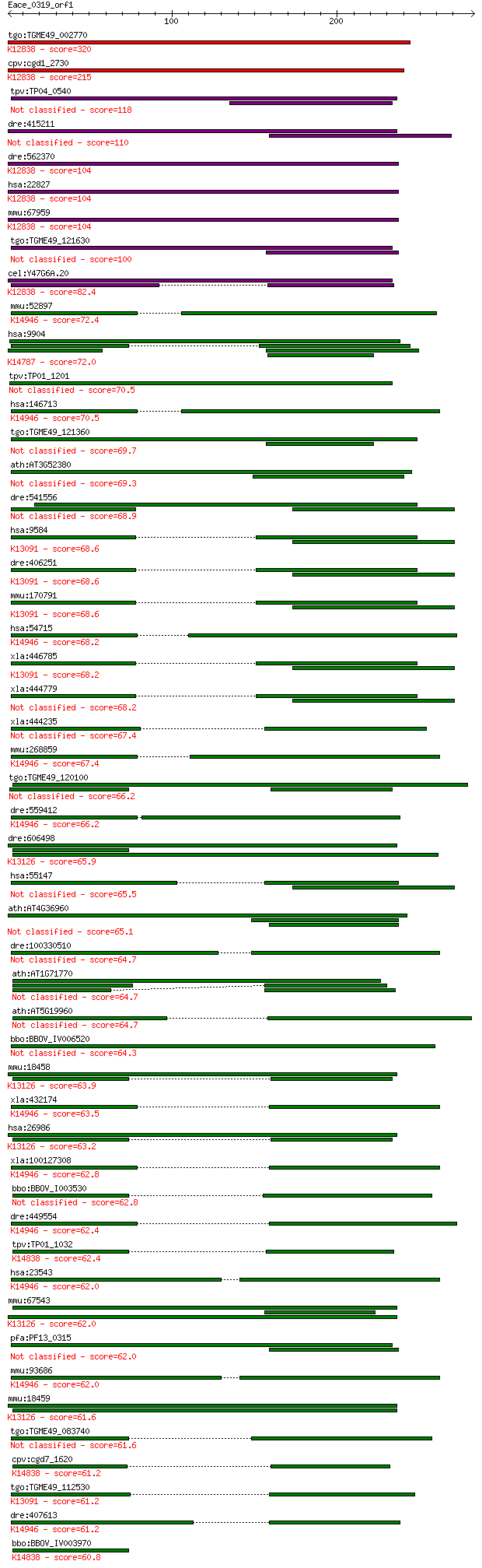
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0319_orf1
Length=282
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_002770 RNA-binding protein, putative ; K12838 poly(... 320 5e-87
cpv:cgd1_2730 Ro ribonucleoprotein-binding protein 1, RNA bind... 215 1e-55
tpv:TP04_0540 hypothetical protein 118 2e-26
dre:415211 puf60b, SIAHBP1, fe37c05, repressor, si:ch211-12p8.... 110 7e-24
dre:562370 puf60a, fc21a05, wu:fc21a05, wu:fi43a08; poly-U bin... 104 3e-22
hsa:22827 PUF60, FIR, FLJ31379, RoBPI, SIAHBP1; poly-U binding... 104 4e-22
mmu:67959 Puf60, 2410104I19Rik, 2810454F19Rik, SIAHBP1; poly-U... 104 4e-22
tgo:TGME49_121630 hypothetical protein 100 9e-21
cel:Y47G6A.20 rnp-6; RNP (RRM RNA binding domain) containing f... 82.4 2e-15
mmu:52897 Rbfox3, D11Bwg0517e, Fox-3, Hrnbp3, NeuN, Neuna60; R... 72.4 2e-12
hsa:9904 RBM19, DKFZp586F1023, KIAA0682; RNA binding motif pro... 72.0 2e-12
tpv:TP01_1201 hypothetical protein 70.5 7e-12
hsa:146713 RBFOX3, FLJ56884, FLJ58356, FOX-3, FOX3, HRNBP3, NE... 70.5 7e-12
tgo:TGME49_121360 RNA recognition motif domain-containing protein 69.7 1e-11
ath:AT3G52380 CP33; CP33; RNA binding 69.3 2e-11
dre:541556 rbm39b, rnpc2l, wu:fa97g07, wu:fb09c08, zgc:112139,... 68.9 2e-11
hsa:9584 RBM39, CAPER, CAPERalpha, CC1.3, DKFZp781C0423, FLJ44... 68.6 2e-11
dre:406251 rbm39a, rnpc2, zgc:55780; RNA binding motif protein... 68.6 3e-11
mmu:170791 Rbm39, 1500012C14Rik, 2310040E03Rik, B330012G18Rik,... 68.6 3e-11
hsa:54715 RBFOX1, A2BP1, FOX-1, FOX1, HRNBP1; RNA binding prot... 68.2 3e-11
xla:446785 rbm39, MGC80448, rnpc2; RNA binding motif protein 3... 68.2 3e-11
xla:444779 MGC81970 protein 68.2 4e-11
xla:444235 rbm23, MGC80803; RNA binding motif protein 23 67.4
mmu:268859 Rbfox1, A2bp, A2bp1, Hrnbp1, fox-1; RNA binding pro... 67.4 6e-11
tgo:TGME49_120100 RNA recognition motif-containing protein 66.2 1e-10
dre:559412 RNA binding motif protein 9-like; K14946 RNA bindin... 66.2 1e-10
dre:606498 pabpc1a, pabpc1, wu:fb16a02, wu:fi19b08, wu:fj12d09... 65.9 2e-10
hsa:55147 RBM23, CAPERbeta, FLJ10482, MGC4458, RNPC4; RNA bind... 65.5 2e-10
ath:AT4G36960 RNA recognition motif (RRM)-containing protein 65.1 3e-10
dre:100330510 Fox-1 homolog C-like 64.7 3e-10
ath:AT1G71770 PAB5; PAB5 (POLY(A)-BINDING PROTEIN 5); RNA bind... 64.7 3e-10
ath:AT5G19960 RNA recognition motif (RRM)-containing protein 64.7 4e-10
bbo:BBOV_IV006520 23.m06153; RNA binding motif containing protein 64.3 5e-10
mmu:18458 Pabpc1, PABP, Pabp1, PabpI, Pabpl1, ePAB; poly(A) bi... 63.9 7e-10
xla:432174 rbfox2-b, MGC78805, fox-2, fox2, fxh, hnrbp2, hrnbp... 63.5 8e-10
hsa:26986 PABPC1, PAB1, PABP, PABP1, PABPC2, PABPL1; poly(A) b... 63.2 1e-09
xla:100127308 rbfox2-a, fox-2, fox2, fxh, hnrbp2, hrnbp2, rbm9... 62.8 1e-09
bbo:BBOV_I003530 19.m02179; RNA recognition motif. (a.k.a. RRM... 62.8 1e-09
dre:449554 rbfox1, a2bp1, fox1, zgc:103635; RNA binding protei... 62.4 2e-09
tpv:TP01_1032 hypothetical protein; K14838 nucleolar protein 15 62.4
hsa:23543 RBFOX2, FOX2, Fox-2, HNRBP2, HRNBP2, RBM9, RTA, dJ10... 62.0 2e-09
mmu:67543 Pabpc6, 4932702K14Rik, AI428050, MGC132900, Pabpc3; ... 62.0 2e-09
pfa:PF13_0315 RNA binding protein, putative 62.0 2e-09
mmu:93686 Rbfox2, 2810460A15Rik, AA407676, AI118529, Fbm2, Fxh... 62.0 2e-09
mmu:18459 Pabpc2, PABP, PABP+, Pabp2; poly(A) binding protein,... 61.6 3e-09
tgo:TGME49_083740 mRNA processing protein, putative (EC:3.4.21... 61.6 3e-09
cpv:cgd7_1620 nop15p/nopp34; nucleolar protein with 1 RRM doma... 61.2 4e-09
tgo:TGME49_112530 splicing factor protein, putative ; K13091 R... 61.2 4e-09
dre:407613 rbfox1l, Fox-1, a2bp1l, fox1l, hm:zeh0082; RNA bind... 61.2 4e-09
bbo:BBOV_IV003970 23.m05815; RNA recognition motif containing ... 60.8 6e-09
> tgo:TGME49_002770 RNA-binding protein, putative ; K12838 poly(U)-binding-splicing
factor PUF60
Length=532
Score = 320 bits (819), Expect = 5e-87, Method: Compositional matrix adjust.
Identities = 184/264 (69%), Positives = 206/264 (78%), Gaps = 24/264 (9%)
Query 1 MCRIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGDRSKGFCFVEYASPEAAQMALST 60
MCRIYVGSLDYYLTELEIKSVFQAFGT+ SVDMPKEGDRSKGFCFVEYASPEAA+MALST
Sbjct 74 MCRIYVGSLDYYLTELEIKSVFQAFGTIVSVDMPKEGDRSKGFCFVEYASPEAAEMALST 133
Query 61 MHNFVLKGRTIKVGRPTAVGAGQPQQLLARP--LGGYDMNVGAA--------ATPGMSLV 110
M NFVLKGRTIKVGRPTAVG G Q L RP + +VGAA +PG S+V
Sbjct 134 MQNFVLKGRTIKVGRPTAVGTG--GQQLQRPAMMMPTVGSVGAAGGLTSMAGTSPG-SMV 190
Query 111 SSAQAGAAVATAILAGRAGQTPQGDMASLVSMGQ---VTPE--------KQQDSAPGSNR 159
+SAQAGAA ATAILAGR G ++A L+S Q +TPE QQ +NR
Sbjct 191 NSAQAGAAAATAILAGRTGAADPSNLAGLLSTSQTAVLTPEQQQMQQQLLQQQQQMANNR 250
Query 160 IYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAIDT 219
+Y+GNVPFGFSSEDLKKIFV FG IL+CQLLPSQENPQQHRGYGFIE+A+ AAKLAI+T
Sbjct 251 VYIGNVPFGFSSEDLKKIFVVFGPILSCQLLPSQENPQQHRGYGFIEYATADAAKLAIET 310
Query 220 MNGFEVVGKQLKVNYATALRSAAV 243
MNGFEV GKQLKVN+ATA+R++ V
Sbjct 311 MNGFEVAGKQLKVNFATAMRNSPV 334
> cpv:cgd1_2730 Ro ribonucleoprotein-binding protein 1, RNA binding
protein with 3x RRM domains ; K12838 poly(U)-binding-splicing
factor PUF60
Length=693
Score = 215 bits (548), Expect = 1e-55, Method: Compositional matrix adjust.
Identities = 116/240 (48%), Positives = 151/240 (62%), Gaps = 24/240 (10%)
Query 1 MCRIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGDRSKGFCFVEYASPEAAQMALST 60
+ RIYVGSLDY L E ++K VF +FG + ++DMP+EG+RSKGFCF+EY S E+A+MAL+T
Sbjct 289 ISRIYVGSLDYSLNEADLKQVFGSFGPIVNIDMPREGNRSKGFCFIEYTSQESAEMALAT 348
Query 61 MHNFVLKGRTIKVGRPT-AVGAGQPQQLLARPLGGYDMNVGAAATPGMSLVSSAQAGAAV 119
M+ FVLKGR I+VGRPT A + Q GG NV P +++ ++
Sbjct 349 MNRFVLKGRPIRVGRPTNAASSNGNQSGSGGIGGGGSGNV---INPNIAVFNNNHI---- 401
Query 120 ATAILAGRAGQTPQGDMASLVSMGQVTPEKQQDSAPGSNRIYVGNVPFGFSSEDLKKIFV 179
Q Q + G T + NRIY+G+VP+ F+++DL+ IF
Sbjct 402 --------THQNHQIQSETTHGTGIATHSQ--------NRIYIGSVPYSFTTDDLRHIFK 445
Query 180 CFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAIDTMNGFEVVGKQLKVNYATALR 239
FG IL+CQL+PS E P HRGYGFIEF + AKLAI+TMNGFEV GKQLKVN ATAL+
Sbjct 446 TFGVILSCQLIPSIEKPGTHRGYGFIEFGTADQAKLAIETMNGFEVGGKQLKVNVATALK 505
> tpv:TP04_0540 hypothetical protein
Length=486
Score = 118 bits (296), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 78/247 (31%), Positives = 121/247 (48%), Gaps = 37/247 (14%)
Query 3 RIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGD--RSKGFCFVEYASPEAAQMALST 60
+IY+G LD T +I+ +F +FG + ++D+P E D + KGFCFVEY E+A +AL++
Sbjct 162 KIYIGQLDPSCTIEDIRVIFSSFGDILNIDLPTEPDTNKVKGFCFVEYRKKESADLALNS 221
Query 61 MHNFVLKGRTIKVGRPTA-----------VGAGQPQQLLARPLGGYDMNVGAAATPGMSL 109
M F +KG+ IK+ RP +G P L+ PL AA +
Sbjct 222 MQGFHIKGKPIKLARPNISSSGSSSGICPIGYNNP---LSNPL--------AAGAVAAAT 270
Query 110 VSSAQAGAAVATA-ILAGRAGQTPQGDMASLVSMGQVTPEKQQDSAPGSNRIYVGNVPFG 168
++G V T +L + TP ++G PE R+ + NVPF
Sbjct 271 FLQNRSGVRVDTNNVLTKISASTPS------FTIGATPPEL------NGKRVVLENVPFE 318
Query 169 FSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAIDTMNGFEVVGK 228
++ D+++IF FG+I C L + P G+I+F S A+ TMNGFE+ G
Sbjct 319 LAASDIRRIFEPFGNITECVLYSREMLPGAFFALGYIDFVSANVAQTVCSTMNGFEIAGS 378
Query 229 QLKVNYA 235
+++V A
Sbjct 379 KIQVTLA 385
Score = 60.5 bits (145), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 30/98 (30%), Positives = 55/98 (56%), Gaps = 1/98 (1%)
Query 135 DMASLVSMGQVTPEKQQDSAPGSNRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQE 194
D SL++ + K + S +IY+G + + ED++ IF FG IL L P++
Sbjct 138 DTKSLINNKKPEQSKAPEEPQKSYKIYIGQLDPSCTIEDIRVIFSSFGDILNIDL-PTEP 196
Query 195 NPQQHRGYGFIEFASPAAAKLAIDTMNGFEVVGKQLKV 232
+ + +G+ F+E+ +A LA+++M GF + GK +K+
Sbjct 197 DTNKVKGFCFVEYRKKESADLALNSMQGFHIKGKPIKL 234
> dre:415211 puf60b, SIAHBP1, fe37c05, repressor, si:ch211-12p8.2,
si:zc12p8.2, wu:fb33e11, wu:fe37c05, zgc:86806; poly-U
binding splicing factor b
Length=502
Score = 110 bits (274), Expect = 7e-24, Method: Compositional matrix adjust.
Identities = 74/239 (30%), Positives = 110/239 (46%), Gaps = 68/239 (28%)
Query 1 MCRIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGD--RSKGFCFVEYASPEAAQMAL 58
MCR+YVGS+ Y L E I+ F FG + S+DM + + KGF FVEY PEAAQ+AL
Sbjct 68 MCRVYVGSIYYELGEDTIRQAFAPFGPIKSIDMSWDSVTMKHKGFAFVEYEVPEAAQLAL 127
Query 59 STMHNFVLKGRTIKVGRPTAVGAGQPQQLLARPLGGYDMNVGAAATPGMSLVSSAQAGAA 118
M++ +L GG ++ VG PG
Sbjct 128 EQMNSVML--------------------------GGRNIKVG---RPG------------ 146
Query 119 VATAILAGRAGQTPQGDMASLVSMGQVTP--EKQQDSAPGSNRIYVGNVPFGFSSEDLKK 176
S+GQ P E+ + A NRIYV ++ S +D+K
Sbjct 147 ----------------------SIGQAQPIIEQLAEEARAYNRIYVASIHPDLSDDDIKS 184
Query 177 IFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAIDTMNGFEVVGKQLKVNYA 235
+F FG I +C +L + +H+G+GFIE+ P ++ A+ +MN F++ G+ L+V A
Sbjct 185 VFEAFGKIKSC-MLAREPTTGKHKGFGFIEYEKPQSSLDAVSSMNLFDLGGQYLRVGKA 242
Score = 51.2 bits (121), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 32/111 (28%), Positives = 57/111 (51%), Gaps = 17/111 (15%)
Query 159 RIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQEN-PQQHRGYGFIEFASPAAAKLAI 217
R+YVG++ + + +++ F FG I + + S ++ +H+G+ F+E+ P AA+LA+
Sbjct 70 RVYVGSIYYELGEDTIRQAFAPFGPIKSIDM--SWDSVTMKHKGFAFVEYEVPEAAQLAL 127
Query 218 DTMNGFEVVGKQLKVNYATALRSAAVAGSLGVGMGAQMAPIVAATATAATA 268
+ MN + G+ +KV GS+G Q PI+ A A A
Sbjct 128 EQMNSVMLGGRNIKVGR---------PGSIG-----QAQPIIEQLAEEARA 164
> dre:562370 puf60a, fc21a05, wu:fc21a05, wu:fi43a08; poly-U binding
splicing factor a; K12838 poly(U)-binding-splicing factor
PUF60
Length=518
Score = 104 bits (260), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 74/242 (30%), Positives = 108/242 (44%), Gaps = 72/242 (29%)
Query 1 MCRIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGD--RSKGFCFVEYASPEAAQMAL 58
MCR+YVGS+ Y L E I+ F FG + S+DM + + KGF FVEY PEAAQ+AL
Sbjct 91 MCRVYVGSIYYELGEDTIRQAFAPFGPIKSIDMSWDSVTLKHKGFAFVEYEVPEAAQLAL 150
Query 59 STMHNFVLKGRTIKVGRPTAVGAGQPQQLLARPLGGYDMNVGAAATPGMSLVSSAQAGAA 118
M++ +L GG ++ VG +
Sbjct 151 EQMNSVML--------------------------GGRNIKVGRPS--------------- 169
Query 119 VATAILAGRAGQTPQGDMASLVSMGQVTPEKQQ--DSAPGSNRIYVGNVPFGFSSEDLKK 176
++GQ P Q + A NRIYV +V S +D+K
Sbjct 170 ----------------------NIGQAQPIIDQLAEEARAFNRIYVASVHPDLSDDDIKS 207
Query 177 IFVCFGSILTCQLLPSQENPQ--QHRGYGFIEFASPAAAKLAIDTMNGFEVVGKQLKVNY 234
+F FG I +C L +P +H+GYGFIE+ +A+ A+ +MN F++ G+ L+V
Sbjct 208 VFEAFGRIKSCSL---ARDPTTGKHKGYGFIEYDKAQSAQDAVSSMNLFDLGGQYLRVGK 264
Query 235 AT 236
A
Sbjct 265 AV 266
> hsa:22827 PUF60, FIR, FLJ31379, RoBPI, SIAHBP1; poly-U binding
splicing factor 60KDa; K12838 poly(U)-binding-splicing factor
PUF60
Length=516
Score = 104 bits (259), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 73/242 (30%), Positives = 108/242 (44%), Gaps = 72/242 (29%)
Query 1 MCRIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGD--RSKGFCFVEYASPEAAQMAL 58
MCR+YVGS+ Y L E I+ F FG + S+DM + + KGF FVEY PEAAQ+AL
Sbjct 85 MCRVYVGSIYYELGEDTIRQAFAPFGPIKSIDMSWDSVTMKHKGFAFVEYEVPEAAQLAL 144
Query 59 STMHNFVLKGRTIKVGRPTAVGAGQPQQLLARPLGGYDMNVGAAATPGMSLVSSAQAGAA 118
M++ +L GG ++ VG +
Sbjct 145 EQMNSVML--------------------------GGRNIKVGRPS--------------- 163
Query 119 VATAILAGRAGQTPQGDMASLVSMGQVTPEKQQ--DSAPGSNRIYVGNVPFGFSSEDLKK 176
++GQ P Q + A NRIYV +V S +D+K
Sbjct 164 ----------------------NIGQAQPIIDQLAEEARAFNRIYVASVHQDLSDDDIKS 201
Query 177 IFVCFGSILTCQLLPSQENPQ--QHRGYGFIEFASPAAAKLAIDTMNGFEVVGKQLKVNY 234
+F FG I +C L +P +H+GYGFIE+ +++ A+ +MN F++ G+ L+V
Sbjct 202 VFEAFGKIKSCTL---ARDPTTGKHKGYGFIEYEKAQSSQDAVSSMNLFDLGGQYLRVGK 258
Query 235 AT 236
A
Sbjct 259 AV 260
> mmu:67959 Puf60, 2410104I19Rik, 2810454F19Rik, SIAHBP1; poly-U
binding splicing factor 60; K12838 poly(U)-binding-splicing
factor PUF60
Length=499
Score = 104 bits (259), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 73/242 (30%), Positives = 108/242 (44%), Gaps = 72/242 (29%)
Query 1 MCRIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGD--RSKGFCFVEYASPEAAQMAL 58
MCR+YVGS+ Y L E I+ F FG + S+DM + + KGF FVEY PEAAQ+AL
Sbjct 68 MCRVYVGSIYYELGEDTIRQAFAPFGPIKSIDMSWDSVTMKHKGFAFVEYEVPEAAQLAL 127
Query 59 STMHNFVLKGRTIKVGRPTAVGAGQPQQLLARPLGGYDMNVGAAATPGMSLVSSAQAGAA 118
M++ +L GG ++ VG +
Sbjct 128 EQMNSVML--------------------------GGRNIKVGRPS--------------- 146
Query 119 VATAILAGRAGQTPQGDMASLVSMGQVTPEKQQ--DSAPGSNRIYVGNVPFGFSSEDLKK 176
++GQ P Q + A NRIYV +V S +D+K
Sbjct 147 ----------------------NIGQAQPIIDQLAEEARAFNRIYVASVHQDLSDDDIKS 184
Query 177 IFVCFGSILTCQLLPSQENPQ--QHRGYGFIEFASPAAAKLAIDTMNGFEVVGKQLKVNY 234
+F FG I +C L +P +H+GYGFIE+ +++ A+ +MN F++ G+ L+V
Sbjct 185 VFEAFGKIKSCTLA---RDPTTGKHKGYGFIEYEKAQSSQDAVSSMNLFDLGGQYLRVGK 241
Query 235 AT 236
A
Sbjct 242 AV 243
> tgo:TGME49_121630 hypothetical protein
Length=400
Score = 100 bits (248), Expect = 9e-21, Method: Compositional matrix adjust.
Identities = 72/237 (30%), Positives = 111/237 (46%), Gaps = 41/237 (17%)
Query 3 RIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGDRSK--GFCFVEYASPEAAQMALST 60
RIY+ SL ++E E+++VFQ+FG + V +P D K + +++A+ EAAQ A+
Sbjct 95 RIYITSLKVPVSEEELRAVFQSFGEIKQVYIPPREDEKKRTTYALMDFATLEAAQTAVQA 154
Query 61 MHNFVLKGRTIKVGRPTAVGAGQPQQLLARPLGGYDMNVGAAATPGMSLVSSAQAGAAVA 120
M+NF KG+ I+V A P G+ + A G VS ++ A +
Sbjct 155 MNNFRFKGQRIQVAH-------------ASPAMGHHASREAQKDEGPKRVSVSKRAATPS 201
Query 121 T-----AILAGRAGQTPQGDMASLVSMGQVTPEKQQDSAPGSNRIYVGNVPFGFSSEDLK 175
T A A + D+A+ +S GS + V N+P GFS ++K
Sbjct 202 TGLFGVATYQQFATRAESEDLANPLS--------------GSKVVKVTNLPEGFSEAEVK 247
Query 176 KIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAIDTMNGFEVVGKQLKV 232
++F FGSI T P HR ++ F + A A+ TM+GF+V G L V
Sbjct 248 QVFDVFGSITTLDHFP-------HRREAYVHFFEKSHANTAVATMHGFDVGGNTLHV 297
Score = 53.1 bits (126), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 24/80 (30%), Positives = 46/80 (57%), Gaps = 1/80 (1%)
Query 157 SNRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLA 216
RIY+ ++ S E+L+ +F FG I +P +E+ ++ Y ++FA+ AA+ A
Sbjct 93 ERRIYITSLKVPVSEEELRAVFQSFGEIKQV-YIPPREDEKKRTTYALMDFATLEAAQTA 151
Query 217 IDTMNGFEVVGKQLKVNYAT 236
+ MN F G++++V +A+
Sbjct 152 VQAMNNFRFKGQRIQVAHAS 171
> cel:Y47G6A.20 rnp-6; RNP (RRM RNA binding domain) containing
family member (rnp-6); K12838 poly(U)-binding-splicing factor
PUF60
Length=749
Score = 82.4 bits (202), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 69/237 (29%), Positives = 101/237 (42%), Gaps = 62/237 (26%)
Query 1 MCRIYVGSLDYYLTELEIKSVFQAFGTVTSVDM---PKEGDRSKGFCFVEYASPEAAQMA 57
M RIYVGS+ + + E ++ F FG + S++M P G K F FVEY PEAA +A
Sbjct 101 MSRIYVGSISFEIREDMLRRAFDPFGPIKSINMSWDPATG-HHKTFAFVEYEVPEAALLA 159
Query 58 LSTMHNFVLKGRTIKVGRPTAVGAGQPQQLLARPLGGYDMNVGAAATPGMSLVSSAQAGA 117
+M+ +L GG ++ V + M L
Sbjct 160 QESMNGQML--------------------------GGRNLKVNSMMFQEMRL-------- 185
Query 118 AVATAILAGRAGQTPQGDMASLVSMGQVTP--EKQQDSAPGSNRIYVGNVPFGFSSEDLK 175
PQ +M Q P + Q A R+YV +V S DLK
Sbjct 186 --------------PQ-------NMPQAQPIIDMVQKDAKKYFRVYVSSVHPDLSETDLK 224
Query 176 KIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAIDTMNGFEVVGKQLKV 232
+F FG I+ CQL + + HRG+G++EF + + AI MN F++ G+ L+V
Sbjct 225 SVFEAFGEIVKCQLARAPTG-RGHRGFGYLEFNNLTSQSEAIAGMNMFDLGGQYLRV 280
Score = 53.1 bits (126), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 28/78 (35%), Positives = 45/78 (57%), Gaps = 5/78 (6%)
Query 158 NRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQ--QHRGYGFIEFASPAAAKL 215
+RIYVG++ F + L++ F FG I + + +P H+ + F+E+ P AA L
Sbjct 102 SRIYVGSISFEIREDMLRRAFDPFGPIKSINM---SWDPATGHHKTFAFVEYEVPEAALL 158
Query 216 AIDTMNGFEVVGKQLKVN 233
A ++MNG + G+ LKVN
Sbjct 159 AQESMNGQMLGGRNLKVN 176
Score = 49.7 bits (117), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 30/95 (31%), Positives = 54/95 (56%), Gaps = 6/95 (6%)
Query 3 RIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPK--EGDRSKGFCFVEYASPEAAQMALST 60
R+YV S+ L+E ++KSVF+AFG + + + G +GF ++E+ + + A++
Sbjct 208 RVYVSSVHPDLSETDLKSVFEAFGEIVKCQLARAPTGRGHRGFGYLEFNNLTSQSEAIAG 267
Query 61 MHNFVLKGRTIKVGR----PTAVGAGQPQQLLARP 91
M+ F L G+ ++VG+ P A+ QP + A P
Sbjct 268 MNMFDLGGQYLRVGKCVTPPDALTYLQPASVSAIP 302
> mmu:52897 Rbfox3, D11Bwg0517e, Fox-3, Hrnbp3, NeuN, Neuna60;
RNA binding protein, fox-1 homolog (C. elegans) 3; K14946 RNA
binding protein fox-1
Length=313
Score = 72.4 bits (176), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 51/163 (31%), Positives = 81/163 (49%), Gaps = 18/163 (11%)
Query 106 GMSLVSSAQA-----GAAVATAILAGRAGQTPQGDMASLVSMGQVTP----EKQQDSAPG 156
GM+L + AQ G +T +AG PQ D A+ Q+ P EKQQ
Sbjct 44 GMTLYTPAQTHPEQPGTEASTQPIAG-TQTVPQADEAAQTDNQQLHPSDPTEKQQ----- 97
Query 157 SNRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLA 216
R++V N+PF F DL+++F FG IL +++ N + +G+GF+ F + + A A
Sbjct 98 PKRLHVSNIPFRFRDPDLRQMFGQFGKILDVEII---FNERGSKGFGFVTFETSSDADRA 154
Query 217 IDTMNGFEVVGKQLKVNYATALRSAAVAGSLGVGMGAQMAPIV 259
+ +NG V G++++VN ATA G ++ P+V
Sbjct 155 REKLNGTIVEGRKIEVNNATARVMTNKKPGNPYANGWKLNPVV 197
Score = 40.4 bits (93), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 20/76 (26%), Positives = 41/76 (53%), Gaps = 0/76 (0%)
Query 3 RIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGDRSKGFCFVEYASPEAAQMALSTMH 62
R++V ++ + + +++ +F FG + V++ SKGF FV + + A A ++
Sbjct 100 RLHVSNIPFRFRDPDLRQMFGQFGKILDVEIIFNERGSKGFGFVTFETSSDADRAREKLN 159
Query 63 NFVLKGRTIKVGRPTA 78
+++GR I+V TA
Sbjct 160 GTIVEGRKIEVNNATA 175
> hsa:9904 RBM19, DKFZp586F1023, KIAA0682; RNA binding motif protein
19; K14787 multiple RNA-binding domain-containing protein
1
Length=960
Score = 72.0 bits (175), Expect = 2e-12, Method: Composition-based stats.
Identities = 60/242 (24%), Positives = 102/242 (42%), Gaps = 65/242 (26%)
Query 2 CRIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGDR-----SKGFCFVEYASPEAAQM 56
C +++ +L++ TE ++K VF GTV S + K+ ++ S GF FVEY PE AQ
Sbjct 730 CTLFIKNLNFDTTEEKLKEVFSKVGTVKSCSISKKKNKAGVLLSMGFGFVEYRKPEQAQK 789
Query 57 ALSTMHNFVLKGRTIKVGRPTAVGAGQPQQLLARPLGGYDMNVGAAATPGMSLVSSAQAG 116
AL + V+ G ++V ++ R A P ++L Q
Sbjct 790 ALKQLQGHVVDGHKLEV------------RISER-----------ATKPAVTLARKKQ-- 824
Query 117 AAVATAILAGRAGQTPQGDMASLVSMGQVTPEKQQDSAPGSNRIYVGNVPFGFSSEDLKK 176
P KQ S +I V N+PF S ++++
Sbjct 825 -----------------------------VPRKQTTS-----KILVRNIPFQAHSREIRE 850
Query 177 IFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAIDTM-NGFEVVGKQLKVNYA 235
+F FG + T +L HRG+GF++F + AK A + + + + G++L + +A
Sbjct 851 LFSTFGELKTVRLPKKMTGTGTHRGFGFVDFLTKQDAKRAFNALCHSTHLYGRRLVLEWA 910
Query 236 TA 237
+
Sbjct 911 DS 912
Score = 41.2 bits (95), Expect = 0.004, Method: Composition-based stats.
Identities = 22/73 (30%), Positives = 40/73 (54%), Gaps = 2/73 (2%)
Query 3 RIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEG--DRSKGFCFVEYASPEAAQMALST 60
R++V +L Y TE +++ +F +G ++ + P + + KGF F+ + PE A A S
Sbjct 403 RLFVRNLPYTSTEEDLEKLFSKYGPLSELHYPIDSLTKKPKGFAFITFMFPEHAVKAYSE 462
Query 61 MHNFVLKGRTIKV 73
+ V +GR + V
Sbjct 463 VDGQVFQGRMLHV 475
Score = 40.8 bits (94), Expect = 0.005, Method: Composition-based stats.
Identities = 25/93 (26%), Positives = 46/93 (49%), Gaps = 3/93 (3%)
Query 153 SAPGSNRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQ--QHRGYGFIEFASP 210
S PG +++ N+ F + E LK++F G++ +C + + G+GF+E+ P
Sbjct 726 SLPGCT-LFIKNLNFDTTEEKLKEVFSKVGTVKSCSISKKKNKAGVLLSMGFGFVEYRKP 784
Query 211 AAAKLAIDTMNGFEVVGKQLKVNYATALRSAAV 243
A+ A+ + G V G +L+V + AV
Sbjct 785 EQAQKALKQLQGHVVDGHKLEVRISERATKPAV 817
Score = 40.0 bits (92), Expect = 0.009, Method: Composition-based stats.
Identities = 28/93 (30%), Positives = 48/93 (51%), Gaps = 2/93 (2%)
Query 157 SNRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLA 216
S R++V N+P+ + EDL+K+F +G + P ++ +G+ FI F P A A
Sbjct 401 SGRLFVRNLPYTSTEEDLEKLFSKYGPLSELH-YPIDSLTKKPKGFAFITFMFPEHAVKA 459
Query 217 IDTMNGFEVVGKQLKVNYATALRSAAV-AGSLG 248
++G G+ L V +T + A+ A +LG
Sbjct 460 YSEVDGQVFQGRMLHVLPSTIKKEASEDASALG 492
Score = 37.7 bits (86), Expect = 0.054, Method: Composition-based stats.
Identities = 20/64 (31%), Positives = 34/64 (53%), Gaps = 2/64 (3%)
Query 158 NRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAI 217
+R+ V N+P G E +++F FG++ C L +++ + R +GFI F S A+ A
Sbjct 2 SRLIVKNLPNGMKEERFRQLFAAFGTLTDCSLKFTKDG--KFRKFGFIGFKSEEEAQKAQ 59
Query 218 DTMN 221
N
Sbjct 60 KHFN 63
Score = 32.0 bits (71), Expect = 2.6, Method: Composition-based stats.
Identities = 21/59 (35%), Positives = 33/59 (55%), Gaps = 3/59 (5%)
Query 1 MCRIYVGSLDYYLTELEIKSVFQAFGTVT--SVDMPKEGDRSKGFCFVEYASPEAAQMA 57
M R+ V +L + E + +F AFGT+T S+ K+G + + F F+ + S E AQ A
Sbjct 1 MSRLIVKNLPNGMKEERFRQLFAAFGTLTDCSLKFTKDG-KFRKFGFIGFKSEEEAQKA 58
> tpv:TP01_1201 hypothetical protein
Length=268
Score = 70.5 bits (171), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 62/233 (26%), Positives = 115/233 (49%), Gaps = 16/233 (6%)
Query 2 CRIYVGSLDYYLTELEIKSVFQAFGTVTSV-DMPKEGDRSKGFCFVEYASPEAAQMALST 60
+++VGS+ +E E+K +G + S+ MP + ++ G+ FV + S ++A A+
Sbjct 8 AKLFVGSIPSNTSEEELKEELSKYGQLVSLFYMPDQMKQNNGWAFVTFESNQSASNAIDA 67
Query 61 MH-NFVLKGRTIKVGRPTAVGAGQPQQLLARPLGGYDMNVGAAATPGMSLVSSAQAGAAV 119
++ + +G T+ + V A Q + +P A P S++ A
Sbjct 68 LNGKIIFQGTTVGL---EVVYASQRSMVENQP--------SVPAVPASSVLWQQFTTAEG 116
Query 120 ATAILAGRAGQTPQGDMASLVSMGQVTPEKQQDSAPGSNRIYVGNVPFGFSSEDLKKIFV 179
R GQT A L++ + PG+N ++V +VP ++ DL + F
Sbjct 117 VPYYYNVRTGQTQWEKPAELMAPARTVAGGSSFGPPGAN-LFVFHVPANWNDLDLVEHFK 175
Query 180 CFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAIDTMNGFEVVGKQLKV 232
FG++++ ++ +++ ++RG+GFI + +P +A +AI MNGF V GK LKV
Sbjct 176 HFGNVISARV--QRDSAGRNRGFGFISYDNPQSAVVAIKNMNGFSVGGKYLKV 226
> hsa:146713 RBFOX3, FLJ56884, FLJ58356, FOX-3, FOX3, HRNBP3,
NEUN; RNA binding protein, fox-1 homolog (C. elegans) 3; K14946
RNA binding protein fox-1
Length=312
Score = 70.5 bits (171), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 51/165 (30%), Positives = 82/165 (49%), Gaps = 18/165 (10%)
Query 106 GMSLVSSAQA-----GAAVATAILAGRAGQTPQGDMASLVSMGQVTP----EKQQDSAPG 156
GM+L + AQ G+ +T +AG PQ D A+ + P EKQQ
Sbjct 44 GMTLYTPAQTHPEQPGSEASTQPIAGTQ-TVPQTDEAAQTDSQPLHPSDPTEKQQ----- 97
Query 157 SNRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLA 216
R++V N+PF F DL+++F FG IL +++ N + +G+GF+ F + + A A
Sbjct 98 PKRLHVSNIPFRFRDPDLRQMFGQFGKILDVEII---FNERGSKGFGFVTFETSSDADRA 154
Query 217 IDTMNGFEVVGKQLKVNYATALRSAAVAGSLGVGMGAQMAPIVAA 261
+ +NG V G++++VN ATA G ++ P+V A
Sbjct 155 REKLNGTIVEGRKIEVNNATARVMTNKKTGNPYTNGWKLNPVVGA 199
Score = 40.8 bits (94), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 20/76 (26%), Positives = 41/76 (53%), Gaps = 0/76 (0%)
Query 3 RIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGDRSKGFCFVEYASPEAAQMALSTMH 62
R++V ++ + + +++ +F FG + V++ SKGF FV + + A A ++
Sbjct 100 RLHVSNIPFRFRDPDLRQMFGQFGKILDVEIIFNERGSKGFGFVTFETSSDADRAREKLN 159
Query 63 NFVLKGRTIKVGRPTA 78
+++GR I+V TA
Sbjct 160 GTIVEGRKIEVNNATA 175
> tgo:TGME49_121360 RNA recognition motif domain-containing protein
Length=274
Score = 69.7 bits (169), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 78/269 (28%), Positives = 118/269 (43%), Gaps = 38/269 (14%)
Query 3 RIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPK---EGDRSKGFCFVEYASPEAAQMALS 59
+I+VGS+ + +TE E + + G+VT++ K EGDR G+ FV Y + AQ A+
Sbjct 7 KIFVGSIPHTVTEEEFRKKVEEHGSVTALFYMKDQTEGDR--GWAFVTYETVYDAQNAIE 64
Query 60 TMHNFVL----KGRTIKVGRPTAVGAGQPQQLLARPLGGYDMNVGAAATPGMSLVSS--A 113
++ L G ++V +P A+A PG S+
Sbjct 65 ALNEKHLFNDSSGPALEVRFANQKPGSNSTSFQNKP---------ASAAPGPSVWQEYFT 115
Query 114 QAGAAVATAILAGRAGQTPQGDM---ASLV-----SMGQVTPEKQQDSAPGSNRIYVGNV 165
G A G D A +V S+G V + PGSN ++V N+
Sbjct 116 PEGYAYYYNTSTGVTQWEKPEDFDKPAPVVVSRPASVGIVNHAATRFGPPGSN-VFVANL 174
Query 166 PFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAIDTMNGFEV 225
P+ ++ DL + F FG+IL+ ++ E RG+GF+ F + AA AI MNGF
Sbjct 175 PYEWNDIDLIQHFQHFGNILSARIQRGTEG--NSRGFGFVSFDNSQAAVNAIRGMNGFSC 232
Query 226 VGKQLKV-------NYATALRSAAVAGSL 247
G+ L+V +Y TA A VA L
Sbjct 233 GGRYLRVFLKKGEEHYLTAAMQAQVAQYL 261
Score = 33.1 bits (74), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 16/65 (24%), Positives = 34/65 (52%), Gaps = 2/65 (3%)
Query 157 SNRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLA 216
+ +I+VG++P + E+ +K GS+ + Q + RG+ F+ + + A+ A
Sbjct 5 TKKIFVGSIPHTVTEEEFRKKVEEHGSVTALFYMKDQT--EGDRGWAFVTYETVYDAQNA 62
Query 217 IDTMN 221
I+ +N
Sbjct 63 IEALN 67
> ath:AT3G52380 CP33; CP33; RNA binding
Length=329
Score = 69.3 bits (168), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 63/244 (25%), Positives = 99/244 (40%), Gaps = 58/244 (23%)
Query 3 RIYVGSLDYYLTELEIKSVFQAFGTVTSVDM--PKEGDRSKGFCFVEYASPEAAQMALST 60
R+YVG+L Y +T E+ +F GTV V + K DRS+GF FV S E A+ A+
Sbjct 117 RLYVGNLPYTITSSELSQIFGEAGTVVDVQIVYDKVTDRSRGFGFVTMGSIEEAKEAMQM 176
Query 61 MHNFVLKGRTIKVGRPTAVGAGQPQQLLARPLGGYDMNVGAAATPGMSLVSSAQAGAAVA 120
++ + GRT+KV P G+ + + + D N +P
Sbjct 177 FNSSQIGGRTVKVNFPEVPRGGENEVMRTKI---RDNNRSYVDSP--------------- 218
Query 121 TAILAGRAGQTPQGDMASLVSMGQVTPEKQQDSAPGSNRIYVGNVPFGFSSEDLKKIFVC 180
+ AG G + +S+ LK F
Sbjct 219 HKVYAGNLG-------------------------------------WNLTSQGLKDAFGD 241
Query 181 FGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAIDTMNGFEVVGKQLKVNYATALRS 240
+L +++ + N + RG+GFI F S + A+ TMNG EV G+ L++N A+
Sbjct 242 QPGVLGAKVI-YERNTGRSRGFGFISFESAENVQSALATMNGVEVEGRALRLNLASEREK 300
Query 241 AAVA 244
V+
Sbjct 301 PTVS 304
Score = 53.1 bits (126), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 30/92 (32%), Positives = 51/92 (55%), Gaps = 2/92 (2%)
Query 149 KQQDSAPGSN-RIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEF 207
KQ A G R+YVGN+P+ +S +L +IF G+++ Q++ + + RG+GF+
Sbjct 106 KQTTQASGEEGRLYVGNLPYTITSSELSQIFGEAGTVVDVQIVYDKVT-DRSRGFGFVTM 164
Query 208 ASPAAAKLAIDTMNGFEVVGKQLKVNYATALR 239
S AK A+ N ++ G+ +KVN+ R
Sbjct 165 GSIEEAKEAMQMFNSSQIGGRTVKVNFPEVPR 196
> dre:541556 rbm39b, rnpc2l, wu:fa97g07, wu:fb09c08, zgc:112139,
zgc:113117; RNA binding motif protein 39b
Length=539
Score = 68.9 bits (167), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 62/233 (26%), Positives = 93/233 (39%), Gaps = 64/233 (27%)
Query 17 EIKSVFQAFGTVTSVDM--PKEGDRSKGFCFVEYASPEAAQMALSTMHNFVLKGRTIKVG 74
+++ F A G V V M + RSKG +VE+ + +A+ L G+ + +G
Sbjct 182 DLEDFFSAVGKVRDVRMISDRNSRRSKGIAYVEFVDSTSVPLAIG------LTGQRV-LG 234
Query 75 RPTAVGAGQPQQLLARPLGGYDMNVGAAATPGMSLVSSAQAGAAVATAILAGRAGQTPQG 134
P V A Q A+ A A A + R G P
Sbjct 235 VPIIVQASQ-----------------------------AEKNRAAAMASMLQRGGAGPM- 264
Query 135 DMASLVSMGQVTPEKQQDSAPGSNRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQE 194
R+YVG++ F + + L+ IF FG I QL+ E
Sbjct 265 ------------------------RLYVGSLHFNITEDMLRGIFEPFGKIEGIQLMMDSE 300
Query 195 NPQQHRGYGFIEFASPAAAKLAIDTMNGFEVVGKQLKVNYATALRSAAVAGSL 247
+ +GYGFI FA AK A++ +NGFE+ G+ +KV + T A+ A S
Sbjct 301 T-GRSKGYGFISFADAECAKKALEQLNGFELAGRPMKVGHVTERSDASSASSF 352
Score = 63.9 bits (154), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 31/77 (40%), Positives = 47/77 (61%), Gaps = 2/77 (2%)
Query 3 RIYVGSLDYYLTELEIKSVFQAFGTVTSVD--MPKEGDRSKGFCFVEYASPEAAQMALST 60
R+YVGSL + +TE ++ +F+ FG + + M E RSKG+ F+ +A E A+ AL
Sbjct 265 RLYVGSLHFNITEDMLRGIFEPFGKIEGIQLMMDSETGRSKGYGFISFADAECAKKALEQ 324
Query 61 MHNFVLKGRTIKVGRPT 77
++ F L GR +KVG T
Sbjct 325 LNGFELAGRPMKVGHVT 341
Score = 30.8 bits (68), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 27/100 (27%), Positives = 45/100 (45%), Gaps = 4/100 (4%)
Query 173 DLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAIDTMNGFEVVGKQLKV 232
DL+ F G + +++ S N ++ +G ++EF + LAI + G V+G + V
Sbjct 182 DLEDFFSAVGKVRDVRMI-SDRNSRRSKGIAYVEFVDSTSVPLAI-GLTGQRVLGVPIIV 239
Query 233 NYATAL--RSAAVAGSLGVGMGAQMAPIVAATATAATATM 270
+ A R+AA+A L G M V + T M
Sbjct 240 QASQAEKNRAAAMASMLQRGGAGPMRLYVGSLHFNITEDM 279
> hsa:9584 RBM39, CAPER, CAPERalpha, CC1.3, DKFZp781C0423, FLJ44170,
HCC1, RNPC2, fSAP59; RNA binding motif protein 39; K13091
RNA-binding protein 39
Length=524
Score = 68.6 bits (166), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 36/97 (37%), Positives = 56/97 (57%), Gaps = 1/97 (1%)
Query 151 QDSAPGSNRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASP 210
Q + G R+YVG++ F + + L+ IF FG I + QL+ E + +GYGFI F+
Sbjct 243 QKGSAGPMRLYVGSLHFNITEDMLRGIFEPFGRIESIQLMMDSETGRS-KGYGFITFSDS 301
Query 211 AAAKLAIDTMNGFEVVGKQLKVNYATALRSAAVAGSL 247
AK A++ +NGFE+ G+ +KV + T A+ A S
Sbjct 302 ECAKKALEQLNGFELAGRPMKVGHVTERTDASSASSF 338
Score = 63.9 bits (154), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 31/77 (40%), Positives = 48/77 (62%), Gaps = 2/77 (2%)
Query 3 RIYVGSLDYYLTELEIKSVFQAFGTVTSVD--MPKEGDRSKGFCFVEYASPEAAQMALST 60
R+YVGSL + +TE ++ +F+ FG + S+ M E RSKG+ F+ ++ E A+ AL
Sbjct 251 RLYVGSLHFNITEDMLRGIFEPFGRIESIQLMMDSETGRSKGYGFITFSDSECAKKALEQ 310
Query 61 MHNFVLKGRTIKVGRPT 77
++ F L GR +KVG T
Sbjct 311 LNGFELAGRPMKVGHVT 327
Score = 33.1 bits (74), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 27/100 (27%), Positives = 48/100 (48%), Gaps = 4/100 (4%)
Query 173 DLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAIDTMNGFEVVGKQLKV 232
DL++ F G + +++ S N ++ +G ++EF ++ LAI + G V+G + V
Sbjct 168 DLEEFFSTVGKVRDVRMI-SDRNSRRSKGIAYVEFVDVSSVPLAI-GLTGQRVLGVPIIV 225
Query 233 NYATAL--RSAAVAGSLGVGMGAQMAPIVAATATAATATM 270
+ A R+AA+A +L G M V + T M
Sbjct 226 QASQAEKNRAAAMANNLQKGSAGPMRLYVGSLHFNITEDM 265
> dre:406251 rbm39a, rnpc2, zgc:55780; RNA binding motif protein
39a; K13091 RNA-binding protein 39
Length=523
Score = 68.6 bits (166), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 36/97 (37%), Positives = 56/97 (57%), Gaps = 1/97 (1%)
Query 151 QDSAPGSNRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASP 210
Q + G R+YVG++ F + + L+ IF FG I + QL+ E + +GYGFI F+
Sbjct 239 QKGSAGPMRLYVGSLHFNITEDMLRGIFEPFGRIDSIQLMMDSET-GRSKGYGFITFSDA 297
Query 211 AAAKLAIDTMNGFEVVGKQLKVNYATALRSAAVAGSL 247
AK A++ +NGFE+ G+ +KV + T A+ A S
Sbjct 298 ECAKKALEQLNGFELAGRPMKVGHVTERTDASTASSF 334
Score = 64.3 bits (155), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 31/77 (40%), Positives = 48/77 (62%), Gaps = 2/77 (2%)
Query 3 RIYVGSLDYYLTELEIKSVFQAFGTVTSVD--MPKEGDRSKGFCFVEYASPEAAQMALST 60
R+YVGSL + +TE ++ +F+ FG + S+ M E RSKG+ F+ ++ E A+ AL
Sbjct 247 RLYVGSLHFNITEDMLRGIFEPFGRIDSIQLMMDSETGRSKGYGFITFSDAECAKKALEQ 306
Query 61 MHNFVLKGRTIKVGRPT 77
++ F L GR +KVG T
Sbjct 307 LNGFELAGRPMKVGHVT 323
Score = 33.1 bits (74), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 27/100 (27%), Positives = 48/100 (48%), Gaps = 4/100 (4%)
Query 173 DLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAIDTMNGFEVVGKQLKV 232
DL++ F G + +++ S N ++ +G +IEF + LAI ++G ++G + V
Sbjct 164 DLEEFFSAVGKVRDVRII-SDRNSRRSKGIAYIEFVDSTSVPLAI-GLSGQRLLGVPIIV 221
Query 233 NYATAL--RSAAVAGSLGVGMGAQMAPIVAATATAATATM 270
+ A R+AA+A +L G M V + T M
Sbjct 222 QASQAEKNRAAALANNLQKGSAGPMRLYVGSLHFNITEDM 261
> mmu:170791 Rbm39, 1500012C14Rik, 2310040E03Rik, B330012G18Rik,
C79248, R75070, Rnpc2, caper; RNA binding motif protein 39;
K13091 RNA-binding protein 39
Length=530
Score = 68.6 bits (166), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 36/97 (37%), Positives = 56/97 (57%), Gaps = 1/97 (1%)
Query 151 QDSAPGSNRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASP 210
Q + G R+YVG++ F + + L+ IF FG I + QL+ E + +GYGFI F+
Sbjct 243 QKGSAGPMRLYVGSLHFNITEDMLRGIFEPFGRIESIQLMMDSETGRS-KGYGFITFSDS 301
Query 211 AAAKLAIDTMNGFEVVGKQLKVNYATALRSAAVAGSL 247
AK A++ +NGFE+ G+ +KV + T A+ A S
Sbjct 302 ECAKKALEQLNGFELAGRPMKVGHVTERTDASSASSF 338
Score = 63.9 bits (154), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 31/77 (40%), Positives = 48/77 (62%), Gaps = 2/77 (2%)
Query 3 RIYVGSLDYYLTELEIKSVFQAFGTVTSVD--MPKEGDRSKGFCFVEYASPEAAQMALST 60
R+YVGSL + +TE ++ +F+ FG + S+ M E RSKG+ F+ ++ E A+ AL
Sbjct 251 RLYVGSLHFNITEDMLRGIFEPFGRIESIQLMMDSETGRSKGYGFITFSDSECAKKALEQ 310
Query 61 MHNFVLKGRTIKVGRPT 77
++ F L GR +KVG T
Sbjct 311 LNGFELAGRPMKVGHVT 327
Score = 33.1 bits (74), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 27/100 (27%), Positives = 48/100 (48%), Gaps = 4/100 (4%)
Query 173 DLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAIDTMNGFEVVGKQLKV 232
DL++ F G + +++ S N ++ +G ++EF ++ LAI + G V+G + V
Sbjct 168 DLEEFFSTVGKVRDVRMI-SDRNSRRSKGIAYVEFVDVSSVPLAI-GLTGQRVLGVPIIV 225
Query 233 NYATAL--RSAAVAGSLGVGMGAQMAPIVAATATAATATM 270
+ A R+AA+A +L G M V + T M
Sbjct 226 QASQAEKNRAAAMANNLQKGSAGPMRLYVGSLHFNITEDM 265
> hsa:54715 RBFOX1, A2BP1, FOX-1, FOX1, HRNBP1; RNA binding protein,
fox-1 homolog (C. elegans) 1; K14946 RNA binding protein
fox-1
Length=370
Score = 68.2 bits (165), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 45/162 (27%), Positives = 78/162 (48%), Gaps = 5/162 (3%)
Query 110 VSSAQAGAAVATAILAGRAGQTPQGDMASLVSMGQVTPEKQQDSAPGSNRIYVGNVPFGF 169
S Q+ A + ++G A QT D A Q P + ++ R++V N+PF F
Sbjct 71 THSEQSPADTSAQTVSGTATQT--DDAAPTDGQPQTQPSENTENKSQPKRLHVSNIPFRF 128
Query 170 SSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAIDTMNGFEVVGKQ 229
DL+++F FG IL +++ N + +G+GF+ F + A A A + ++G V G++
Sbjct 129 RDPDLRQMFGQFGKILDVEII---FNERGSKGFGFVTFENSADADRAREKLHGTVVEGRK 185
Query 230 LKVNYATALRSAAVAGSLGVGMGAQMAPIVAATATAATATMP 271
++VN ATA G ++ P+V A + +P
Sbjct 186 IEVNNATARVMTNKKTVNPYTNGWKLNPVVGAVYSPEFYAVP 227
Score = 44.3 bits (103), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 22/76 (28%), Positives = 41/76 (53%), Gaps = 0/76 (0%)
Query 3 RIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGDRSKGFCFVEYASPEAAQMALSTMH 62
R++V ++ + + +++ +F FG + V++ SKGF FV + + A A +H
Sbjct 118 RLHVSNIPFRFRDPDLRQMFGQFGKILDVEIIFNERGSKGFGFVTFENSADADRAREKLH 177
Query 63 NFVLKGRTIKVGRPTA 78
V++GR I+V TA
Sbjct 178 GTVVEGRKIEVNNATA 193
> xla:446785 rbm39, MGC80448, rnpc2; RNA binding motif protein
39; K13091 RNA-binding protein 39
Length=540
Score = 68.2 bits (165), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 36/97 (37%), Positives = 55/97 (56%), Gaps = 1/97 (1%)
Query 151 QDSAPGSNRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASP 210
Q G R+YVG++ F + + L+ IF FG I + QL+ E + +GYGFI F+
Sbjct 242 QKGTAGPMRLYVGSLHFNITEDMLRGIFEPFGRIESIQLMMDSETGRS-KGYGFITFSDS 300
Query 211 AAAKLAIDTMNGFEVVGKQLKVNYATALRSAAVAGSL 247
AK A++ +NGFE+ G+ +KV + T A+ A S
Sbjct 301 ECAKKALEQLNGFELAGRPMKVGHVTERTDASNASSF 337
Score = 64.7 bits (156), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 31/77 (40%), Positives = 48/77 (62%), Gaps = 2/77 (2%)
Query 3 RIYVGSLDYYLTELEIKSVFQAFGTVTSVD--MPKEGDRSKGFCFVEYASPEAAQMALST 60
R+YVGSL + +TE ++ +F+ FG + S+ M E RSKG+ F+ ++ E A+ AL
Sbjct 250 RLYVGSLHFNITEDMLRGIFEPFGRIESIQLMMDSETGRSKGYGFITFSDSECAKKALEQ 309
Query 61 MHNFVLKGRTIKVGRPT 77
++ F L GR +KVG T
Sbjct 310 LNGFELAGRPMKVGHVT 326
Score = 33.9 bits (76), Expect = 0.77, Method: Compositional matrix adjust.
Identities = 27/100 (27%), Positives = 49/100 (49%), Gaps = 4/100 (4%)
Query 173 DLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAIDTMNGFEVVGKQLKV 232
DL++ F G + +++ S N ++ +G ++EF ++ LAI + G +V+G + V
Sbjct 167 DLEEFFSTVGKVRDVRMI-SDRNSRRSKGIAYVEFLDQSSVPLAI-GLTGQKVLGVPIIV 224
Query 233 NYATAL--RSAAVAGSLGVGMGAQMAPIVAATATAATATM 270
+ A R+AA+A +L G M V + T M
Sbjct 225 QASQAEKNRAAALANNLQKGTAGPMRLYVGSLHFNITEDM 264
> xla:444779 MGC81970 protein
Length=512
Score = 68.2 bits (165), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 36/97 (37%), Positives = 55/97 (56%), Gaps = 1/97 (1%)
Query 151 QDSAPGSNRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASP 210
Q G R+YVG++ F + + L+ IF FG I + QL+ E + +GYGFI F+
Sbjct 216 QKGTAGPMRLYVGSLHFNITEDMLRGIFEPFGRIESIQLMMDSET-GRSKGYGFITFSDS 274
Query 211 AAAKLAIDTMNGFEVVGKQLKVNYATALRSAAVAGSL 247
AK A++ +NGFE+ G+ +KV + T A+ A S
Sbjct 275 ECAKKALEQLNGFELAGRPMKVGHVTERTDASNASSF 311
Score = 64.3 bits (155), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 31/77 (40%), Positives = 48/77 (62%), Gaps = 2/77 (2%)
Query 3 RIYVGSLDYYLTELEIKSVFQAFGTVTSVD--MPKEGDRSKGFCFVEYASPEAAQMALST 60
R+YVGSL + +TE ++ +F+ FG + S+ M E RSKG+ F+ ++ E A+ AL
Sbjct 224 RLYVGSLHFNITEDMLRGIFEPFGRIESIQLMMDSETGRSKGYGFITFSDSECAKKALEQ 283
Query 61 MHNFVLKGRTIKVGRPT 77
++ F L GR +KVG T
Sbjct 284 LNGFELAGRPMKVGHVT 300
Score = 33.9 bits (76), Expect = 0.65, Method: Compositional matrix adjust.
Identities = 27/100 (27%), Positives = 48/100 (48%), Gaps = 4/100 (4%)
Query 173 DLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAIDTMNGFEVVGKQLKV 232
DL++ F G + +++ S N ++ +G ++EF ++ LAI + G V+G + V
Sbjct 141 DLEEFFSTVGKVRDVRMI-SDRNSRRSKGIAYVEFVDQSSVPLAIG-LTGQRVLGVPIIV 198
Query 233 NYATAL--RSAAVAGSLGVGMGAQMAPIVAATATAATATM 270
+ A R+AA+A +L G M V + T M
Sbjct 199 QASQAEKNRAAALANNLQKGTAGPMRLYVGSLHFNITEDM 238
> xla:444235 rbm23, MGC80803; RNA binding motif protein 23
Length=416
Score = 67.4 bits (163), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 32/80 (40%), Positives = 50/80 (62%), Gaps = 2/80 (2%)
Query 3 RIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGD--RSKGFCFVEYASPEAAQMALST 60
R+YVGSL + +TE ++ +F+ FG + ++ + KE D RSKGF F+ + E A+ AL
Sbjct 251 RLYVGSLHFNITEEMLRGIFEPFGKIENIQLLKEPDTGRSKGFGFITFTDAECARRALEQ 310
Query 61 MHNFVLKGRTIKVGRPTAVG 80
++ F L G+ +KVG T G
Sbjct 311 LNGFELAGKPMKVGHVTGGG 330
Score = 66.6 bits (161), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 38/103 (36%), Positives = 58/103 (56%), Gaps = 6/103 (5%)
Query 156 GSNRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKL 215
G R+YVG++ F + E L+ IF FG I QLL + + + +G+GFI F A+
Sbjct 248 GPMRLYVGSLHFNITEEMLRGIFEPFGKIENIQLL-KEPDTGRSKGFGFITFTDAECARR 306
Query 216 AIDTMNGFEVVGKQLKVNYATALRSAAVAGS-----LGVGMGA 253
A++ +NGFE+ GK +KV + T A+ S G+G+GA
Sbjct 307 ALEQLNGFELAGKPMKVGHVTGGGDASFLDSDELERNGIGLGA 349
> mmu:268859 Rbfox1, A2bp, A2bp1, Hrnbp1, fox-1; RNA binding protein,
fox-1 homolog (C. elegans) 1; K14946 RNA binding protein
fox-1
Length=396
Score = 67.4 bits (163), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 43/151 (28%), Positives = 75/151 (49%), Gaps = 5/151 (3%)
Query 111 SSAQAGAAVATAILAGRAGQTPQGDMASLVSMGQVTPEKQQDSAPGSNRIYVGNVPFGFS 170
+ ++ A + ++G A QT D A Q P + +S R++V N+PF F
Sbjct 71 THSEQSADTSAQTVSGTATQT--DDAAPTDGQPQTQPSENTESKSQPKRLHVSNIPFRFR 128
Query 171 SEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAIDTMNGFEVVGKQL 230
DL+++F FG IL +++ N + +G+GF+ F + A A A + ++G V G+++
Sbjct 129 DPDLRQMFGQFGKILDVEII---FNERGSKGFGFVTFENSADADRAREKLHGTVVEGRKI 185
Query 231 KVNYATALRSAAVAGSLGVGMGAQMAPIVAA 261
+VN ATA G ++ P+V A
Sbjct 186 EVNNATARVMTNKKTVNPYTNGWKLNPVVGA 216
Score = 44.3 bits (103), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 22/76 (28%), Positives = 41/76 (53%), Gaps = 0/76 (0%)
Query 3 RIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGDRSKGFCFVEYASPEAAQMALSTMH 62
R++V ++ + + +++ +F FG + V++ SKGF FV + + A A +H
Sbjct 117 RLHVSNIPFRFRDPDLRQMFGQFGKILDVEIIFNERGSKGFGFVTFENSADADRAREKLH 176
Query 63 NFVLKGRTIKVGRPTA 78
V++GR I+V TA
Sbjct 177 GTVVEGRKIEVNNATA 192
> tgo:TGME49_120100 RNA recognition motif-containing protein
Length=1216
Score = 66.2 bits (160), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 75/287 (26%), Positives = 127/287 (44%), Gaps = 37/287 (12%)
Query 4 IYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGD-RSKGFCFVEYASPEAAQMALSTMH 62
++V + E ++ F +GT+TSV + ++ D R++G+ FV ++ E+AQ A+ M
Sbjct 247 VFVFQIPLSWAEDDLHQQFSEWGTITSVRVERKSDGRNRGYGFVCFSDAESAQRAVEGMD 306
Query 63 NFVLKGRTIKVGRPTAVGAGQPQQLLARPLGGYDMNVGAAA---TPGMSLVSSAQAGAAV 119
V +G+ +KV ++ + Q+ AR D + G A+ +PG VS V
Sbjct 307 GRVFEGKQLKV----SLKKPRQQEPSAR-----DDDRGQASHAVSPGRGGVSQKARNDEV 357
Query 120 ATAILAGRAGQTPQGDMASLVSMGQVTPEKQQDSAP-GSNR-----IYVGNVPFGFSSED 173
+ DM SL + E+ + A G R ++V +VP + E
Sbjct 358 RE-----------ENDMESLPGTTRPDCERGRKGAGVGPTRGVKCSLFVFHVPPLWGDEQ 406
Query 174 LKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAIDTMNGFEVVGKQLKVN 233
L + F +G + ++ ++ +GYGF++F +A A+ N V GK+LKV
Sbjct 407 LLQHFELYGRCASAVVVRRRDG--TSKGYGFVDFEDAESALCALQQANQAHVDGKRLKVL 464
Query 234 YAT--ALRSAAVAGSLGVGMGAQMAPIVAATATAATATMPSRSRSRS 278
T R AGS A+ AP + A+ +SR RS
Sbjct 465 LKTEPKKRPYVHAGSC---ASAERAPYPRKSMEPASGVSSLQSRERS 508
Score = 55.5 bits (132), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 30/73 (41%), Positives = 42/73 (57%), Gaps = 1/73 (1%)
Query 160 IYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAIDT 219
++V +VP +S DL++ F G I + +++ Q RG+GFI F SPAAA A+
Sbjct 729 VFVFHVPPEWSDGDLRRHFRHLGRIRAATIQRDKDD-GQSRGFGFITFGSPAAALNAVAG 787
Query 220 MNGFEVVGKQLKV 232
MNGF K LKV
Sbjct 788 MNGFHTGSKYLKV 800
Score = 38.1 bits (87), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 18/74 (24%), Positives = 41/74 (55%), Gaps = 2/74 (2%)
Query 2 CRIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGD--RSKGFCFVEYASPEAAQMALS 59
C ++V + ++ +++ F+ G + + + ++ D +S+GF F+ + SP AA A++
Sbjct 727 CTVFVFHVPPEWSDGDLRRHFRHLGRIRAATIQRDKDDGQSRGFGFITFGSPAAALNAVA 786
Query 60 TMHNFVLKGRTIKV 73
M+ F + +KV
Sbjct 787 GMNGFHTGSKYLKV 800
> dre:559412 RNA binding motif protein 9-like; K14946 RNA binding
protein fox-1
Length=318
Score = 66.2 bits (160), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 50/164 (30%), Positives = 80/164 (48%), Gaps = 19/164 (11%)
Query 82 GQPQQLLARPLGGYDMNVGAAATP--GMSLVSSAQAGAAV------ATAILAGRAGQTPQ 133
G P + A P Y G + P G++L + AQ + + TAI G
Sbjct 16 GIPAEFTALPSQDY---TGQSRVPDHGLTLYTPAQTHSDLNNTDSQTTAISTGSNTAPQT 72
Query 134 GDMASLVSMGQVTPEKQQDSAPGSNRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQ 193
D+ + + EKQQ R++V N+PF F DL+++F FG IL +++
Sbjct 73 EDVTQTDVLISESTEKQQ-----PKRLHVSNIPFRFRDPDLRQMFGQFGKILDVEII--- 124
Query 194 ENPQQHRGYGFIEFASPAAAKLAIDTMNGFEVVGKQLKVNYATA 237
N + +G+GF+ F + A A A + +NG V G++++VN ATA
Sbjct 125 FNERGSKGFGFVTFETSADADRAREKLNGTIVEGRKIEVNNATA 168
Score = 40.4 bits (93), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 20/76 (26%), Positives = 41/76 (53%), Gaps = 0/76 (0%)
Query 3 RIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGDRSKGFCFVEYASPEAAQMALSTMH 62
R++V ++ + + +++ +F FG + V++ SKGF FV + + A A ++
Sbjct 93 RLHVSNIPFRFRDPDLRQMFGQFGKILDVEIIFNERGSKGFGFVTFETSADADRAREKLN 152
Query 63 NFVLKGRTIKVGRPTA 78
+++GR I+V TA
Sbjct 153 GTIVEGRKIEVNNATA 168
> dre:606498 pabpc1a, pabpc1, wu:fb16a02, wu:fi19b08, wu:fj12d09,
wu:fj61f06, zgc:109879; poly A binding protein, cytoplasmic
1 a; K13126 polyadenylate-binding protein
Length=634
Score = 65.9 bits (159), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 66/264 (25%), Positives = 116/264 (43%), Gaps = 36/264 (13%)
Query 1 MCRIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKE--GDRSKGFCFVEYASPEAAQMAL 58
M +YVG L +TE + F G + S+ + ++ RS G+ +V + P A+ AL
Sbjct 10 MASLYVGDLHPDVTEAMLYEKFSPAGPILSIRVCRDMMTRRSLGYAYVNFQQPADAERAL 69
Query 59 STMHNFVLKGRTIKVG--------RPTAVGAGQPQQLLARPLGG-------YDM-----N 98
TM+ V+KGR +++ R + VG + + L YD N
Sbjct 70 DTMNFDVIKGRPVRIMWSQRDPSLRKSGVG-----NIFIKNLDKSIDNKALYDTFSAFGN 124
Query 99 VGAAATPGMSLVSSAQAGAAVATAILAGRAGQTPQGDMAS--LVSMGQVTPEKQQDSAPG 156
+ + S T A RA + G + + V +G+ K++++ G
Sbjct 125 ILSCKVVCDENGSKGYGFVHFETHEAAERAIEKMNGMLLNDRKVFVGRFKSRKEREAEMG 184
Query 157 SN-----RIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPA 211
+ +Y+ N E LK+IF +G L+ +++ ++ + +G+GF+ F
Sbjct 185 ARAKEFTNVYIKNFGEDMDDEKLKEIFCKYGPALSIRVM--TDDSGKSKGFGFVSFERHE 242
Query 212 AAKLAIDTMNGFEVVGKQLKVNYA 235
A+ A+D MNG E+ GKQ+ V A
Sbjct 243 DAQRAVDEMNGKEMNGKQVYVGRA 266
Score = 53.5 bits (127), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 27/70 (38%), Positives = 43/70 (61%), Gaps = 0/70 (0%)
Query 4 IYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGDRSKGFCFVEYASPEAAQMALSTMHN 63
+YV +LD L + ++ F FGT+TS + EG RSKGF FV ++SPE A A++ M+
Sbjct 296 LYVKNLDDGLDDERLRKEFSPFGTITSAKVMMEGGRSKGFGFVCFSSPEEATKAVTEMNG 355
Query 64 FVLKGRTIKV 73
++ + + V
Sbjct 356 RIVATKPLYV 365
Score = 50.4 bits (119), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 59/258 (22%), Positives = 95/258 (36%), Gaps = 60/258 (23%)
Query 4 IYVGSLDYYLTELEIKSVFQAFGTVTSVD-MPKEGDRSKGFCFVEYASPEAAQMALSTMH 62
+Y+ + + + ++K +F +G S+ M + +SKGF FV + E AQ A+ M+
Sbjct 193 VYIKNFGEDMDDEKLKEIFCKYGPALSIRVMTDDSGKSKGFGFVSFERHEDAQRAVDEMN 252
Query 63 NFVLKGRTIKVGRPTAVGAGQPQQLLARPLGGYDMNVGAAATPGMSLVSSAQAGAAVATA 122
+ + G + VG A G
Sbjct 253 --------------------------GKEMNGKQVYVGRAQKKGERQTE----------- 275
Query 123 ILAGRAGQTPQGDMASLVSMGQVTPEKQQDSAPGSNRIYVGNVPFGFSSEDLKKIFVCFG 182
L + Q Q M + +YV N+ G E L+K F FG
Sbjct 276 -LKRKFEQMKQDRMTRYQGVN----------------LYVKNLDDGLDDERLRKEFSPFG 318
Query 183 SILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAIDTMNGFEVVGKQLKVNYATALRSAA 242
+I + +++ + +G+GF+ F+SP A A+ MNG V K L V A A R
Sbjct 319 TITSAKVM---MEGGRSKGFGFVCFSSPEEATKAVTEMNGRIVATKPLYV--ALAQRKEE 373
Query 243 VAGSLGVGMGAQMAPIVA 260
L +MA + A
Sbjct 374 RQAHLTSQYMQRMASVRA 391
> hsa:55147 RBM23, CAPERbeta, FLJ10482, MGC4458, RNPC4; RNA binding
motif protein 23
Length=439
Score = 65.5 bits (158), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 34/102 (33%), Positives = 61/102 (59%), Gaps = 4/102 (3%)
Query 3 RIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGD--RSKGFCFVEYASPEAAQMALST 60
R+YVGSL + +TE ++ +F+ FG + ++ + K+ D RSKG+ F+ ++ E A+ AL
Sbjct 264 RLYVGSLHFNITEDMLRGIFEPFGKIDNIVLMKDSDTGRSKGYGFITFSDSECARRALEQ 323
Query 61 MHNFVLKGRTIKVGRPTAVGAGQPQQLLARPLGGYDMNVGAA 102
++ F L GR ++VG T G + P G ++++G+A
Sbjct 324 LNGFELAGRPMRVGHVTERLDGGTD--ITFPDGDQELDLGSA 363
Score = 60.1 bits (144), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 48/81 (59%), Gaps = 1/81 (1%)
Query 156 GSNRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKL 215
G R+YVG++ F + + L+ IF FG I L+ + + +GYGFI F+ A+
Sbjct 261 GPMRLYVGSLHFNITEDMLRGIFEPFGKIDNIVLMKDSDTGRS-KGYGFITFSDSECARR 319
Query 216 AIDTMNGFEVVGKQLKVNYAT 236
A++ +NGFE+ G+ ++V + T
Sbjct 320 ALEQLNGFELAGRPMRVGHVT 340
Score = 32.3 bits (72), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 27/100 (27%), Positives = 46/100 (46%), Gaps = 4/100 (4%)
Query 173 DLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAIDTMNGFEVVGKQLKV 232
DL+ F G + +++ S N ++ +G ++EF + LAI + G ++G + V
Sbjct 181 DLEDFFSAVGKVRDVRII-SDRNSRRSKGIAYVEFCEIQSVPLAI-GLTGQRLLGVPIIV 238
Query 233 NYATAL--RSAAVAGSLGVGMGAQMAPIVAATATAATATM 270
+ A R AA+A +L G G M V + T M
Sbjct 239 QASQAEKNRLAAMANNLQKGNGGPMRLYVGSLHFNITEDM 278
> ath:AT4G36960 RNA recognition motif (RRM)-containing protein
Length=379
Score = 65.1 bits (157), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 61/248 (24%), Positives = 105/248 (42%), Gaps = 24/248 (9%)
Query 1 MCRIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGD--RSKGFCFVEYASPEAAQMAL 58
+ RI+V + ++E + +S F+ +G +T + MPK+ + + +G F+ ++S ++ + +
Sbjct 90 VTRIFVARIPSSVSESDFRSHFERYGEITDLYMPKDYNSKQHRGIGFITFSSADSVEDLM 149
Query 59 STMHNFVLKGRTIKVGRPTAVGAGQPQQ-----LLARPLGGYDMNVGAAATPGMSLVSSA 113
H+ L G T+ V R T P + ++RP V A G A
Sbjct 150 EDTHD--LGGTTVAVDRATPKEDDHPPRPPPVARMSRP------PVAIAGGFGAPGGYGA 201
Query 114 QAGAAVATAILAGRAGQTPQGDMASLVSMGQVTPEKQQDSAPGSNRIYVGNVPFGFSSED 173
A A T + A+ G+ T N+I+VG +P S +D
Sbjct 202 YDAYISAATRYAALGAPTLYDNPATFYGRGEPTTRGI------GNKIFVGRLPQEASVDD 255
Query 174 LKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAIDTMNGFEVVGKQLKVN 233
L+ F FG I +P HRG+GF+ FA A E+ G+++ ++
Sbjct 256 LRDYFGRFGHIQDA-YIPKDPKRSGHRGFGFVTFAENGVADRV--ARRSHEICGQEVAID 312
Query 234 YATALRSA 241
AT L A
Sbjct 313 SATPLDEA 320
Score = 45.1 bits (105), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 27/89 (30%), Positives = 45/89 (50%), Gaps = 3/89 (3%)
Query 148 EKQQDSAPGSNRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEF 207
E+ + A RI+V +P S D + F +G I T +P N +QHRG GFI F
Sbjct 81 EEMRQPAKKVTRIFVARIPSSVSESDFRSHFERYGEI-TDLYMPKDYNSKQHRGIGFITF 139
Query 208 ASPAAAKLAIDTMNGFEVVGKQLKVNYAT 236
+S + + ++ + ++ G + V+ AT
Sbjct 140 SSADSVEDLMEDTH--DLGGTTVAVDRAT 166
Score = 35.0 bits (79), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 40/79 (50%), Gaps = 5/79 (6%)
Query 159 RIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAID 218
++ V +P+ S+ LK FG + C ++ + + RG+G++ FAS AK A
Sbjct 4 KLVVLGIPWDIDSDGLKDYMSKFGDLEDCIVMKDRST-GRSRGFGYVTFASAEDAKNA-- 60
Query 219 TMNGFEVVGKQ-LKVNYAT 236
+ G +G + L+V AT
Sbjct 61 -LKGEHFLGNRILEVKVAT 78
> dre:100330510 Fox-1 homolog C-like
Length=450
Score = 64.7 bits (156), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 40/116 (34%), Positives = 65/116 (56%), Gaps = 12/116 (10%)
Query 148 EKQQDSAPGSNRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEF 207
EKQQ R++V N+PF F DL+++F FG IL +++ N + +G+GF+ F
Sbjct 135 EKQQ-----PKRLHVSNIPFRFRDPDLRQMFGQFGKILDVEII---FNERGSKGFGFVTF 186
Query 208 ASPAAAKLAIDTMNGFEVVGKQLKVNYATA--LRSAAVAGSLGVGMGAQMAPIVAA 261
+ A A A + +NG V G++++VN ATA + + VA G ++ P+V A
Sbjct 187 ETSADADRAREKLNGTIVEGRKIEVNNATARVMTNKKVANPYTNGW--KLNPVVGA 240
Score = 41.2 bits (95), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 37/133 (27%), Positives = 63/133 (47%), Gaps = 10/133 (7%)
Query 3 RIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGDRSKGFCFVEYASPEAAQMALSTMH 62
R++V ++ + + +++ +F FG + V++ SKGF FV + + A A ++
Sbjct 141 RLHVSNIPFRFRDPDLRQMFGQFGKILDVEIIFNERGSKGFGFVTFETSADADRAREKLN 200
Query 63 NFVLKGRTIKVGRPTAVGAGQPQQLLARPL-GGYDMN--VGAAATPGMSLVSS---AQAG 116
+++GR I+V TA + +A P G+ +N VGA P V+ G
Sbjct 201 GTIVEGRKIEVNNATA--RVMTNKKVANPYTNGWKLNPVVGAVYGPEFYAVTGFPYPTTG 258
Query 117 AAVAT--AILAGR 127
A VA A L GR
Sbjct 259 ATVAYRGAHLRGR 271
> ath:AT1G71770 PAB5; PAB5 (POLY(A)-BINDING PROTEIN 5); RNA binding
/ poly(A) binding / translation initiation factor
Length=668
Score = 64.7 bits (156), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 59/252 (23%), Positives = 111/252 (44%), Gaps = 38/252 (15%)
Query 4 IYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKE-GDRSKGFCFVEYASPEAAQMALSTMH 62
+YVG LD + E + +F V ++ + ++ RS G+ +V +A+PE A A+ +++
Sbjct 47 LYVGDLDPSVNESHLLDLFNQVAPVHNLRVCRDLTHRSLGYAYVNFANPEDASRAMESLN 106
Query 63 NFVLKGRTIKVGRPTAVGAGQPQQLLARPLGGYDMNVGAAATPGMSLVSSAQAGAAVATA 122
++ R I++ + P L+ + N+ A+ + + G ++
Sbjct 107 YAPIRDRPIRI----MLSNRDPSTRLSGKGNVFIKNLDASIDNKALYETFSSFGTILSCK 162
Query 123 I---LAGRAG----------QTPQGDMASLVSMGQVTPEKQ---------QDSA------ 154
+ + GR+ +T Q + L G + +KQ QD A
Sbjct 163 VAMDVVGRSKGYGFVQFEKEETAQAAIDKL--NGMLLNDKQVFVGHFVRRQDRARSESGA 220
Query 155 -PGSNRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAA 213
P +YV N+P + ++LKK F +G I + ++ Q R +GF+ F SP AA
Sbjct 221 VPSFTNVYVKNLPKEITDDELKKTFGKYGDISSAVVMKDQSG--NSRSFGFVNFVSPEAA 278
Query 214 KLAIDTMNGFEV 225
+A++ MNG +
Sbjct 279 AVAVEKMNGISL 290
Score = 48.1 bits (113), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 26/73 (35%), Positives = 41/73 (56%), Gaps = 1/73 (1%)
Query 4 IYVGSLDYYLTELEIKSVFQAFGTVTS-VDMPKEGDRSKGFCFVEYASPEAAQMALSTMH 62
+YV +L +T+ E+K F +G ++S V M + S+ F FV + SPEAA +A+ M+
Sbjct 227 VYVKNLPKEITDDELKKTFGKYGDISSAVVMKDQSGNSRSFGFVNFVSPEAAAVAVEKMN 286
Query 63 NFVLKGRTIKVGR 75
L + VGR
Sbjct 287 GISLGEDVLYVGR 299
Score = 47.8 bits (112), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 23/74 (31%), Positives = 49/74 (66%), Gaps = 4/74 (5%)
Query 156 GSNRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKL 215
GSN +Y+ N+ + E LK++F +G++ +C+++ + + RG+GF+ +++P A L
Sbjct 327 GSN-LYLKNLDDSVNDEKLKEMFSEYGNVTSCKVMMNSQG--LSRGFGFVAYSNPEEALL 383
Query 216 AIDTMNGFEVVGKQ 229
A+ MNG +++G++
Sbjct 384 AMKEMNG-KMIGRK 396
Score = 47.8 bits (112), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 45/79 (56%), Gaps = 2/79 (2%)
Query 156 GSNRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKL 215
G +++ N+ ++ L + F FG+IL+C++ + + + +GYGF++F A+
Sbjct 130 GKGNVFIKNLDASIDNKALYETFSSFGTILSCKV--AMDVVGRSKGYGFVQFEKEETAQA 187
Query 216 AIDTMNGFEVVGKQLKVNY 234
AID +NG + KQ+ V +
Sbjct 188 AIDKLNGMLLNDKQVFVGH 206
Score = 43.5 bits (101), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 23/61 (37%), Positives = 39/61 (63%), Gaps = 3/61 (4%)
Query 4 IYVGSLDYYLTELEIKSVFQAFGTVTS--VDMPKEGDRSKGFCFVEYASPEAAQMALSTM 61
+Y+ +LD + + ++K +F +G VTS V M +G S+GF FV Y++PE A +A+ M
Sbjct 330 LYLKNLDDSVNDEKLKEMFSEYGNVTSCKVMMNSQG-LSRGFGFVAYSNPEEALLAMKEM 388
Query 62 H 62
+
Sbjct 389 N 389
> ath:AT5G19960 RNA recognition motif (RRM)-containing protein
Length=337
Score = 64.7 bits (156), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 38/127 (29%), Positives = 64/127 (50%), Gaps = 8/127 (6%)
Query 158 NRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRG--YGFIEFASPAAAKL 215
N +YVG +P+ + E ++++F +GS+LT +++ N + RG YGF+ F++ +A
Sbjct 7 NSVYVGGLPYDITEEAVRRVFSIYGSVLTVKIV----NDRSVRGKCYGFVTFSNRRSADD 62
Query 216 AIDTMNGFEVVGKQLKVNYATAL--RSAAVAGSLGVGMGAQMAPIVAATATAATATMPSR 273
AI+ M+G + G+ ++VN T R G L G +P + R
Sbjct 63 AIEDMDGKSIGGRAVRVNDVTTRGGRMNPGPGRLQPHGGWDRSPDRRSDGNYERDRYSDR 122
Query 274 SRSRSRS 280
SR R RS
Sbjct 123 SRERDRS 129
Score = 46.2 bits (108), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 32/97 (32%), Positives = 51/97 (52%), Gaps = 6/97 (6%)
Query 4 IYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGDRS-KGFC--FVEYASPEAAQMALST 60
+YVG L Y +TE ++ VF +G+V +V + DRS +G C FV +++ +A A+
Sbjct 9 VYVGGLPYDITEEAVRRVFSIYGSVLTVKIV--NDRSVRGKCYGFVTFSNRRSADDAIED 66
Query 61 MHNFVLKGRTIKVGRPTAVGAG-QPQQLLARPLGGYD 96
M + GR ++V T G P +P GG+D
Sbjct 67 MDGKSIGGRAVRVNDVTTRGGRMNPGPGRLQPHGGWD 103
> bbo:BBOV_IV006520 23.m06153; RNA binding motif containing protein
Length=278
Score = 64.3 bits (155), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 62/261 (23%), Positives = 113/261 (43%), Gaps = 15/261 (5%)
Query 3 RIYVGSLDYYLTELEIKSVFQAFGTVTSV-DMPKEGDRSKGFCFVEYASPEAAQMALSTM 61
+I++G + + E +++ FG + S+ MP G S G+ F Y + A++ +
Sbjct 14 KIFIGCIPGDVLEDQLRWELAKFGNLKSIFYMPDLGHESMGWAFATYNDHYSGIAAVNAI 73
Query 62 HNFVLKGRTIKVGRPTAVGAGQPQQLLARPLGGYDMNVGAAATPGMSLVSSAQAGAAVAT 121
++ + + + V Q+ L G N AT ++
Sbjct 74 NDELFFAGSNVPCKAQFVTPRSAQEYTK--LKGNTRNWPVTATTEWQQFTNTDG----ML 127
Query 122 AILAGRAGQTPQGDMASLVSMGQVTPEKQQDSA----PGSNRIYVGNVPFGFSSEDLKKI 177
R G+T L + + P++ +SA PGSN ++V ++P +S DL
Sbjct 128 YYYNKRTGETQWQRPYELPNFS-LLPKRVTNSASYGPPGSN-LFVFHLPPEWSDSDLLLH 185
Query 178 FVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAIDTMNGFEVVGKQLKVNYATA 237
F FG+I++ ++ + ++RGYGF+ + +P +A AI MNG+ V GK LKV
Sbjct 186 FQSFGTIVSARV--QLDTVGRNRGYGFVSYDNPTSALTAIKNMNGYSVCGKYLKVQLKRG 243
Query 238 LRSAAVAGSLGVGMGAQMAPI 258
A+ L + + P+
Sbjct 244 EEQMALTNELEAPFSSDIRPV 264
> mmu:18458 Pabpc1, PABP, Pabp1, PabpI, Pabpl1, ePAB; poly(A)
binding protein, cytoplasmic 1; K13126 polyadenylate-binding
protein
Length=636
Score = 63.9 bits (154), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 65/264 (24%), Positives = 115/264 (43%), Gaps = 36/264 (13%)
Query 1 MCRIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKE--GDRSKGFCFVEYASPEAAQMAL 58
M +YVG L +TE + F G + S+ + ++ RS G+ +V + P A+ AL
Sbjct 10 MASLYVGDLHPDVTEAMLYEKFSPAGPILSIRVCRDMITRRSLGYAYVNFQQPADAERAL 69
Query 59 STMHNFVLKGRTIKVG--------RPTAVGAGQPQQLLARPLGG-------YDM-----N 98
TM+ V+KG+ +++ R + VG + + L YD N
Sbjct 70 DTMNFDVIKGKPVRIMWSQRDPSLRKSGVG-----NIFIKNLDKSIDNKALYDTFSAFGN 124
Query 99 VGAAATPGMSLVSSAQAGAAVATAILAGRAGQTPQGDMAS--LVSMGQVTPEKQQDSAPG 156
+ + S T A RA + G + + V +G+ K++++ G
Sbjct 125 ILSCKVVCDENGSKGYGFVHFETQEAAERAIEKMNGMLLNDRKVFVGRFKSRKEREAELG 184
Query 157 SN-----RIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPA 211
+ +Y+ N E LK++F FG L+ +++ + + +G+GF+ F
Sbjct 185 ARAKEFTNVYIKNFGEDMDDERLKELFGKFGPALSVKVMTDESG--KSKGFGFVSFERHE 242
Query 212 AAKLAIDTMNGFEVVGKQLKVNYA 235
A+ A+D MNG E+ GKQ+ V A
Sbjct 243 DAQKAVDEMNGKELNGKQIYVGRA 266
Score = 52.8 bits (125), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 26/70 (37%), Positives = 43/70 (61%), Gaps = 0/70 (0%)
Query 4 IYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGDRSKGFCFVEYASPEAAQMALSTMHN 63
+YV +LD + + ++ F FGT+TS + EG RSKGF FV ++SPE A A++ M+
Sbjct 296 LYVKNLDDGIDDERLRKEFSPFGTITSAKVMMEGGRSKGFGFVCFSSPEEATKAVTEMNG 355
Query 64 FVLKGRTIKV 73
++ + + V
Sbjct 356 RIVATKPLYV 365
Score = 45.1 bits (105), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 26/73 (35%), Positives = 40/73 (54%), Gaps = 3/73 (4%)
Query 160 IYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAIDT 219
+YV N+ G E L+K F FG+I + +++ + +G+GF+ F+SP A A+
Sbjct 296 LYVKNLDDGIDDERLRKEFSPFGTITSAKVM---MEGGRSKGFGFVCFSSPEEATKAVTE 352
Query 220 MNGFEVVGKQLKV 232
MNG V K L V
Sbjct 353 MNGRIVATKPLYV 365
> xla:432174 rbfox2-b, MGC78805, fox-2, fox2, fxh, hnrbp2, hrnbp2,
rbfox2, rbm9, rbm9-b, rbm9b, rta, xrbm9; RNA binding protein,
fox-1 homolog (C. elegans) 2; K14946 RNA binding protein
fox-1
Length=411
Score = 63.5 bits (153), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 33/103 (32%), Positives = 58/103 (56%), Gaps = 3/103 (2%)
Query 159 RIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAID 218
R++V N+PF F DL+++F FG IL +++ N + +G+GF+ F + A A A +
Sbjct 174 RLHVSNIPFRFRDPDLRQMFGQFGKILDVEII---FNERGSKGFGFVTFETSADADRARE 230
Query 219 TMNGFEVVGKQLKVNYATALRSAAVAGSLGVGMGAQMAPIVAA 261
++ V G++++VN ATA G G +++P+V A
Sbjct 231 KLHSTVVEGRKIEVNNATARVMTNKKSVTPYGNGWKLSPVVGA 273
Score = 45.4 bits (106), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 22/76 (28%), Positives = 42/76 (55%), Gaps = 0/76 (0%)
Query 3 RIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGDRSKGFCFVEYASPEAAQMALSTMH 62
R++V ++ + + +++ +F FG + V++ SKGF FV + + A A +H
Sbjct 174 RLHVSNIPFRFRDPDLRQMFGQFGKILDVEIIFNERGSKGFGFVTFETSADADRAREKLH 233
Query 63 NFVLKGRTIKVGRPTA 78
+ V++GR I+V TA
Sbjct 234 STVVEGRKIEVNNATA 249
> hsa:26986 PABPC1, PAB1, PABP, PABP1, PABPC2, PABPL1; poly(A)
binding protein, cytoplasmic 1; K13126 polyadenylate-binding
protein
Length=636
Score = 63.2 bits (152), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 65/264 (24%), Positives = 114/264 (43%), Gaps = 36/264 (13%)
Query 1 MCRIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKE--GDRSKGFCFVEYASPEAAQMAL 58
M +YVG L +TE + F G + S+ + ++ RS G+ +V + P A+ AL
Sbjct 10 MASLYVGDLHPDVTEAMLYEKFSPAGPILSIRVCRDMITRRSLGYAYVNFQQPADAERAL 69
Query 59 STMHNFVLKGRTIKVG--------RPTAVGAGQPQQLLARPLGG-------YDM-----N 98
TM+ V+KG+ +++ R + VG + + L YD N
Sbjct 70 DTMNFDVIKGKPVRIMWSQRDPSLRKSGVG-----NIFIKNLDKSIDNKALYDTFSAFGN 124
Query 99 VGAAATPGMSLVSSAQAGAAVATAILAGRAGQTPQGDMAS--LVSMGQVTPEKQQDSAPG 156
+ + S T A RA + G + + V +G+ K++++ G
Sbjct 125 ILSCKVVCDENGSKGYGFVHFETQEAAERAIEKMNGMLLNDRKVFVGRFKSRKEREAELG 184
Query 157 SN-----RIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPA 211
+ +Y+ N E LK +F FG L+ +++ + + +G+GF+ F
Sbjct 185 ARAKEFTNVYIKNFGEDMDDERLKDLFGKFGPALSVKVMTDESG--KSKGFGFVSFERHE 242
Query 212 AAKLAIDTMNGFEVVGKQLKVNYA 235
A+ A+D MNG E+ GKQ+ V A
Sbjct 243 DAQKAVDEMNGKELNGKQIYVGRA 266
Score = 52.8 bits (125), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 26/70 (37%), Positives = 43/70 (61%), Gaps = 0/70 (0%)
Query 4 IYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGDRSKGFCFVEYASPEAAQMALSTMHN 63
+YV +LD + + ++ F FGT+TS + EG RSKGF FV ++SPE A A++ M+
Sbjct 296 LYVKNLDDGIDDERLRKEFSPFGTITSAKVMMEGGRSKGFGFVCFSSPEEATKAVTEMNG 355
Query 64 FVLKGRTIKV 73
++ + + V
Sbjct 356 RIVATKPLYV 365
Score = 45.1 bits (105), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 26/73 (35%), Positives = 40/73 (54%), Gaps = 3/73 (4%)
Query 160 IYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAIDT 219
+YV N+ G E L+K F FG+I + +++ + +G+GF+ F+SP A A+
Sbjct 296 LYVKNLDDGIDDERLRKEFSPFGTITSAKVM---MEGGRSKGFGFVCFSSPEEATKAVTE 352
Query 220 MNGFEVVGKQLKV 232
MNG V K L V
Sbjct 353 MNGRIVATKPLYV 365
> xla:100127308 rbfox2-a, fox-2, fox2, fxh, hnrbp2, hrnbp2, rbm9,
rbm9-a, rbm9a, rta, xrbm9; RNA binding protein, fox-1 homolog
(C. elegans) 2; K14946 RNA binding protein fox-1
Length=381
Score = 62.8 bits (151), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 33/103 (32%), Positives = 58/103 (56%), Gaps = 3/103 (2%)
Query 159 RIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAID 218
R++V N+PF F DL+++F FG IL +++ N + +G+GF+ F + A A A +
Sbjct 111 RLHVSNIPFRFRDPDLRQMFGQFGKILDVEII---FNERGSKGFGFVTFETSADADQARE 167
Query 219 TMNGFEVVGKQLKVNYATALRSAAVAGSLGVGMGAQMAPIVAA 261
++ V G++++VN ATA G G +++P+V A
Sbjct 168 KLHSTVVEGRKIEVNNATARVMTNKKSVTPYGNGWKLSPVVGA 210
Score = 45.4 bits (106), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 22/76 (28%), Positives = 42/76 (55%), Gaps = 0/76 (0%)
Query 3 RIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGDRSKGFCFVEYASPEAAQMALSTMH 62
R++V ++ + + +++ +F FG + V++ SKGF FV + + A A +H
Sbjct 111 RLHVSNIPFRFRDPDLRQMFGQFGKILDVEIIFNERGSKGFGFVTFETSADADQAREKLH 170
Query 63 NFVLKGRTIKVGRPTA 78
+ V++GR I+V TA
Sbjct 171 STVVEGRKIEVNNATA 186
> bbo:BBOV_I003530 19.m02179; RNA recognition motif. (a.k.a. RRM,
RBD, or RNP) domain containing protein
Length=420
Score = 62.8 bits (151), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 37/104 (35%), Positives = 61/104 (58%), Gaps = 4/104 (3%)
Query 155 PGSNRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAK 214
PG+N +++ ++P ++ DL F FG IL+ ++ S + +H+GY F+ + +P +A
Sbjct 285 PGAN-LFIFHIPNEWTHHDLVHTFSQFGKILSSRIA-SDRSTGRHKGYAFVSYDTPESAA 342
Query 215 LAIDTMNGFEVVGKQLKVNYATALRSAA-VAGSL-GVGMGAQMA 256
AI +NGF V+GK+LKV S VA S+ G M A++A
Sbjct 343 QAIQHLNGFTVLGKRLKVTIKKGDESTVPVASSVPGASMQARLA 386
Score = 39.7 bits (91), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 19/72 (26%), Positives = 37/72 (51%), Gaps = 2/72 (2%)
Query 4 IYVGSLDYYLTELEIKSVFQAFGTVTS--VDMPKEGDRSKGFCFVEYASPEAAQMALSTM 61
+++ + T ++ F FG + S + + R KG+ FV Y +PE+A A+ +
Sbjct 289 LFIFHIPNEWTHHDLVHTFSQFGKILSSRIASDRSTGRHKGYAFVSYDTPESAAQAIQHL 348
Query 62 HNFVLKGRTIKV 73
+ F + G+ +KV
Sbjct 349 NGFTVLGKRLKV 360
> dre:449554 rbfox1, a2bp1, fox1, zgc:103635; RNA binding protein,
fox-1 homolog (C. elegans) 1; K14946 RNA binding protein
fox-1
Length=373
Score = 62.4 bits (150), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 35/113 (30%), Positives = 60/113 (53%), Gaps = 3/113 (2%)
Query 159 RIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAID 218
R++V N+PF F DL+++F FG IL +++ N + +G+GF+ F S A A A +
Sbjct 120 RLHVSNIPFRFRDPDLRQMFGQFGKILDVEII---FNERGSKGFGFVTFESSADADRARE 176
Query 219 TMNGFEVVGKQLKVNYATALRSAAVAGSLGVGMGAQMAPIVAATATAATATMP 271
++G V G++++VN ATA G ++ P+V A + +P
Sbjct 177 KLHGTVVEGRKIEVNNATARVMTNKKTVNPYANGWKLNPVVGAVYSPEFYAVP 229
Score = 45.1 bits (105), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 41/76 (53%), Gaps = 0/76 (0%)
Query 3 RIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGDRSKGFCFVEYASPEAAQMALSTMH 62
R++V ++ + + +++ +F FG + V++ SKGF FV + S A A +H
Sbjct 120 RLHVSNIPFRFRDPDLRQMFGQFGKILDVEIIFNERGSKGFGFVTFESSADADRAREKLH 179
Query 63 NFVLKGRTIKVGRPTA 78
V++GR I+V TA
Sbjct 180 GTVVEGRKIEVNNATA 195
> tpv:TP01_1032 hypothetical protein; K14838 nucleolar protein
15
Length=195
Score = 62.4 bits (150), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 30/77 (38%), Positives = 47/77 (61%), Gaps = 1/77 (1%)
Query 157 SNRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLA 216
+N IYVGN+P + E LK F FG ++ +L S++ + RGY F++F AK+A
Sbjct 12 NNVIYVGNLPKQLTEEQLKTYFNQFGDVIKIRLFKSRK-TNRSRGYAFVQFEDHEIAKIA 70
Query 217 IDTMNGFEVVGKQLKVN 233
+TM+ + + GK LKV+
Sbjct 71 AETMDKYLIDGKSLKVH 87
Score = 57.8 bits (138), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 27/72 (37%), Positives = 47/72 (65%), Gaps = 2/72 (2%)
Query 4 IYVGSLDYYLTELEIKSVFQAFGTVTSVDMPK--EGDRSKGFCFVEYASPEAAQMALSTM 61
IYVG+L LTE ++K+ F FG V + + K + +RS+G+ FV++ E A++A TM
Sbjct 15 IYVGNLPKQLTEEQLKTYFNQFGDVIKIRLFKSRKTNRSRGYAFVQFEDHEIAKIAAETM 74
Query 62 HNFVLKGRTIKV 73
+++ G+++KV
Sbjct 75 DKYLIDGKSLKV 86
> hsa:23543 RBFOX2, FOX2, Fox-2, HNRBP2, HRNBP2, RBM9, RTA, dJ106I20.3,
fxh; RNA binding protein, fox-1 homolog (C. elegans)
2; K14946 RNA binding protein fox-1
Length=380
Score = 62.0 bits (149), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 37/121 (30%), Positives = 64/121 (52%), Gaps = 5/121 (4%)
Query 141 SMGQVTPEKQQDSAPGSNRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHR 200
S Q + + S P R++V N+PF F DL+++F FG IL +++ N + +
Sbjct 96 SQTQSSENSESKSTP--KRLHVSNIPFRFRDPDLRQMFGQFGKILDVEII---FNERGSK 150
Query 201 GYGFIEFASPAAAKLAIDTMNGFEVVGKQLKVNYATALRSAAVAGSLGVGMGAQMAPIVA 260
G+GF+ F + A A A + ++G V G++++VN ATA G +++P+V
Sbjct 151 GFGFVTFENSADADRAREKLHGTVVEGRKIEVNNATARVMTNKKMVTPYANGWKLSPVVG 210
Query 261 A 261
A
Sbjct 211 A 211
Score = 47.8 bits (112), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 36/132 (27%), Positives = 66/132 (50%), Gaps = 6/132 (4%)
Query 3 RIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGDRSKGFCFVEYASPEAAQMALSTMH 62
R++V ++ + + +++ +F FG + V++ SKGF FV + + A A +H
Sbjct 112 RLHVSNIPFRFRDPDLRQMFGQFGKILDVEIIFNERGSKGFGFVTFENSADADRAREKLH 171
Query 63 NFVLKGRTIKVGRPTAVGAGQPQQLLARPLGGYDMN--VGAAATPGMSLVSSAQAGAAV- 119
V++GR I+V TA ++++ G+ ++ VGA P + SS QA ++
Sbjct 172 GTVVEGRKIEVNNATARVMTN-KKMVTPYANGWKLSPVVGAVYGPELYAASSFQADVSLG 230
Query 120 --ATAILAGRAG 129
A L+GR G
Sbjct 231 NDAAVPLSGRGG 242
> mmu:67543 Pabpc6, 4932702K14Rik, AI428050, MGC132900, Pabpc3;
poly(A) binding protein, cytoplasmic 6; K13126 polyadenylate-binding
protein
Length=643
Score = 62.0 bits (149), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 64/233 (27%), Positives = 97/233 (41%), Gaps = 48/233 (20%)
Query 4 IYVGSLDYYLTELEIKSVFQAFGTVTSVD-MPKEGDRSKGFCFVEYASPEAAQMALSTMH 62
+Y+ +L + + ++ +F FG SV M E +SKGF FV + E A+ A+ M+
Sbjct 193 VYIKNLGEDMDDERLQDLFGRFGPALSVKVMTDESGKSKGFGFVSFERHEDARKAVEEMN 252
Query 63 NFVLKGRTIKVGRPTAVGAGQPQQLLARPLGGYDMNVGAAATPGMSLVSSAQAGAAVATA 122
L G+ I VGR AQ T
Sbjct 253 GKDLNGKQIYVGR-------------------------------------AQKKVERQTE 275
Query 123 ILAGRAGQTPQGDMASLVSMGQVTPEKQQDSAPGSNRIYVGNVPFGFSSEDLKKIFVCFG 182
L + GQ Q D + + P+ + G N +YV N+ G E L+K F FG
Sbjct 276 -LKHKFGQMKQ-DKHKI----ERVPQDRSVRCKGVN-LYVKNLDDGIDDERLRKEFSPFG 328
Query 183 SILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAIDTMNGFEVVGKQLKVNYA 235
+I + ++ + E + +G+GF+ F+SP A A+ MNG V K L V A
Sbjct 329 TITSAKV--TMEGGRS-KGFGFVCFSSPEEATKAVTEMNGKIVATKPLYVALA 378
Score = 48.1 bits (113), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 37/67 (55%), Gaps = 3/67 (4%)
Query 156 GSNRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKL 215
G I+V N+ S+ L F FG+IL+C+++ + +GYGF+ F + A+
Sbjct 97 GVGNIFVKNLDRSIDSKTLYDTFSAFGNILSCKVVCDENGS---KGYGFVHFETQEEAER 153
Query 216 AIDTMNG 222
AI+ MNG
Sbjct 154 AIEKMNG 160
Score = 46.6 bits (109), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 61/261 (23%), Positives = 111/261 (42%), Gaps = 30/261 (11%)
Query 1 MCRIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKE--GDRSKGFCFVEYASPEAAQMAL 58
+ +YVG L +TE + F G + S+ + ++ RS G+ V + E A+ AL
Sbjct 10 LASLYVGDLHPDVTEAMLYEKFSPAGPILSIRVYRDRITRRSLGYASVNFQQLEDAERAL 69
Query 59 STMHNFVLKGRTIKVG--------RPTAVG----AGQPQQLLARPLGGYDM-----NVGA 101
TM+ V+KG+ +++ R + VG + + ++ L YD N+ +
Sbjct 70 DTMNFDVIKGKPVRIMWSQRDPSLRKSGVGNIFVKNLDRSIDSKTL--YDTFSAFGNILS 127
Query 102 AATPGMSLVSSAQAGAAVATAILAGRAGQTPQGDMAS--LVSMGQVTPEKQQDSAPGSNR 159
S T A RA + G + V +G+ + + + G+
Sbjct 128 CKVVCDENGSKGYGFVHFETQEEAERAIEKMNGMFLNDHKVFVGRFKSRRDRQAELGARA 187
Query 160 IYVGNVPFGFSSED-----LKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAK 214
NV ED L+ +F FG L+ +++ + + +G+GF+ F A+
Sbjct 188 KEFTNVYIKNLGEDMDDERLQDLFGRFGPALSVKVMTDESG--KSKGFGFVSFERHEDAR 245
Query 215 LAIDTMNGFEVVGKQLKVNYA 235
A++ MNG ++ GKQ+ V A
Sbjct 246 KAVEEMNGKDLNGKQIYVGRA 266
> pfa:PF13_0315 RNA binding protein, putative
Length=509
Score = 62.0 bits (149), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 70/264 (26%), Positives = 119/264 (45%), Gaps = 40/264 (15%)
Query 3 RIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGDRS--KGFCFVEYASPEAAQMALST 60
++++GSL +TE IK +F +GTV V + K+ KG FV+++ E A A+ +
Sbjct 185 KLFIGSLPKNITEDNIKEMFSPYGTVEEVFIMKDNSTGLGKGCSFVKFSYKEQALYAIKS 244
Query 61 MH-NFVLKG--RTIKVGRPTAVGAGQPQ-QLLARPLGGYDMNVGAAATPGMSLVSSA--- 113
++ L+G R ++V + QPQ L +P+ N A P S+ S
Sbjct 245 LNGKKTLEGCTRPVEVRFAEPKSSKQPQIPLTLQPM----QNPPHAMAPQPSISSPNNIN 300
Query 114 -------------QAGAAVATAILAGR------AGQTPQGDM----ASLVSMGQVTPEKQ 150
Q G GR T Q +M +L
Sbjct 301 FGNNFSVNNNYPRQVGPWKEYYSGEGRPYYYNEQTNTTQWEMPKEFETLFMNNSANMHNL 360
Query 151 QDSA--PGSNRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFA 208
+S+ PG+N +++ +VP + DL + F FG +L+ ++ +++N ++RG+ F+ +
Sbjct 361 SESSGPPGAN-LFIFHVPNEWQQTDLIQAFSPFGELLSARIA-TEKNTGRNRGFAFVSYD 418
Query 209 SPAAAKLAIDTMNGFEVVGKQLKV 232
S +A AI MNGF + K+LKV
Sbjct 419 SLESAAAAISQMNGFMALNKKLKV 442
Score = 31.2 bits (69), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 21/81 (25%), Positives = 41/81 (50%), Gaps = 4/81 (4%)
Query 159 RIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAID 218
++++G VP + L+ IF +G I+ ++ + H+ F++ AS + A AI
Sbjct 86 KLFIGRVPKNIEEDQLRPIFEEYG-IVNEVVIIRDKITNVHKSSAFVKMASISEADNAIR 144
Query 219 TMNGFEVVGKQ---LKVNYAT 236
+N + + Q L+V YA+
Sbjct 145 LLNNQKTLDAQLGSLQVKYAS 165
> mmu:93686 Rbfox2, 2810460A15Rik, AA407676, AI118529, Fbm2, Fxh,
Hrnbp2, Rbm9; RNA binding protein, fox-1 homolog (C. elegans)
2; K14946 RNA binding protein fox-1
Length=445
Score = 62.0 bits (149), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 37/121 (30%), Positives = 64/121 (52%), Gaps = 5/121 (4%)
Query 141 SMGQVTPEKQQDSAPGSNRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHR 200
S Q + + S P R++V N+PF F DL+++F FG IL +++ N + +
Sbjct 165 SQTQSSENSESKSTP--KRLHVSNIPFRFRDPDLRQMFGQFGKILDVEII---FNERGSK 219
Query 201 GYGFIEFASPAAAKLAIDTMNGFEVVGKQLKVNYATALRSAAVAGSLGVGMGAQMAPIVA 260
G+GF+ F + A A A + ++G V G++++VN ATA G +++P+V
Sbjct 220 GFGFVTFENSADADRAREKLHGTVVEGRKIEVNNATARVMTNKKMVTPYANGWKLSPVVG 279
Query 261 A 261
A
Sbjct 280 A 280
Score = 47.4 bits (111), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 36/132 (27%), Positives = 66/132 (50%), Gaps = 6/132 (4%)
Query 3 RIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGDRSKGFCFVEYASPEAAQMALSTMH 62
R++V ++ + + +++ +F FG + V++ SKGF FV + + A A +H
Sbjct 181 RLHVSNIPFRFRDPDLRQMFGQFGKILDVEIIFNERGSKGFGFVTFENSADADRAREKLH 240
Query 63 NFVLKGRTIKVGRPTAVGAGQPQQLLARPLGGYDMN--VGAAATPGMSLVSSAQAGAAV- 119
V++GR I+V TA ++++ G+ ++ VGA P + SS QA ++
Sbjct 241 GTVVEGRKIEVNNATARVMTN-KKMVTPYANGWKLSPVVGAVYGPELYAASSFQADVSLG 299
Query 120 --ATAILAGRAG 129
A L+GR G
Sbjct 300 NEAAVPLSGRGG 311
> mmu:18459 Pabpc2, PABP, PABP+, Pabp2; poly(A) binding protein,
cytoplasmic 2; K13126 polyadenylate-binding protein
Length=628
Score = 61.6 bits (148), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 64/264 (24%), Positives = 114/264 (43%), Gaps = 36/264 (13%)
Query 1 MCRIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKE--GDRSKGFCFVEYASPEAAQMAL 58
M +YVG L +TE + F + G + S+ + ++ RS G+ V + P A+ AL
Sbjct 10 MASLYVGDLHPDVTEAMLYEKFSSAGPILSIRVYRDVITRRSLGYASVNFEQPADAERAL 69
Query 59 STMHNFVLKGRTIKVG--------RPTAVGAGQPQQLLARPLGG-------YDM-----N 98
TM+ V+KG+ +++ R + VG + + L YD N
Sbjct 70 DTMNFDVIKGKPVRIMWSQRDPSLRRSGVG-----NVFIKNLNKTIDNKALYDTFSAFGN 124
Query 99 VGAAATPGMSLVSSAQAGAAVATAILAGRAGQTPQGDMAS--LVSMGQVTPEKQQDSAPG 156
+ + S T A RA + G + + V +G+ +K++++ G
Sbjct 125 ILSCKVVSDENGSKGHGFVHFETEEAAERAIEKMNGMLLNDRKVFVGRFKSQKEREAELG 184
Query 157 S-----NRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPA 211
+ +Y+ N E L +F FG IL+ +++ + + +G+GF+ F
Sbjct 185 TGTKEFTNVYIKNFGDRMDDETLNGLFGRFGQILSVKVMTDEGG--KSKGFGFVSFERHE 242
Query 212 AAKLAIDTMNGFEVVGKQLKVNYA 235
A+ A+D MNG E+ GK + V A
Sbjct 243 DAQKAVDEMNGKELNGKHIYVGRA 266
Score = 61.2 bits (147), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 68/273 (24%), Positives = 109/273 (39%), Gaps = 46/273 (16%)
Query 4 IYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGDRSKGFCFVEYASPEAAQMALSTMHN 63
+++ +L+ + + F AFG + S + + + SKG FV + + EAA+ A+ M+
Sbjct 101 VFIKNLNKTIDNKALYDTFSAFGNILSCKVVSDENGSKGHGFVHFETEEAAERAIEKMNG 160
Query 64 FVLKGRTIKVG-------RPTAVGAGQPQ--QLLARPLG---------------GYDMNV 99
+L R + VG R +G G + + + G G ++V
Sbjct 161 MLLNDRKVFVGRFKSQKEREAELGTGTKEFTNVYIKNFGDRMDDETLNGLFGRFGQILSV 220
Query 100 GAAATPG--------MSLVSSAQAGAAV---------ATAILAGRAGQTPQGDMASLVSM 142
G +S A AV I GRA +
Sbjct 221 KVMTDEGGKSKGFGFVSFERHEDAQKAVDEMNGKELNGKHIYVGRAQKKDDRHTELKHKF 280
Query 143 GQVTPEKQQDSAPGSNRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGY 202
QVT +K G N +YV N+ G E L+K F FG+I + +++ + +G+
Sbjct 281 EQVTQDKSIRYQ-GIN-LYVKNLDDGIDDERLQKEFSPFGTITSTKVMTEG---GRSKGF 335
Query 203 GFIEFASPAAAKLAIDTMNGFEVVGKQLKVNYA 235
GF+ F+SP A A+ MNG V K L V A
Sbjct 336 GFVCFSSPEEATKAVSEMNGRIVATKPLYVALA 368
> tgo:TGME49_083740 mRNA processing protein, putative (EC:3.4.21.72)
Length=615
Score = 61.6 bits (148), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 37/125 (29%), Positives = 62/125 (49%), Gaps = 18/125 (14%)
Query 148 EKQQDSAPGSNRIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEF 207
E+ + G+ ++++GN+PF + EDLK + G +L ++ + Q +G+ F EF
Sbjct 6 ERNSSPSSGAYQMWLGNIPFDATEEDLKTLLSRVGRVLQVRI--KYDEGGQSKGFAFCEF 63
Query 208 ASPAAAKLAIDTMNGFEVVGKQLKVNYAT----------------ALRSAAVAGSLGVGM 251
P LA T+N ++ G++LK+++AT A R A AGSL
Sbjct 64 PDPETCYLAYVTLNNADLGGRKLKIDFATDELRQRYGSSGGGAARAERRKAPAGSLAGTS 123
Query 252 GAQMA 256
GA+ A
Sbjct 124 GAEEA 128
Score = 53.1 bits (126), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 24/72 (33%), Positives = 43/72 (59%), Gaps = 1/72 (1%)
Query 3 RIYVGSLDYYLTELEIKSVFQAFGTVTSVDMP-KEGDRSKGFCFVEYASPEAAQMALSTM 61
++++G++ + TE ++K++ G V V + EG +SKGF F E+ PE +A T+
Sbjct 17 QMWLGNIPFDATEEDLKTLLSRVGRVLQVRIKYDEGGQSKGFAFCEFPDPETCYLAYVTL 76
Query 62 HNFVLKGRTIKV 73
+N L GR +K+
Sbjct 77 NNADLGGRKLKI 88
> cpv:cgd7_1620 nop15p/nopp34; nucleolar protein with 1 RRM domain
; K14838 nucleolar protein 15
Length=156
Score = 61.2 bits (147), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 31/72 (43%), Positives = 43/72 (59%), Gaps = 1/72 (1%)
Query 160 IYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAIDT 219
+YVG++PFG LK+ F FG IL +L S++N RGY FIEF S A++A T
Sbjct 18 VYVGHIPFGLFEPQLKEYFSQFGKILRIKLSRSKKNGHS-RGYAFIEFESMEVAQIAAST 76
Query 220 MNGFEVVGKQLK 231
MN + + + LK
Sbjct 77 MNNYIIFKRSLK 88
Score = 54.7 bits (130), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 45/71 (63%), Gaps = 2/71 (2%)
Query 4 IYVGSLDYYLTELEIKSVFQAFGTVTSVDMP--KEGDRSKGFCFVEYASPEAAQMALSTM 61
+YVG + + L E ++K F FG + + + K+ S+G+ F+E+ S E AQ+A STM
Sbjct 18 VYVGHIPFGLFEPQLKEYFSQFGKILRIKLSRSKKNGHSRGYAFIEFESMEVAQIAASTM 77
Query 62 HNFVLKGRTIK 72
+N+++ R++K
Sbjct 78 NNYIIFKRSLK 88
> tgo:TGME49_112530 splicing factor protein, putative ; K13091
RNA-binding protein 39
Length=633
Score = 61.2 bits (147), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 36/92 (39%), Positives = 53/92 (57%), Gaps = 5/92 (5%)
Query 159 RIYVGNVPFGFS---SEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKL 215
R+YVG + + EDL+ +F FG I ++ P N RGYGF+ +AS A A
Sbjct 325 RVYVGGLVDNLAKIQEEDLRVLFGPFGRINEVEI-PKDANTGGLRGYGFVTYASAADAHE 383
Query 216 AIDTMNGFEVVGKQLKVNY-ATALRSAAVAGS 246
A+ MN FE++G+QL+V Y A +R +A G+
Sbjct 384 AMQHMNNFELLGQQLRVGYAADGVRGSATEGT 415
Score = 51.2 bits (121), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 28/77 (36%), Positives = 45/77 (58%), Gaps = 5/77 (6%)
Query 3 RIYVGSLDYYLT---ELEIKSVFQAFGTVTSVDMPKEGDRS--KGFCFVEYASPEAAQMA 57
R+YVG L L E +++ +F FG + V++PK+ + +G+ FV YAS A A
Sbjct 325 RVYVGGLVDNLAKIQEEDLRVLFGPFGRINEVEIPKDANTGGLRGYGFVTYASAADAHEA 384
Query 58 LSTMHNFVLKGRTIKVG 74
+ M+NF L G+ ++VG
Sbjct 385 MQHMNNFELLGQQLRVG 401
> dre:407613 rbfox1l, Fox-1, a2bp1l, fox1l, hm:zeh0082; RNA binding
protein, fox-1 homolog (C. elegans) 1-like; K14946 RNA
binding protein fox-1
Length=382
Score = 61.2 bits (147), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 30/79 (37%), Positives = 49/79 (62%), Gaps = 3/79 (3%)
Query 159 RIYVGNVPFGFSSEDLKKIFVCFGSILTCQLLPSQENPQQHRGYGFIEFASPAAAKLAID 218
R++V N+PF F DL+++F FG IL +++ N + +G+GF+ F S A A +
Sbjct 148 RLHVSNIPFRFRDPDLRQMFGQFGKILDVEII---FNERGSKGFGFVTFESAVEADRARE 204
Query 219 TMNGFEVVGKQLKVNYATA 237
+NG V G++++VN ATA
Sbjct 205 KLNGTIVEGRKIEVNNATA 223
Score = 44.7 bits (104), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 32/113 (28%), Positives = 58/113 (51%), Gaps = 4/113 (3%)
Query 3 RIYVGSLDYYLTELEIKSVFQAFGTVTSVDMPKEGDRSKGFCFVEYASPEAAQMALSTMH 62
R++V ++ + + +++ +F FG + V++ SKGF FV + S A A ++
Sbjct 148 RLHVSNIPFRFRDPDLRQMFGQFGKILDVEIIFNERGSKGFGFVTFESAVEADRAREKLN 207
Query 63 NFVLKGRTIKVGRPTA-VGAGQPQQLLARPLGGYDMN--VGAAATPGMSLVSS 112
+++GR I+V TA V +PQ L G+ +N +GA P + V+S
Sbjct 208 GTIVEGRKIEVNNATARVVTKKPQTPLVN-AAGWKINPVMGAMYAPELYTVAS 259
> bbo:BBOV_IV003970 23.m05815; RNA recognition motif containing
protein; K14838 nucleolar protein 15
Length=187
Score = 60.8 bits (146), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 28/72 (38%), Positives = 45/72 (62%), Gaps = 2/72 (2%)
Query 4 IYVGSLDYYLTELEIKSVFQAFGTVTSVDM--PKEGDRSKGFCFVEYASPEAAQMALSTM 61
IYVG+L L E I+ F+ FGTV + + K+ S+G+CF+++ S E A++A M
Sbjct 12 IYVGNLPKALNESNIRKYFEQFGTVKKIRLMKSKKTGNSRGYCFLQFESNEIAKIAAEAM 71
Query 62 HNFVLKGRTIKV 73
+N+ + GR +KV
Sbjct 72 NNYFIDGRVLKV 83
Lambda K H
0.317 0.130 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 10911908772
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40