bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
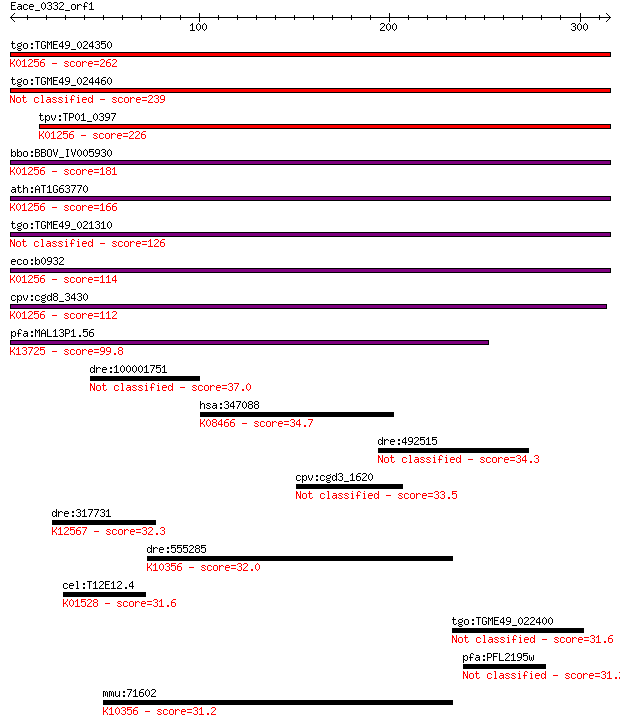
Query= Eace_0332_orf1
Length=315
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_024350 aminopeptidase N, putative (EC:3.4.11.2); K0... 262 1e-69
tgo:TGME49_024460 aminopeptidase N, putative (EC:3.4.11.2) 239 1e-62
tpv:TP01_0397 alpha-aminoacylpeptide hydrolase (EC:3.4.11.2); ... 226 9e-59
bbo:BBOV_IV005930 23.m06519; aminopeptidase (EC:3.4.11.2); K01... 181 3e-45
ath:AT1G63770 peptidase M1 family protein; K01256 aminopeptida... 166 9e-41
tgo:TGME49_021310 aminopeptidase N, putative (EC:3.4.11.2) 126 1e-28
eco:b0932 pepN, ECK0923, JW0915; aminopeptidase N (EC:3.4.11.2... 114 5e-25
cpv:cgd8_3430 zincin/aminopeptidase N like metalloprotease ; K... 112 2e-24
pfa:MAL13P1.56 M1-family aminopeptidase (EC:3.4.11.-); K13725 ... 99.8 1e-20
dre:100001751 sned1; sushi, nidogen and EGF-like domains 1 37.0
hsa:347088 GPR144, PGR24; G protein-coupled receptor 144; K084... 34.7 0.54
dre:492515 ftr72, zgc:103602; finTRIM family, member 72 34.3
cpv:cgd3_1620 hypothetical protein 33.5 1.1
dre:317731 ttna, fi20g08, si:busm1-258d18.1, si:dz258d18.1, tt... 32.3 2.1
dre:555285 myo1e, fe49h01, wu:fc15g04, wu:fe49h01; myosin IE; ... 32.0 2.9
cel:T12E12.4 drp-1; Dynamin Related Protein family member (drp... 31.6 3.7
tgo:TGME49_022400 hypothetical protein 31.6 4.2
pfa:PFL2195w clathrin coat assembly protein AP180, putative 31.2 5.3
mmu:71602 Myo1e, 2310020N23Rik, 9130023P14Rik, AA407778, myosi... 31.2 5.6
> tgo:TGME49_024350 aminopeptidase N, putative (EC:3.4.11.2);
K01256 aminopeptidase N [EC:3.4.11.2]
Length=1419
Score = 262 bits (669), Expect = 1e-69, Method: Compositional matrix adjust.
Identities = 143/318 (44%), Positives = 202/318 (63%), Gaps = 4/318 (1%)
Query 1 TPRLEVLRSEYKEDAHTFEISFRQHTPPTPGQETKLPQVIPIKIGLIGKDSHRDLLQP-S 59
TP + V + ++ D F+++ +Q TPPTPGQ K P IPIK+GLIGK S +D+L P +
Sbjct 983 TPEVTVSEAVFQPDRKKFKLTLKQRTPPTPGQVEKHPFHIPIKVGLIGKTSKKDILSPPT 1042
Query 60 MVLEMTEEEQTFRVRGVDEDCVPSILRGFSAPVRLINRRTAEENAFLLAYDTDPVTKWFS 119
VLE+TE EQTF + EDCV S LR FSAPV++ + +T E+ AFL+A+D+D KW +
Sbjct 1043 KVLELTEAEQTFELDAA-EDCVLSFLRDFSAPVKVKHEQTDEDIAFLMAHDSDDFAKWQA 1101
Query 120 SRALAAPIVISRCEKIAADKNVQ-HFAGLPDEYVEALRTALTDQATDSALKALILQLPDW 178
+ LA+ ++ R E+ + FA LP YVEA + L +Q D +++A L+LPD
Sbjct 1102 AHTLASGLLKHRAEQWREKQGEDVEFARLPKIYVEAFKQTLLEQGRDRSIQAYTLRLPDR 1161
Query 179 NVLAAEMRPIDPEALHKAIRTVKADLATALKSEMLELYRQLTLPAGQ-EEKVTEGEAGRR 237
+ +A EM PIDP AL +A +V+ ++ LKS++L++Y L+ P + EE + E RR
Sbjct 1162 DGVAQEMEPIDPLALKEATESVRREVGQLLKSDLLKVYASLSAPESEAEESRDQSEVSRR 1221
Query 238 KLRNALLHFLSAARDEEAVERAFMHFTEAKCMTDKYAGLIALSNMPTEQREHAFSHFYDD 297
+LRN +L+FL+ RD+EA A HF AK MT+KYA L L ++ +R A FY D
Sbjct 1222 RLRNVILYFLTGERDKEAAALAMNHFKSAKGMTEKYAALSILCDIEGPERTAALEQFYRD 1281
Query 298 AAGDPLVVDKWFKAQALS 315
A GDPLV+DKWF QALS
Sbjct 1282 AKGDPLVLDKWFAVQALS 1299
> tgo:TGME49_024460 aminopeptidase N, putative (EC:3.4.11.2)
Length=970
Score = 239 bits (609), Expect = 1e-62, Method: Compositional matrix adjust.
Identities = 133/329 (40%), Positives = 203/329 (61%), Gaps = 14/329 (4%)
Query 1 TPRLEVLRSEYKEDAHTFEISFRQHTPPTPGQETKLPQVIPIKIGLIGKDSHRDLLQP-S 59
TP + V Y ++ +QHTPPTPGQ+ KLP IP+ +G IGK S RD+L P +
Sbjct 524 TPHVTVSSFIYDAAERKMHLTLKQHTPPTPGQDKKLPLQIPVAVGCIGKTSKRDVLSPPT 583
Query 60 MVLEMTEEEQTFRVRGVDEDCVPSILRGFSAPVRLIN-RRTAEENAFLLAYDTDPVTKWF 118
+LE+TEEEQ F + V EDCV S+LR FSAPV+LI +T E+ AFL+ YD+D V +W
Sbjct 584 QILELTEEEQAFVLVDVGEDCVLSVLRDFSAPVKLIQPLQTDEDLAFLVTYDSDNVNRWQ 643
Query 119 SSRALAAPIVISR-------CEKIAADKNVQH--FAGLPDEYVEALRTALT--DQATDSA 167
+++ L+ ++++R E+I + + F+ LP +YV+ +R + + +
Sbjct 644 AAQTLSRKVLLTRISEYQQRVEEIEEGRVSESDLFSKLPTQYVQTVRDIVIAPESSMGKD 703
Query 168 LKALILQLPDWNVLAAEMRPIDPEALHKAIRTVKADLATALKSEMLELYRQLTLPAGQEE 227
+K+L+L LP L + IDP+A++ A+ +V+ D+ AL EML+LY +LTLPAG EE
Sbjct 704 IKSLLLSLPTKAQLELAVDSIDPDAINAALASVRRDIVDALGEEMLQLYTELTLPAGTEE 763
Query 228 KVTEGEA-GRRKLRNALLHFLSAARDEEAVERAFMHFTEAKCMTDKYAGLIALSNMPTEQ 286
+ E GRR LRN LL FL+A+ D+++ + A HF A M+DK A L L+ +P ++
Sbjct 764 SGADIEHWGRRALRNELLRFLTASFDQKSAKLASAHFDRAMVMSDKVAALTVLTEIPGQE 823
Query 287 REHAFSHFYDDAAGDPLVVDKWFKAQALS 315
R+ AF F + AG+ ++++KW QA+S
Sbjct 824 RDEAFERFRQEVAGNAVLMEKWMSLQAMS 852
> tpv:TP01_0397 alpha-aminoacylpeptide hydrolase (EC:3.4.11.2);
K01256 aminopeptidase N [EC:3.4.11.2]
Length=1020
Score = 226 bits (576), Expect = 9e-59, Method: Compositional matrix adjust.
Identities = 117/300 (39%), Positives = 183/300 (61%), Gaps = 10/300 (3%)
Query 16 HTFEISFRQHTPPTPGQETKLPQVIPIKIGLIGKDSHRDLLQPSMVLEMTEEEQTFRVRG 75
+ +++ RQ TPPTP Q K P IP+++GLIGK + DLL ++++E+ EE+Q F
Sbjct 617 NKYKLVLRQSTPPTPKQPVKQPFHIPLRLGLIGKSTGGDLLG-NVLVELKEEQQEFEFGP 675
Query 76 VDEDCVPSILRGFSAPVRLINRRTAEENAFLLAYDTDPVTKWFSSRALAAPIVISRCEKI 135
V EDCV S+ RGFSAPV++ ++ EE FL+ YDTD + +W ++++LA +V+ +
Sbjct 676 VTEDCVLSLNRGFSAPVKVKYEQSPEELLFLMLYDTDGLNRWNAAQSLATKVVLDLTKDP 735
Query 136 AADKNVQHFAGLPDEYVEALRTALTDQATDSALKALILQLPDWNVLAAEMRPIDPEALHK 195
A+ +P Y+EA + L + D K L + +PD ++LA++M+P DP L
Sbjct 736 TAE--------VPKTYLEAYK-KLLNSDMDHNEKGLCMSMPDVDILASKMKPYDPGLLFA 786
Query 196 AIRTVKADLATALKSEMLELYRQLTLPAGQEEKVTEGEAGRRKLRNALLHFLSAARDEEA 255
++R +K +L + E+Y+ LTL GQ++++T+ + RR LRN + FL + RD E+
Sbjct 787 SLRKLKQELGRTFRPTFTEMYKSLTLREGQKDELTKEDMARRFLRNTVFSFLVSMRDMES 846
Query 256 VERAFMHFTEAKCMTDKYAGLIALSNMPTEQREHAFSHFYDDAAGDPLVVDKWFKAQALS 315
VE A H+ +AK M DKY + L +M + R+ FY+ A GD +VDKWFKAQA+S
Sbjct 847 VELALKHYRDAKVMNDKYTAFVQLMHMEFQDRQKVVDDFYNFANGDAALVDKWFKAQAVS 906
> bbo:BBOV_IV005930 23.m06519; aminopeptidase (EC:3.4.11.2); K01256
aminopeptidase N [EC:3.4.11.2]
Length=846
Score = 181 bits (459), Expect = 3e-45, Method: Compositional matrix adjust.
Identities = 114/315 (36%), Positives = 164/315 (52%), Gaps = 70/315 (22%)
Query 1 TPRLEVLRSEYKEDAHTFEISFRQHTPPTPGQETKLPQVIPIKIGLIGKDSHRDLLQPSM 60
TP +EVL E D TF + RQ+TPPTP QETKLP IPIKIGL+GK S RDL + S+
Sbjct 482 TPEVEVL--EAVRDGTTFRLRLRQYTPPTPRQETKLPFHIPIKIGLLGKSSKRDL-KGSI 538
Query 61 VLEMTEEEQTFRVRGVDEDCVPSILRGFSAPVRLINRRTAEENAFLLAYDTDPVTKWFSS 120
VLE+ E EQTF + V+EDCV S RGFSAP+++ +++ E+ FL+++DTD + +W
Sbjct 539 VLELRESEQTFEINDVNEDCVLSFNRGFSAPIKVKFQQSDEDLLFLMSHDTDGLNRW--- 595
Query 121 RALAAPIVISRCEKIAADKNVQHFAGLPDEYVEALRTALTDQATDSALKALILQLPDWNV 180
E + +RT KA++ +
Sbjct 596 -----------------------------EAGQCMRT-----------KAILSNI----- 610
Query 181 LAAEMRPIDPEALHKAIRTVKADLATALKSEMLELYRQLTLPAGQEEKVTEGEAGRRKLR 240
++PID + + +I ++ L S+ L +++ + + + RR LR
Sbjct 611 --GSVKPID-DMIFSSIESI-------LSSD---------LDNNEKDTLEKDDMARRYLR 651
Query 241 NALLHFLSAARDEEAVERAFMHFTEAKCMTDKYAGLIALSNMPTEQREHAFSHFYDDAAG 300
N LL +L D AVE A H+ A+CMTD+Y + L NM ++ + FY+ AAG
Sbjct 652 NTLLGYLVCRSDASAVELALGHYRAARCMTDRYYAFVQLMNMDFAGKDDVIADFYERAAG 711
Query 301 DPLVVDKWFKAQALS 315
D LV+DKWF AQA S
Sbjct 712 DALVIDKWFSAQASS 726
> ath:AT1G63770 peptidase M1 family protein; K01256 aminopeptidase
N [EC:3.4.11.2]
Length=987
Score = 166 bits (421), Expect = 9e-41, Method: Compositional matrix adjust.
Identities = 107/328 (32%), Positives = 170/328 (51%), Gaps = 20/328 (6%)
Query 1 TPRLEVLRSEYKEDAHTFEISFRQHTPPTPGQETKLPQVIPIKIGLI---GKD-----SH 52
TP ++V+ S Y DA TF + F Q PPTPGQ TK P IP+ +GL+ GKD H
Sbjct 553 TPVVKVV-SSYNADARTFSLKFSQEIPPTPGQPTKEPTFIPVVVGLLDSSGKDITLSSVH 611
Query 53 RD-----LLQPSMVLEMTEEEQTFRVRGVDEDCVPSILRGFSAPVRLINRRTAEENAFLL 107
D + S +L +T++E+ F + E VPS+ RGFSAPVR+ + ++ FLL
Sbjct 612 HDGTVQTISGSSTILRVTKKEEEFVFSDIPERPVPSLFRGFSAPVRVETDLSNDDLFFLL 671
Query 108 AYDTDPVTKWFSSRALAAPIVISRCEKIAADKNVQHFAGLPDEYVEALRTALTDQATDSA 167
A+D+D +W + + LA ++++ +K + L ++V+ L + L+D + D
Sbjct 672 AHDSDEFNRWEAGQVLARKLMLNLVSDFQQNKPL----ALNPKFVQGLGSVLSDSSLDKE 727
Query 168 LKALILQLPDWNVLAAEMRPIDPEALHKAIRTVKADLATALKSEMLELYRQLTLPAGQEE 227
A + LP + M DP+A+H + V+ LA+ LK E+L++ + +
Sbjct 728 FIAKAITLPGEGEIMDMMAVADPDAVHAVRKFVRKQLASELKEELLKIVENNR--STEAY 785
Query 228 KVTEGEAGRRKLRNALLHFLSAARDEEAVERAFMHFTEAKCMTDKYAGLIALSNMPTEQR 287
RR L+N L +L++ D +E A + A +TD++A L ALS P + R
Sbjct 786 VFDHSNMARRALKNTALAYLASLEDPAYMELALNEYKMATNLTDQFAALAALSQNPGKTR 845
Query 288 EHAFSHFYDDAAGDPLVVDKWFKAQALS 315
+ + FY+ D LVV+KWF Q+ S
Sbjct 846 DDILADFYNKWQDDYLVVNKWFLLQSTS 873
> tgo:TGME49_021310 aminopeptidase N, putative (EC:3.4.11.2)
Length=966
Score = 126 bits (316), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 105/362 (29%), Positives = 170/362 (46%), Gaps = 51/362 (14%)
Query 1 TPRLEVLRSEYKEDAHTFEISFRQHTPPTPGQETKLPQV------IPIKIGLIGKDSHRD 54
TP +E+ + + F + Q PP E ++ IPI++ I + + +
Sbjct 494 TPEVEITGFTFDKTRKQFSFTVTQKPPPMSDYEIRMYGQKAQFLHIPIRVSFINRRTKKP 553
Query 55 LL---QPSMVLEMTEEEQTFRVRGVDEDCVPSILRGFSAPVRL-INRRTAEENAFLLAYD 110
L Q + VLE+ E QTF +E+ + + L+ FSAPV+L + +T E+ A L+
Sbjct 554 ELYVGQRTTVLELRGETQTFTFTDTEEEPLVAPLQSFSAPVKLTVPSQTEEDLAILIGAS 613
Query 111 TDPVTKWFSSRALAAPIVISRCEKIAADKNVQHFAGLPDEYVEALRTALTDQATDSALKA 170
DP T+W + + LA I+ R A K + L + + L AL D A A
Sbjct 614 FDPFTRWNAFQFLALQILKERM----ATKKKETSGVLSPAFQQGLHFALRDMPLSPAFTA 669
Query 171 LILQLPDWNVLAAEM-RPIDPEALHKAIRTVKADLATALKSEMLELYRQLTLP-AGQEEK 228
L+L LP ++ L + RP+DP+A+ A R++ D+ ++ + E Y T+P E+
Sbjct 670 LLLTLPSYSRLEQDAPRPLDPDAIISARRSLLRDIYYFHRNALDEAYVATTIPKVDDRER 729
Query 229 VTEGEAG-------RRKLRNALLHFLSAARDEEAVERAFMHFTEAKCMTDKYAGLIALSN 281
+ E+ RR LR+ LL +++A RDE + + A HF +A+ MTDK A L L +
Sbjct 730 DRQLESAEDPEQWQRRALRSILLEYVTANRDERSAKLALKHFKDARVMTDKIAALHVLVD 789
Query 282 MP-TEQREHAFSHFYDDAAG---------------------------DPLVVDKWFKAQA 313
+P ++RE A FY++A G +P ++ KWF QA
Sbjct 790 LPFNKEREEALHLFYEEARGTTRGNTARTEEQEAEAENEIKSIAVLCNPQLLTKWFALQA 849
Query 314 LS 315
S
Sbjct 850 RS 851
> eco:b0932 pepN, ECK0923, JW0915; aminopeptidase N (EC:3.4.11.2);
K01256 aminopeptidase N [EC:3.4.11.2]
Length=870
Score = 114 bits (285), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 100/320 (31%), Positives = 147/320 (45%), Gaps = 13/320 (4%)
Query 1 TPRLEVLRSEYKEDAHTFEISFRQHTPPTPGQETKLPQVIPIKIGLIGKDSHRDLLQP-- 58
TP + V + +Y + + ++ Q TP TP Q K P IP I L + LQ
Sbjct 445 TPIVTV-KDDYNPETEQYTLTISQRTPATPDQAEKQPLHIPFAIELYDNEGKVIPLQKGG 503
Query 59 ---SMVLEMTEEEQTFRVRGVDEDCVPSILRGFSAPVRLINRRTAEENAFLLAYDTDPVT 115
+ VL +T+ EQTF V VP++L FSAPV+L + + ++ FL+ + + +
Sbjct 504 HPVNSVLNVTQAEQTFVFDNVYFQPVPALLCEFSAPVKLEYKWSDQQLTFLMRHARNDFS 563
Query 116 KWFSSRALAAPIVISRCEKIAADKNVQHFAGLPDEYVEALRTALTDQATDSALKALILQL 175
+W ++++L A + +A + Q + LP +A R L D+ D AL A IL L
Sbjct 564 RWDAAQSLLATYIKL---NVARHQQGQPLS-LPVHVADAFRAVLLDEKIDPALAAEILTL 619
Query 176 PDWNVLAAEMRPIDPEALHKAIRTVKADLATALKSEMLELYRQLTLPAGQEEKVTEGEAG 235
P N +A IDP A+ + + LAT L E+L +Y E +V +
Sbjct 620 PSVNEMAELFDIIDPIAIAEVREALTRTLATELADELLAIYNA---NYQSEYRVEHEDIA 676
Query 236 RRKLRNALLHFLSAARDEEAVERAFMHFTEAKCMTDKYAGLIALSNMPTEQREHAFSHFY 295
+R LRNA L FL+ A F EA MTD A L A R+ +
Sbjct 677 KRTLRNACLRFLAFGETHLADVLVSKQFHEANNMTDALAALSAAVAAQLPCRDALMQEYD 736
Query 296 DDAAGDPLVVDKWFKAQALS 315
D + LV+DKWF QA S
Sbjct 737 DKWHQNGLVMDKWFILQATS 756
> cpv:cgd8_3430 zincin/aminopeptidase N like metalloprotease ;
K01256 aminopeptidase N [EC:3.4.11.2]
Length=936
Score = 112 bits (280), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 93/336 (27%), Positives = 164/336 (48%), Gaps = 24/336 (7%)
Query 1 TPRLEVLRSEYKEDAHTFEISFRQHTPPTPGQETKLPQVIPIKIGLIGKDSHRDLLQPSM 60
TP +EV+ ++ + T I+ RQ PTP Q K P IP+ +GLIGKDS +++ Q S
Sbjct 484 TPEVEVVSIDHNKAEGTCSITLRQRCSPTPEQPKKRPFYIPVIVGLIGKDSCKEIRQ-SE 542
Query 61 VLEMTEEEQTFRVRGVDEDCVPSILRGFSAPV--RLINRRTAEENAFLLAYDTDPVTKWF 118
L + E++QTF + GV E V SILRGFSAPV R R+ EE AFL A+DTD ++
Sbjct 543 TLILKEQQQTFILDGVWETPVASILRGFSAPVNLRFKTPRSDEELAFLFAFDTDEYNRFD 602
Query 119 SSRALAAPIVI----SRCEKIAADKNV-----QHFAGLPDEYVEALRTALTDQATDSALK 169
+++ L I+I + ++I+ + Q + ++ ++T+ T S +
Sbjct 603 AAQTLYKKILIQASTNSSQEISPSSVIESILSQTILSCFESFISKIKTSKNPNGTVSPMV 662
Query 170 A-LILQLPDWNVLAAEMRPIDPEALHKAIRTVKADLATALKSEMLELYRQLT-----LPA 223
A L++P ++ + A + + + + ++ + A+ K+++++L++ LT LP+
Sbjct 663 ASYTLRIPSYSQVLASIAEPNFDFVVESFNALTVAFASRFKTQIVDLFQVLTEDLSALPS 722
Query 224 G----QEEKVTEGEAGRRKLRNALLHFLSAARDEEAVERAFMHFTEAKCMTDKYAGLIAL 279
+ V R+L N LL F++ + AF MT K + AL
Sbjct 723 QSPFIHGKGVNVEAIAIRRLLNILLDFIAKLDPSLGTKLAFEQHNRFDYMTSKLGAISAL 782
Query 280 --SNMPTEQREHAFSHFYDDAAGDPLVVDKWFKAQA 313
++ + + A + D ++++F QA
Sbjct 783 QIADHKSSECIQALDSLLLATSDDVSTLNQYFSIQA 818
> pfa:MAL13P1.56 M1-family aminopeptidase (EC:3.4.11.-); K13725
M1-family aminopeptidase [EC:3.4.11.-]
Length=1085
Score = 99.8 bits (247), Expect = 1e-20, Method: Composition-based stats.
Identities = 68/261 (26%), Positives = 119/261 (45%), Gaps = 12/261 (4%)
Query 1 TPRLEVLRSEYKEDAHTFEISFRQHTPPTPGQETKLPQVIPIKIGLIGKDSHRDLLQPSM 60
TP + + Y + + I Q+T P Q+ K P IPI +GLI ++ ++++ +
Sbjct 651 TPHVS-FKYNYDAEKKQYSIHVNQYTKPDENQKEKKPLFIPISVGLINPENGKEMISQT- 708
Query 61 VLEMTEEEQTFRVRGVDEDCVPSILRGFSAPVRLINRRTAEENAFLLAYDTDPVTKWFSS 120
LE+T+E TF + +PS+ RGFSAPV + + T EE LL YD+D ++ S
Sbjct 709 TLELTKESDTFVFNNIAVKPIPSLFRGFSAPVYIEDNLTDEERILLLKYDSDAFVRYNSC 768
Query 121 RALAAPIVISRCEKIAADKN--VQHFAGLP--DEYVEALRTALTDQATDSALKALILQLP 176
+ ++ + KN ++ F P ++++A++ L D D+ K+ I+ LP
Sbjct 769 TNIYMKQILMNYNEFLKAKNEKLESFNLTPVNAQFIDAIKYLLEDPHADAGFKSYIVSLP 828
Query 177 DWNVLAAEMRPIDPEALHKAIRTVKADLATALKSEMLELYRQLTLPA------GQEEKVT 230
+ + +D + L + + L ++++ L A E V
Sbjct 829 QDRYIINFVSNLDTDVLADTKEYIYKQIGDKLNDVYYKMFKSLEAKADDLTYFNDESHVD 888
Query 231 EGEAGRRKLRNALLHFLSAAR 251
+ R LRN LL LS A+
Sbjct 889 FDQMNMRTLRNTLLSLLSKAQ 909
> dre:100001751 sned1; sushi, nidogen and EGF-like domains 1
Length=1369
Score = 37.0 bits (84), Expect = 0.10, Method: Composition-based stats.
Identities = 20/60 (33%), Positives = 32/60 (53%), Gaps = 3/60 (5%)
Query 43 KIGLIGKDSHRDLLQPSMVLEMTEEEQTFRVRGVDEDCVPSILRGFS---APVRLINRRT 99
++G G+D RDLL PS + + EE +R D +++ GF+ APV + R+T
Sbjct 873 RVGHTGRDCERDLLPPSGLNVLRVEENEVELRWDQSDVTQNLISGFAVAFAPVGRVPRKT 932
> hsa:347088 GPR144, PGR24; G protein-coupled receptor 144; K08466
G protein-coupled receptor 144
Length=963
Score = 34.7 bits (78), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 32/101 (31%), Positives = 45/101 (44%), Gaps = 12/101 (11%)
Query 101 EENAFLLAYDTDPVTKWFSSRALAAPIVISRCEKIAADKNVQHFAGLPDEYVEALRTALT 160
E FL Y T+P + +++ VISR +A D + LPD E + AL+
Sbjct 349 EPQPFLCCYRTEPYRRLQDAQSWPGQDVISRVNALANDIVL-----LPDPLSE-VHGALS 402
Query 161 DQATDSALKALILQLPDWNVLAAEMRPIDPEALHKAIRTVK 201
S L L +VLA EM P+ P AL +R +K
Sbjct 403 PAEASSFLGLLE------HVLAMEMAPLGPAALLAVVRFLK 437
> dre:492515 ftr72, zgc:103602; finTRIM family, member 72
Length=550
Score = 34.3 bits (77), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 29/79 (36%), Positives = 38/79 (48%), Gaps = 7/79 (8%)
Query 194 HKAIRTVKADLATALKSEMLELYRQLTLPAGQEEKVTEGEAGRRKLRNALLHFLSAARDE 253
HK V D K +LE ++ L + + E E G+R+LR A H S+A E
Sbjct 176 HKTHSVVSVDSEWTSKKNVLE---EMNLTCVK--LIKEREKGQRQLRTATKHLKSSA--E 228
Query 254 EAVERAFMHFTEAKCMTDK 272
EAVER FTE C +K
Sbjct 229 EAVERMEKVFTELICSLEK 247
> cpv:cgd3_1620 hypothetical protein
Length=345
Score = 33.5 bits (75), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 16/56 (28%), Positives = 30/56 (53%), Gaps = 0/56 (0%)
Query 151 YVEALRTALTDQATDSALKALILQLPDWNVLAAEMRPIDPEALHKAIRTVKADLAT 206
Y+ AL ++++DS KA+ +P+ +RP AL+ AI +V+ D+ +
Sbjct 149 YINALSRGPCNESSDSWSKAIFSSVPNQPYDLVSVRPTTQNALNSAISSVECDIIS 204
> dre:317731 ttna, fi20g08, si:busm1-258d18.1, si:dz258d18.1,
ttn, wu:fi04b11, wu:fi20g08; titin a; K12567 titin [EC:2.7.11.1]
Length=32757
Score = 32.3 bits (72), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 31/54 (57%), Gaps = 1/54 (1%)
Query 23 RQHTPPTPGQETKLPQVIPIKIGLIGKDSHRDLLQPSMVLEMTEEEQTFRVRGV 76
++ PP+P ++ +PQ IP K + H + P+ + E TEE++TF V+ V
Sbjct 8714 KKDVPPSPKKDDIVPQKIP-KQDISQTKPHMKEMIPTQIPEKTEEKETFSVQVV 8766
> dre:555285 myo1e, fe49h01, wu:fc15g04, wu:fe49h01; myosin IE;
K10356 myosin I
Length=1163
Score = 32.0 bits (71), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 39/171 (22%), Positives = 70/171 (40%), Gaps = 16/171 (9%)
Query 73 VRGVDEDCVPSILRGF----SAPVRLINRRTAEEN-------AFLLAYDTDPVTKWFSSR 121
+ G+D V I+ G + R A EN AFLL D + + + +SR
Sbjct 324 ISGIDRTMVLQIVAGILHLGNINFREAGNYAAVENDEFLAFPAFLLGIDQERLKEKLTSR 383
Query 122 ALAAPIVISRCEKIAADKNVQHFAGLPDEYVEALRTALTDQATDSALKALILQLPDWNVL 181
+ + E I NV+ D +AL + + D +S KA++ + + N+
Sbjct 384 KMDGKWG-GKSESIDVTLNVEQACFTRDALSKALHSRVFDYLVESINKAMVKEHQELNIG 442
Query 182 AAEMRPIDPEALHKAIRTVKADLATALKSEMLELYRQLTLPAGQEEKVTEG 232
++ E K + + ++ +++ +LTL A Q+E V EG
Sbjct 443 VLDIYGF--EIFQK--NGFEQFCINFVNEKLQQIFIELTLKAEQDEYVQEG 489
> cel:T12E12.4 drp-1; Dynamin Related Protein family member (drp-1);
K01528 dynamin GTPase [EC:3.6.5.5]
Length=712
Score = 31.6 bits (70), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 13/45 (28%), Positives = 29/45 (64%), Gaps = 2/45 (4%)
Query 29 TPGQETKLPQVIPIKIGLIG--KDSHRDLLQPSMVLEMTEEEQTF 71
T + + +VIP+K+G+IG S +++L ++++ ++EQ+F
Sbjct 226 TDAMDVLMGKVIPVKLGIIGVVNRSQQNILDNKLIVDAVKDEQSF 270
> tgo:TGME49_022400 hypothetical protein
Length=582
Score = 31.6 bits (70), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 36/77 (46%), Gaps = 12/77 (15%)
Query 233 EAGRRKLRNALLHFLSAARDEEAVERAFMHFTEAKCMTDKYAGLIALSNMPT-------- 284
+ GRR RN FL R+ +A+E+ H A + +G +LS P
Sbjct 416 QVGRRTSRN----FLEEEREADALEQGTRHACSAGTQENVKSGEGSLSFSPATTKTAGVD 471
Query 285 EQREHAFSHFYDDAAGD 301
E+RE +FS +Y AG+
Sbjct 472 EEREASFSSYYQIPAGE 488
> pfa:PFL2195w clathrin coat assembly protein AP180, putative
Length=431
Score = 31.2 bits (69), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 13/43 (30%), Positives = 25/43 (58%), Gaps = 0/43 (0%)
Query 239 LRNALLHFLSAARDEEAVERAFMHFTEAKCMTDKYAGLIALSN 281
++N +F+S +D+E + + HFT + + DK G+ +SN
Sbjct 92 MKNGCEYFISDVKDKEELIKKLTHFTHLEDLKDKGIGIRDISN 134
> mmu:71602 Myo1e, 2310020N23Rik, 9130023P14Rik, AA407778, myosin-1e,
myr_3; myosin IE; K10356 myosin I
Length=1107
Score = 31.2 bits (69), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 45/193 (23%), Positives = 78/193 (40%), Gaps = 20/193 (10%)
Query 50 DSHRDL---LQPSMVLEMTEEEQTFRVRGVDEDCVPSILRGFSAPVRLINRRTAEEN--- 103
D RD L V+ + EEQT ++ V IL + + + A E+
Sbjct 250 DDKRDFQETLHAMNVIGIFSEEQTLVLQ-----IVAGILHLGNISFKEVGNYAAVESEEF 304
Query 104 ----AFLLAYDTDPVTKWFSSRALAAPIVISRCEKIAADKNVQHFAGLPDEYVEALRTAL 159
A+LL + D + + +SR + + + E I NV+ D +AL +
Sbjct 305 LAFPAYLLGINQDRLKEKLTSRQMDSKWG-GKSESIHVTLNVEQACYTRDALAKALHARV 363
Query 160 TDQATDSALKALILQLPDWNVLAAEMRPIDPEALHKAIRTVKADLATALKSEMLELYRQL 219
D DS KA+ ++N+ ++ E K + + ++ +++ +L
Sbjct 364 FDFLVDSINKAMEKDHEEYNIGVLDIYGF--EIFQK--NGFEQFCINFVNEKLQQIFIEL 419
Query 220 TLPAGQEEKVTEG 232
TL A QEE V EG
Sbjct 420 TLKAEQEEYVQEG 432
Lambda K H
0.318 0.133 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 12920510708
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40