bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0334_orf1
Length=138
Score E
Sequences producing significant alignments: (Bits) Value
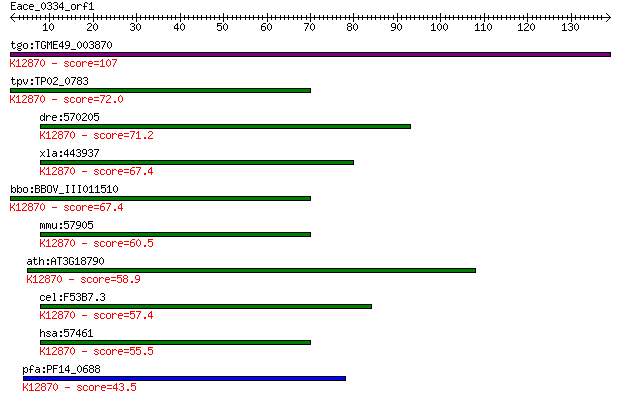
tgo:TGME49_003870 isy1-like splicing family domain-containing ... 107 1e-23
tpv:TP02_0783 hypothetical protein; K12870 pre-mRNA-splicing f... 72.0 5e-13
dre:570205 rab43, MGC136785, fi21h07, wu:fi21h07, zgc:136785; ... 71.2 8e-13
xla:443937 isy1, MGC80278, fsap33; ISY1 splicing factor homolo... 67.4 1e-11
bbo:BBOV_III011510 17.m07982; isy1-like splicing family protei... 67.4 1e-11
mmu:57905 Isy1, 5830446M03Rik, AI181014, AU020769; ISY1 splici... 60.5 2e-09
ath:AT3G18790 hypothetical protein; K12870 pre-mRNA-splicing f... 58.9 5e-09
cel:F53B7.3 hypothetical protein; K12870 pre-mRNA-splicing fac... 57.4 1e-08
hsa:57461 ISY1, FSAP33, KIAA1160; ISY1 splicing factor homolog... 55.5 4e-08
pfa:PF14_0688 Pre-mRNA-splicing factor ISY1 homolog, putative;... 43.5 2e-04
> tgo:TGME49_003870 isy1-like splicing family domain-containing
protein ; K12870 pre-mRNA-splicing factor ISY1
Length=285
Score = 107 bits (266), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 65/139 (46%), Positives = 87/139 (62%), Gaps = 10/139 (7%)
Query 1 ISVAGGD-YYYFGAAKELPGVRELFAGRAEEINEPRKTRAQLFQRITPDYFGWRDEENED 59
+S AG D Y YFGAAKEL GVREL A E +PRKTRA LF+ ITPDY+GWRDEE+ +
Sbjct 115 MSAAGADGYMYFGAAKELKGVRELLEREASEQQKPRKTRAMLFKNITPDYYGWRDEEDGE 174
Query 60 LLLAEQQQEQLWQQQQEEELQQQQELLESVGAASAAAAAKGMAAAAERDKLRRKHKQDGE 119
+LLAE + +EEEL+Q+ L S G+ +A A ++ D+ ++ K+D +
Sbjct 175 ILLAE--------KVREEELRQEA-LGASRGSKRVRSAPLAGDADSDSDEESKRRKRDQD 225
Query 120 DSSSSSSSSKKFQVYVDLP 138
SS S +FQ YVD+P
Sbjct 226 ASSQPSGGVPEFQSYVDVP 244
> tpv:TP02_0783 hypothetical protein; K12870 pre-mRNA-splicing
factor ISY1
Length=182
Score = 72.0 bits (175), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 35/71 (49%), Positives = 50/71 (70%), Gaps = 2/71 (2%)
Query 1 ISVAGGDYYYFGAAKELPGVRELFAGRAEEINEPRK--TRAQLFQRITPDYFGWRDEENE 58
+ + G Y YFGAAK LPGVRELF + +E+ + K +RA+L++ I PDY+G+ DE +E
Sbjct 104 LKIGGSGYKYFGAAKNLPGVRELFEQQQKEVEQSLKDVSRAELYRMIRPDYYGFNDERDE 163
Query 59 DLLLAEQQQEQ 69
+LLL E +EQ
Sbjct 164 ELLLQEYNKEQ 174
> dre:570205 rab43, MGC136785, fi21h07, wu:fi21h07, zgc:136785;
RAB43, member RAS oncogene family; K12870 pre-mRNA-splicing
factor ISY1
Length=285
Score = 71.2 bits (173), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 40/85 (47%), Positives = 56/85 (65%), Gaps = 2/85 (2%)
Query 8 YYYFGAAKELPGVRELFAGRAEEINEPRKTRAQLFQRITPDYFGWRDEENEDLLLAEQQQ 67
Y YFGAAK+LPGVRELF +E + PRKTRA+L + I DY+G+RDE++ LL EQ+
Sbjct 120 YKYFGAAKDLPGVRELF--ESEPVPPPRKTRAELMKDIDADYYGYRDEDDGVLLPLEQEY 177
Query 68 EQLWQQQQEEELQQQQELLESVGAA 92
E+ + E+ + +E +VG A
Sbjct 178 EKQVMAEAVEKWKADKEARLAVGGA 202
> xla:443937 isy1, MGC80278, fsap33; ISY1 splicing factor homolog;
K12870 pre-mRNA-splicing factor ISY1
Length=284
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 37/80 (46%), Positives = 52/80 (65%), Gaps = 10/80 (12%)
Query 8 YYYFGAAKELPGVRELFAGRAEEINEPRKTRAQLFQRITPDYFGWRDEENEDLLLAEQQQ 67
Y YFGAAK+LPGVRELF E + PRKTR +L + I +Y+G+RDE++ L+ EQ+Q
Sbjct 120 YKYFGAAKDLPGVRELF--EKEPLPPPRKTRTELMKSIDAEYYGYRDEDDGVLVPLEQEQ 177
Query 68 E--------QLWQQQQEEEL 79
E + W+ ++EE L
Sbjct 178 EKKAIAEALEKWRLEKEERL 197
> bbo:BBOV_III011510 17.m07982; isy1-like splicing family protein;
K12870 pre-mRNA-splicing factor ISY1
Length=228
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 35/73 (47%), Positives = 51/73 (69%), Gaps = 6/73 (8%)
Query 1 ISVAGGDYYYFGAAKELPGVRELFAGRAEEINEPR----KTRAQLFQRITPDYFGWRDEE 56
+ + GG Y YFGAAK LPGV+ELF +E++E R TRA+L+++I PDY+G+RD+E
Sbjct 117 LKIGGGGYRYFGAAKNLPGVQELF--EKQEMDERRVDTYVTRAELYRKINPDYYGFRDDE 174
Query 57 NEDLLLAEQQQEQ 69
+ L AE + E+
Sbjct 175 DGMLSAAEAELER 187
> mmu:57905 Isy1, 5830446M03Rik, AI181014, AU020769; ISY1 splicing
factor homolog (S. cerevisiae); K12870 pre-mRNA-splicing
factor ISY1
Length=285
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 31/62 (50%), Positives = 44/62 (70%), Gaps = 2/62 (3%)
Query 8 YYYFGAAKELPGVRELFAGRAEEINEPRKTRAQLFQRITPDYFGWRDEENEDLLLAEQQQ 67
Y YFGAAK+LPGVRELF E + PRKTRA+L + I +Y+G+ DE++ ++ EQ+
Sbjct 120 YKYFGAAKDLPGVRELF--EKEPLPPPRKTRAELMKAIDFEYYGYLDEDDGVIVPLEQEY 177
Query 68 EQ 69
E+
Sbjct 178 EK 179
> ath:AT3G18790 hypothetical protein; K12870 pre-mRNA-splicing
factor ISY1
Length=300
Score = 58.9 bits (141), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 40/103 (38%), Positives = 63/103 (61%), Gaps = 2/103 (1%)
Query 5 GGDYYYFGAAKELPGVRELFAGRAEEINEPRKTRAQLFQRITPDYFGWRDEENEDLLLAE 64
G Y YFGAAK+LPGVRELF + E+ + RKTR +++RI Y+G+RD+E+ L E
Sbjct 124 GPGYRYFGAAKKLPGVRELFE-KPPELRK-RKTRYDIYKRIDASYYGYRDDEDGILEKLE 181
Query 65 QQQEQLWQQQQEEELQQQQELLESVGAASAAAAAKGMAAAAER 107
++ E +++ EE ++ E+ + ++ + G AAAA R
Sbjct 182 RKSEGGMRKRSVEEWRRLDEVRKEARKGASEVVSVGAAAAAAR 224
> cel:F53B7.3 hypothetical protein; K12870 pre-mRNA-splicing factor
ISY1
Length=267
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 35/81 (43%), Positives = 52/81 (64%), Gaps = 6/81 (7%)
Query 8 YYYFGAAKELPGVRELFAGRAEEINEPRKTRAQLFQRITPDYFGWRDEENEDL-----LL 62
Y YFGAAK+LPGVRELF ++ E E R+ RA L + I YFG+ D+E+ L L+
Sbjct 121 YKYFGAAKDLPGVRELFE-KSTEGEEQRRHRADLLRNIDAHYFGYLDDEDGRLIPLEKLI 179
Query 63 AEQQQEQLWQQQQEEELQQQQ 83
E+ E++ ++ E++ Q+QQ
Sbjct 180 EEKNIERINKEFAEKQAQKQQ 200
> hsa:57461 ISY1, FSAP33, KIAA1160; ISY1 splicing factor homolog
(S. cerevisiae); K12870 pre-mRNA-splicing factor ISY1
Length=307
Score = 55.5 bits (132), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 32/82 (39%), Positives = 47/82 (57%), Gaps = 20/82 (24%)
Query 8 YYYFGAAKELPGVRELF--------------------AGRAEEINEPRKTRAQLFQRITP 47
Y YFGAAK+LPGVRELF AG ++ + PRKTRA+L + I
Sbjct 120 YKYFGAAKDLPGVRELFEKERQVRWLMPVIPALWEAEAGGSQALPPPRKTRAELMKAIDF 179
Query 48 DYFGWRDEENEDLLLAEQQQEQ 69
+Y+G+ DE++ ++ EQ+ E+
Sbjct 180 EYYGYLDEDDGVIVPLEQEYEK 201
> pfa:PF14_0688 Pre-mRNA-splicing factor ISY1 homolog, putative;
K12870 pre-mRNA-splicing factor ISY1
Length=201
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 31/78 (39%), Positives = 42/78 (53%), Gaps = 8/78 (10%)
Query 4 AGGDYYYFGAAKELPGVRELFAGRAEEINEPRKTR---AQLFQR-ITPDYFGWRDEENED 59
+Y YFGAAK L GVREL E+ + + A+ F++ I YFG+ DE NE
Sbjct 117 GSSNYKYFGAAKNLKGVRELLFKENEDKKQLNIKKKKDARNFEKVINIHYFGYCDEANEH 176
Query 60 LLLAEQQQEQLWQQQQEE 77
LL QQE Q++ E+
Sbjct 177 LL----QQEVKIQKKLEK 190
Lambda K H
0.310 0.126 0.345
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2421919804
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40