bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
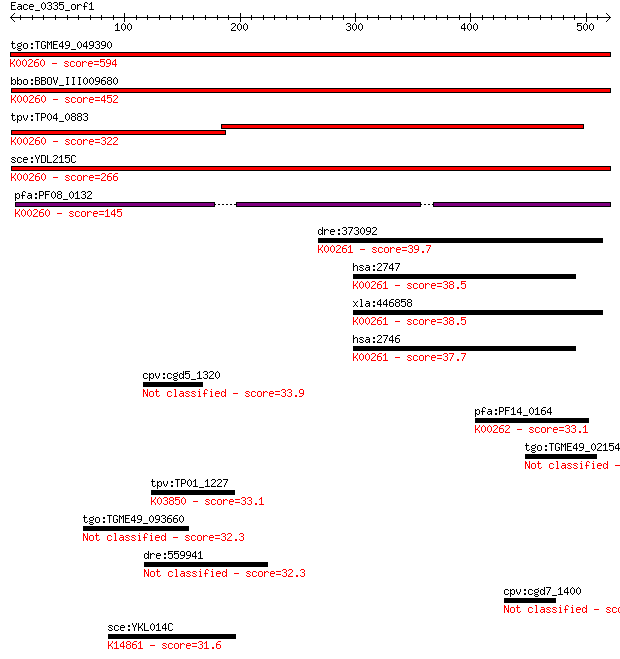
Query= Eace_0335_orf1
Length=520
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_049390 NAD-specific glutamate dehydrogenase, putati... 594 3e-169
bbo:BBOV_III009680 17.m07840; Glutamate/Leucine/Phenylalanine/... 452 1e-126
tpv:TP04_0883 NAD-specific glutamate dehydrogenase (EC:1.4.1.2... 322 2e-87
sce:YDL215C GDH2; Gdh2p (EC:1.4.1.2); K00260 glutamate dehydro... 266 1e-70
pfa:PF08_0132 glutamate dehydrogenase, putative (EC:1.4.1.2); ... 145 5e-34
dre:373092 glud1b, MGC192851, cb719, glud1, wu:fb16e02, wu:fb5... 39.7 0.028
hsa:2747 GLUD2, GDH2, GLUDP1; glutamate dehydrogenase 2 (EC:1.... 38.5 0.069
xla:446858 glud1, MGC80801; glutamate dehydrogenase 1 (EC:1.4.... 38.5 0.071
hsa:2746 GLUD1, GDH, GDH1, GLUD, MGC132003; glutamate dehydrog... 37.7 0.12
cpv:cgd5_1320 actin-like protein 33.9 1.5
pfa:PF14_0164 NADP-specific glutamate dehydrogenase; K00262 gl... 33.1 2.6
tgo:TGME49_021540 hypothetical protein 33.1 2.6
tpv:TP01_1227 hypothetical protein; K03850 alpha-1,2-glucosylt... 33.1 2.7
tgo:TGME49_093660 hypothetical protein 32.3 4.1
dre:559941 si:ch211-1o14.5 32.3 4.2
cpv:cgd7_1400 signal peptide containing protein 32.0 6.5
sce:YKL014C URB1, NPA1; Nucleolar protein required for the nor... 31.6 8.3
> tgo:TGME49_049390 NAD-specific glutamate dehydrogenase, putative
(EC:1.4.1.2); K00260 glutamate dehydrogenase [EC:1.4.1.2]
Length=1113
Score = 594 bits (1531), Expect = 3e-169, Method: Compositional matrix adjust.
Identities = 304/528 (57%), Positives = 377/528 (71%), Gaps = 10/528 (1%)
Query 1 HRIDLAFRRGPHTEEFFSRFGDCLTMWGFYSQRKYVEPLANGSSIITTFVELLPEGILV- 59
+R+DLAFRRG HT+EFFSR GD +TMWGFYSQRKYVEPLANG SIITTF+E LPE L
Sbjct 311 YRMDLAFRRGLHTKEFFSRLGDSVTMWGFYSQRKYVEPLANGVSIITTFIEELPEDQLTD 370
Query 60 FPSMPIQERIDNLLSAVRLQFVMAKSKYADFARDRLLTLHEAAYAMNASHFALHFSGSVG 119
FP M + R++ L+ A+R+Q++M K+ + A++R LT+HEAAYA S F +HFSG+VG
Sbjct 371 FPEMSMSRRVELLVKAIRMQYIMQPCKFTELAQERRLTVHEAAYAWAVSKFIVHFSGTVG 430
Query 120 HSFPRIEEIVRQFGGKS-LTMQEIYELRTRLKTCPFSEDLIFKVVGENANIVKELYKEFA 178
+F IE++V+ FG + L+ QE+YELRTRLK PFSED + KVV + +++K LY+EF
Sbjct 431 PAFGAIEKMVKHFGSATHLSHQELYELRTRLKLPPFSEDTVLKVVDSHPDMIKRLYEEFQ 490
Query 179 SLHCPRVK----RVL--FKETNELKERILRLESSEAVAILLKFREFTKSILCTNFYMPFK 232
+H PR VL ++E LK I L+S +A ILLKFR F K I+ TNF+ K
Sbjct 491 EMHHPRAYAERGHVLKNWEEETSLKRDIQCLDSPDAPPILLKFRLFNKHIVRTNFWKDVK 550
Query 233 QGFAFRIRGSTLPSSDFPSTPCPIFFQIGGLAVGLHIRFAEVSRGGVRLVFSVGTAAHET 292
Q AFR+ S LP +D+P P I F +G G H RFA+V+RGGVR+V S T +++
Sbjct 551 QALAFRMDTSFLPEADYPERPYAILFTVGTNFTGFHARFADVARGGVRVVQSFTTQSYQR 610
Query 293 NRRSLLDEAYKLAFTQQFKNKDISEGGSKGIILLNKTQTLAEAKRQAPLAFKAYIDNMLD 352
NR + DE YKLA TQ KNKDI EGGSKG+ILL+KT + EA AFKAYID+MLD
Sbjct 611 NRDTAFDEVYKLASTQNLKNKDIPEGGSKGVILLSKTDSRDEANALTKSAFKAYIDSMLD 670
Query 353 LLLPHHDVDDGLGISEVWFLGPDENTGTGGLLDWAAQRAKERGSVWWKAFTTGKLIQHGG 412
+LL V D LG EV FLGPDE+TGTGGL+DWAA RAKER + +WKAFTTGKL GG
Sbjct 671 VLLTDPRVVDRLGKEEVCFLGPDEHTGTGGLMDWAAMRAKERNAWFWKAFTTGKLPAMGG 730
Query 413 IPHDRFGMTTASVEAYVKDAGIYNKLGLKEEEMTRIQTGGPDGDLGCNALLQTKSKTIAV 472
IPHD +GMTTAS+E Y+ GI K LKEEE+TR GGPDGDLG NALL++ +KT ++
Sbjct 731 IPHDTYGMTTASIETYIH--GILEKKNLKEEEVTRQLVGGPDGDLGSNALLKSNTKTTSI 788
Query 473 VDGSGVLYDPNGLDVGELHRLCSLRFEGKPTNAMLYDSSLLSPLGFKV 520
VDGSGVL+DP GLD+ EL RL RFEG T+AMLYD LLSP+GFKV
Sbjct 789 VDGSGVLHDPEGLDINELRRLAKRRFEGLQTSAMLYDEKLLSPMGFKV 836
> bbo:BBOV_III009680 17.m07840; Glutamate/Leucine/Phenylalanine/Valine
dehydrogenase family protein (EC:1.4.1.2); K00260 glutamate
dehydrogenase [EC:1.4.1.2]
Length=1025
Score = 452 bits (1164), Expect = 1e-126, Method: Compositional matrix adjust.
Identities = 241/522 (46%), Positives = 329/522 (63%), Gaps = 8/522 (1%)
Query 2 RIDLAFRRGPHTEEFFSRFGDCLTMWGFYSQRKYVEPLANGSSIITTFVELLPEGILVFP 61
RID+AFR+ +F+SRFGDC+T +G YS+ KYV+PL+N IIT F+ LP L P
Sbjct 247 RIDVAFRQDYVQSDFYSRFGDCVTYYGCYSKSKYVDPLSNNVCIITAFITSLPSTELDNP 306
Query 62 SMPIQERIDNLLSAVRLQFVMAKSKYADFARDRLLTLHEAAYAMNASHFALHFSGSVGHS 121
+ + +R ++++A+RL ++ S Y +RLL E AYA AS F HFSGSVG
Sbjct 307 DLSLLDRAHSIMTAIRLCGILPHSHYLSILTERLLNGSEMAYAYCASIFVEHFSGSVG-- 364
Query 122 FPRIEEIVRQFGGKSLTMQEIYELRTRLKTCPFSEDLIFKVVGENANIVKELYKEFASLH 181
P + I R + + E+Y++R++L + + IF+ V EN I+K+L+K F LH
Sbjct 365 -PHMALIERLATREQMAPSELYDIRSKLMIPSYLPNQIFEAVQENIPILKQLHKNFCILH 423
Query 182 CPRVK---RVLFKETNELKERILRLESSEAVAILLKFREFTKSILCTNFYMPFKQGFAFR 238
P + ++N LKE I L++ IL F F S L TNF++ K FAFR
Sbjct 424 NPDINPNGTNTDPDSNALKETIKTLDNQVHAKILSLFLTFNSSTLRTNFFVTEKSSFAFR 483
Query 239 IRGSTLPSSDFPSTPCPIFFQIGGLAVGLHIRFAEVSRGGVRLVFSVGTAAHETNRRSLL 298
+ S L +D+P TP I +G G HIRF+E+SRGG+R+V S A N+ +
Sbjct 484 LDPSFLSKNDYPETPYGIVMLMGPFFRGFHIRFSEISRGGIRVVQSFSHEAFTRNKLQVF 543
Query 299 DEAYKLAFTQQFKNKDISEGGSKGIILLNKTQTLAEAKRQAPLAFKAYIDNMLDLLLPHH 358
DEAY L++TQ KNKDI EGGSKG+ILL+K A+ +F Y+D +LD+++P
Sbjct 544 DEAYNLSYTQSLKNKDIPEGGSKGVILLDKAPNADLAQIYTRNSFMCYVDGLLDVMMPCA 603
Query 359 DVDDGLGISEVWFLGPDENTGTGGLLDWAAQRAKERGSVWWKAFTTGKLIQHGGIPHDRF 418
+ D L E++FLGPDE+TGTG L+DWAA AK RG +W++FTTGK GGIPHD +
Sbjct 604 QMVDHLHQDEIYFLGPDEHTGTGRLMDWAANHAKLRGFPFWRSFTTGKEPVMGGIPHDTY 663
Query 419 GMTTASVEAYVKDAGIYNKLGLKEEEMTRIQTGGPDGDLGCNALLQTKSKTIAVVDGSGV 478
GMTTAS+EAY+ + + N L EEE+TR TGGPDGDLG NALL +K+KT+ V+D SGV
Sbjct 664 GMTTASIEAYIHE--LLNIFHLNEEEVTRFLTGGPDGDLGSNALLCSKTKTLTVIDKSGV 721
Query 479 LYDPNGLDVGELHRLCSLRFEGKPTNAMLYDSSLLSPLGFKV 520
L+DP GLD+ EL RL + R +G PT+AM Y+ +LLS GFKV
Sbjct 722 LHDPEGLDINELQRLAANRLKGLPTSAMHYNEALLSDKGFKV 763
> tpv:TP04_0883 NAD-specific glutamate dehydrogenase (EC:1.4.1.2);
K00260 glutamate dehydrogenase [EC:1.4.1.2]
Length=1178
Score = 322 bits (826), Expect = 2e-87, Method: Compositional matrix adjust.
Identities = 169/318 (53%), Positives = 213/318 (66%), Gaps = 8/318 (2%)
Query 184 RVKRVLFKETNELKERILR----LESSEAVAILLKFREFTKSILCTNFYMPFKQGFAFRI 239
R K VL E L ++I+R LE+ E + ILL F +F L TNF++P K +FR
Sbjct 515 RGKSVL--EAESLAKKIIRTIKQLENLEEIDILLYFIKFNNHTLRTNFFVPNKISLSFRF 572
Query 240 RGSTLPSSDFPSTPCPIFFQIGGLAVGLHIRFAEVSRGGVRLVFSVGTAAHETNRRSLLD 299
L D+P P I IG +G HIRF+E+SRGGVR+V S A+ N+ + D
Sbjct 573 NCGFLSKLDYPKVPYGIALIIGPHFMGFHIRFSEISRGGVRVVQSFSEEAYTRNKLQIFD 632
Query 300 EAYKLAFTQQFKNKDISEGGSKGIILLNKTQTLAEAKRQAPLAFKAYIDNMLDLLLPHHD 359
EAY L+FTQ KNKDI EGGSKG+ILL KT T +A +F YID +LDL+LP+
Sbjct 633 EAYNLSFTQSLKNKDIPEGGSKGVILLEKTSTKYKADLYTRTSFMCYIDGILDLILPNKH 692
Query 360 VDDGLGISEVWFLGPDENTGTGGLLDWAAQRAKERGSVWWKAFTTGKLIQHGGIPHDRFG 419
+ D LG +++FLGPDE TGTGGL+DWA+Q AK +G +W++FTTGK Q GGIPHD +G
Sbjct 693 IVDLLGKDDIYFLGPDEFTGTGGLMDWASQYAKFKGLKYWRSFTTGKAPQLGGIPHDIYG 752
Query 420 MTTASVEAYVKDAGIYNKLGLKEEEMTRIQTGGPDGDLGCNALLQTKSKTIAVVDGSGVL 479
MTT S+EAYV GI NK GLKEEE+TR TGGPDGDLG NA+ + +KT+ V+D SGVL
Sbjct 753 MTTTSIEAYV--TGILNKYGLKEEEVTRFLTGGPDGDLGSNAIKVSNTKTLTVLDKSGVL 810
Query 480 YDPNGLDVGELHRLCSLR 497
+DPNGLD+ EL RL LR
Sbjct 811 HDPNGLDLNELRRLAFLR 828
Score = 124 bits (312), Expect = 6e-28, Method: Compositional matrix adjust.
Identities = 62/185 (33%), Positives = 106/185 (57%), Gaps = 4/185 (2%)
Query 2 RIDLAFRRGPHTEEFFSRFGDCLTMWGFYSQRKYVEPLANGSSIITTFVELLPEGILVFP 61
R+D+AF+RG + +SR D +T YS+ KY++PL+N I T+F+ LP
Sbjct 246 RLDIAFKRGHVHRDLYSRISDAITFHDLYSKSKYLDPLSNKVCIFTSFLTCLPPNQSKI- 304
Query 62 SMPIQERIDNLLSAVRLQFVMAKSKYADFARDRLLTLHEAAYAMNASHFALHFSGSVGHS 121
+PI+ER+ +L+ ++ L ++ SK + DR+L+ +E+ Y+ S F HFSGSVG
Sbjct 305 DLPIKERMSSLIDSIVLSSIVPSSKISILNVDRVLSFYESTYSYCVSTFIQHFSGSVG-- 362
Query 122 FPRIEEIVRQFGGKSLTMQEIYELRTRLKTCPFSEDLIFKVVGENANIVKELYKEFASLH 181
P + + ++ E++E++++LK P++ I+ + N IVK LYK F LH
Sbjct 363 -PYVSTVDNMAKNNQVSQSELHEIKSKLKIQPYTPLQIYTAISNNPEIVKMLYKHFEILH 421
Query 182 CPRVK 186
P+++
Sbjct 422 NPKLQ 426
> sce:YDL215C GDH2; Gdh2p (EC:1.4.1.2); K00260 glutamate dehydrogenase
[EC:1.4.1.2]
Length=1092
Score = 266 bits (680), Expect = 1e-70, Method: Compositional matrix adjust.
Identities = 178/544 (32%), Positives = 276/544 (50%), Gaps = 38/544 (6%)
Query 2 RIDLAFRRGPHTEEFFSRFGDCLTMWGFYSQRKYVEPLANGSSIITTFVELLPEGILVFP 61
R+ +A++R T+ ++S + + Y+E I F L E +
Sbjct 288 RLLVAYKRFT-TKRYYSALNSLFHYYKLKPSKFYLESFNVKDDDIIIFSVYLNENQQLED 346
Query 62 SM--PIQERIDNLLSAVRLQFVMAKSKYADFARDRLLTLHEAAYAMNASHFALHFSGSVG 119
+ ++ + + L + + + + + + R + EA YA + F HF +G
Sbjct 347 VLLHDVEAALKQVEREASLLYAIPNNSFHEVYQRRQFSPKEAIYAHIGAIFINHFVNRLG 406
Query 120 HSFPRIEEIVRQFGGKSLTMQEIYELRTRLKTCPFSEDLIFKVVGENANIVKELYKEFAS 179
+ + + + ++ + L+ +L+ ++ I ++ ++ I+ +LYK FA
Sbjct 407 SDYQNLLSQITIKRNDTTLLEIVENLKRKLRNETLTQQTIINIMSKHYTIISKLYKNFAQ 466
Query 180 LHC--------------PRVKRVL-FKETNELKERILRL--ESSEAVAILLKFREFTKSI 222
+H R+++V FK E + + + S + IL F KSI
Sbjct 467 IHYYHNSTKDMEKTLSFQRLEKVEPFKNDQEFEAYLNKFIPNDSPDLLILKTLNIFNKSI 526
Query 223 LCTNFYMPFKQGFAFRIRGSTLPSS-DFPSTPCPIFFQIGGLAVGLHIRFAEVSRGGVRL 281
L TNF++ K +FR+ S + + ++P TP IFF +G G HIRF +++RGG+R+
Sbjct 527 LKTNFFITRKVAISFRLDPSLVMTKFEYPETPYGIFFVVGNTFKGFHIRFRDIARGGIRI 586
Query 282 VFSVGTAAHETNRRSLLDEAYKLAFTQQFKNKDISEGGSKGIILLNKTQTLAEAKRQAPL 341
V S ++ N ++++DE Y+LA TQQ KNKDI EGGSKG+ILLN L E Q +
Sbjct 587 VCSRNQDIYDLNSKNVIDENYQLASTQQRKNKDIPEGGSKGVILLNP--GLVEHD-QTFV 643
Query 342 AFKAYIDNMLDLLLPHHDVDDGLGI---SEVWFLGPDENTGTGGLLDWAAQRAKERGSVW 398
AF Y+D M+D+L+ ++ + + E+ F GPDE GT G +DWA A+ R W
Sbjct 644 AFSQYVDAMIDILINDPLKENYVNLLPKEEILFFGPDE--GTAGFVDWATNHARVRNCPW 701
Query 399 WKAFTTGKLIQHGGIPHDRFGMTTASVEAYVKDAGIYNKLGLKEEEMTRIQTGGPDGDLG 458
WK+F TGK GGIPHD +GMT+ V AYV IY L L + + QTGGPDGDLG
Sbjct 702 WKSFLTGKSPSLGGIPHDEYGMTSLGVRAYVN--KIYETLNLTNSTVYKFQTGGPDGDLG 759
Query 459 CNALLQTKSKT--IAVVDGSGVLYDPNGLDVGELHRLCSLRFEGKPTNAMLYDSSLLSPL 516
N +L + +A++DGSGVL DP GLD E LC L E K + +D+S LS
Sbjct 760 SNEILLSSPNECYLAILDGSGVLCDPKGLDKDE---LCRLAHERKMISD--FDTSKLSNN 814
Query 517 GFKV 520
GF V
Sbjct 815 GFFV 818
> pfa:PF08_0132 glutamate dehydrogenase, putative (EC:1.4.1.2);
K00260 glutamate dehydrogenase [EC:1.4.1.2]
Length=1397
Score = 145 bits (365), Expect = 5e-34, Method: Compositional matrix adjust.
Identities = 77/157 (49%), Positives = 106/157 (67%), Gaps = 6/157 (3%)
Query 368 EVWFLGPDENTGTGGLLDWAAQRAKERGSVWWKAFTTGKLIQHGGIPHDRFGMTTASVEA 427
++ FLGPDENTG+ L+DWA AK+R +WK F+TGKL ++GG+PHD +GMTT +E
Sbjct 902 DLIFLGPDENTGSDQLMDWACIIAKKRKYPYWKTFSTGKLRKNGGVPHDMYGMTTLGIET 961
Query 428 YVKDAGIYNKLGLKEEEMTRIQTGGPDGDLGCNALLQTKSKTIAVVDGSGVLYDPNGLDV 487
Y+ + + KL +KEE ++R GGPDGDLG NA+LQ+K+K I+++DGSG+LYD GL+
Sbjct 962 YI--SKLCEKLNIKEESISRSLVGGPDGDLGSNAILQSKTKIISIIDGSGILYDKQGLNK 1019
Query 488 GELHRLCSLRFEGKPTNAM----LYDSSLLSPLGFKV 520
EL RL R + A+ LYD S GFK+
Sbjct 1020 EELIRLAKRRNNKDKSKAITCCTLYDEKYFSKDGFKI 1056
Score = 124 bits (310), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 69/160 (43%), Positives = 95/160 (59%), Gaps = 1/160 (0%)
Query 197 KERILRLESSEAVAILLKFREFTKSILCTNFYMPFKQGFAFRIRGSTLPSSDFPSTPCPI 256
K+ I +E + IL F F K L TNF++ K A G+ L S + + P I
Sbjct 515 KDIIDEIEDNHDKKILQYFYMFEKYALKTNFFLTHKISLAVAFDGALLKDSIYEAQPYSI 574
Query 257 FFQIGGLAVGLHIRFAEVSRGGVRLVFSVGTAAHETNRRSLLDEAYKLAFTQQFKNKDIS 316
+G VG HIRF+++SRGGVR+V S ++ N +L DEAY LA+TQ FKNKDI
Sbjct 575 IMILGLHFVGFHIRFSKISRGGVRIVISNNVNSYMHNSDNLFDEAYNLAYTQNFKNKDIP 634
Query 317 EGGSKGIILLN-KTQTLAEAKRQAPLAFKAYIDNMLDLLL 355
EGGSKGIILL+ +A K L+F +Y++++LDLL+
Sbjct 635 EGGSKGIILLDADVCNVANTKYIKNLSFYSYVNSILDLLI 674
Score = 63.5 bits (153), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 42/189 (22%), Positives = 84/189 (44%), Gaps = 16/189 (8%)
Query 5 LAFRRGPHTEEFFSRFGDCLTMWGFYSQRKYVEPLANGSSIITTFVELLPEGILVFP--S 62
+A +R FS GDCL M +S KYVEPL NG +I V+++ +
Sbjct 248 IAVKRSNVISSIFSLIGDCLNMHRCFSYSKYVEPLKNGVLLIILNVKVIVNNEMEREKQK 307
Query 63 MPIQERIDNLLSAVRLQFVMAKSKYADFARDRLLTLHEAAYAMNASHFALHFSGSVGHSF 122
+ ++++I ++ +++ + SK+ + R T E+AY F FS + S+
Sbjct 308 LDLKDKIYKVVKSLKTLCLFNDSKFIQLSVKRTFTAQESAYLFMIIKFITFFSTNTLSSY 367
Query 123 PRIEEI--VRQFGGKSL------------TMQEIYELRTRLKTCPFSEDLIFKVVGENAN 168
+E +R + + + ++Y ++ +LK+ ++++ I + N
Sbjct 368 KNVEHALNLRNYNNNIMDTTTNSSSSPSSVLNDVYIIKEKLKSSKYTKEEILRCAQSNVR 427
Query 169 IVKELYKEF 177
+K L+ F
Sbjct 428 TIKMLFANF 436
> dre:373092 glud1b, MGC192851, cb719, glud1, wu:fb16e02, wu:fb58f12,
wu:fe37f03, wu:fj43f02, zgc:192851, zgc:55630; glutamate
dehydrogenase 1b (EC:1.4.1.3); K00261 glutamate dehydrogenase
(NAD(P)+) [EC:1.4.1.3]
Length=542
Score = 39.7 bits (91), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 64/267 (23%), Positives = 106/267 (39%), Gaps = 54/267 (20%)
Query 268 HIRFAEVSRGGVRLVFSVGTAAHETNRRSLLDEAYKLAFTQQFKNK--DISEGGSKGIIL 325
H + +GG+R V +DE LA +K D+ GG+K +
Sbjct 123 HSQHRTPCKGGIRYSMDVS-----------VDEVKALASLMTYKCAVVDVPFGGAKAGVK 171
Query 326 LNKTQTLAEAKRQAPLAFKAYIDNMLDLLLPHHDVDDGLGISEVWFLGP-------DENT 378
+N + Y DN L+ + ++ +++ F+GP D +T
Sbjct 172 INP---------------RNYSDNELEKITRRFTIE----LAKKGFIGPGIDVPAPDMST 212
Query 379 GTGGLLDWAAQRAKE---RGSVWWKAFTTGKLIQHGGIPHDRFGMTTASV----EAYVKD 431
G + W A + A TGK I GGI H R T V E ++ +
Sbjct 213 GER-EMSWIADTYANTIAHTDINAHACVTGKPISQGGI-HGRISATGRGVFHGIENFINE 270
Query 432 AGIYNKLGLK---EEEMTRIQTGGPDGDLGCNALLQTKSKTIAVVDGSGVLYDPNGLDVG 488
A +KLGL ++ IQ G G L + +K + + + G +++PNG+D
Sbjct 271 ASYMSKLGLTPGFADKTFIIQGFGNVGLHSMRYLHRYGAKCVGIAEIDGSIWNPNGMDPK 330
Query 489 EL--HRLCSLRFEGKPTNAMLYDSSLL 513
EL ++L G P N+ Y+ ++L
Sbjct 331 ELEDYKLQHGTIVGFP-NSQPYEGNIL 356
> hsa:2747 GLUD2, GDH2, GLUDP1; glutamate dehydrogenase 2 (EC:1.4.1.3);
K00261 glutamate dehydrogenase (NAD(P)+) [EC:1.4.1.3]
Length=558
Score = 38.5 bits (88), Expect = 0.069, Method: Compositional matrix adjust.
Identities = 57/208 (27%), Positives = 87/208 (41%), Gaps = 32/208 (15%)
Query 298 LDEAYKLAFTQQFKNK--DISEGGSKGIILLN-KTQTLAEAKR-----QAPLAFKAYIDN 349
+DE LA +K D+ GG+K + +N K T E ++ LA K +I
Sbjct 158 VDEVKALASLMTYKCAVVDVPFGGAKAGVKINPKNYTENELEKITRRFTMELAKKGFIGP 217
Query 350 MLDLLLPHHDVDDGLGISEVWFLGPDENTGTGGLLDWAAQRAKERGSVWWKAFTTGKLIQ 409
+D+ P D G E+ ++ D T G D A A TGK I
Sbjct 218 GVDVPAP----DMNTGEREMSWIA-DTYASTIGHYDINAH-----------ACVTGKPIS 261
Query 410 HGGIPHDRFGMTTASV----EAYVKDAGIYNKLGLK---EEEMTRIQTGGPDGDLGCNAL 462
GGI H R T V E ++ +A + LG+ ++ +Q G G L
Sbjct 262 QGGI-HGRISATGRGVFHGIENFINEASYMSILGMTPGFRDKTFVVQGFGNVGLHSMRYL 320
Query 463 LQTKSKTIAVVDGSGVLYDPNGLDVGEL 490
+ +K IAV + G +++P+G+D EL
Sbjct 321 HRFGAKCIAVGESDGSIWNPDGIDPKEL 348
> xla:446858 glud1, MGC80801; glutamate dehydrogenase 1 (EC:1.4.1.3);
K00261 glutamate dehydrogenase (NAD(P)+) [EC:1.4.1.3]
Length=540
Score = 38.5 bits (88), Expect = 0.071, Method: Compositional matrix adjust.
Identities = 62/233 (26%), Positives = 99/233 (42%), Gaps = 35/233 (15%)
Query 298 LDEAYKLAFTQQFKNK--DISEGGSKGIILLN-KTQTLAEAKR-----QAPLAFKAYIDN 349
+DE LA +K D+ GG+K + +N + + AE ++ LA K +I
Sbjct 140 VDEVKALASLMTYKCAVVDVPFGGAKAGVKINPRNFSDAELEKITRRFTIELAKKGFIGP 199
Query 350 MLDLLLPHHDVDDGLGISEVWFLGPDENTGTGGLLDWAAQRAKERGSVWWKAFTTGKLIQ 409
+D+ P D G E+ ++ D T G D A A TGK I
Sbjct 200 GIDVPAP----DMSTGEREMSWIA-DTYANTIGHTDINAH-----------ACVTGKPIS 243
Query 410 HGGIPHDRFGMTTASV----EAYVKDAGIYNKLGLKE---EEMTRIQTGGPDGDLGCNAL 462
GGI H R T V E ++ +A ++LG+ ++ IQ G G L
Sbjct 244 QGGI-HGRISATGRGVFHGIENFINEASYMSQLGMTPGFGDKTFVIQGFGNVGLHSMRYL 302
Query 463 LQTKSKTIAVVDGSGVLYDPNGLDVGEL--HRLCSLRFEGKPTNAMLYDSSLL 513
+ +K + + + G +++PNG+D EL ++L G P A YD ++L
Sbjct 303 HRFGAKCVGIGEIDGTIWNPNGIDPKELEDYKLQHGTIVGFP-KAQPYDGNIL 354
> hsa:2746 GLUD1, GDH, GDH1, GLUD, MGC132003; glutamate dehydrogenase
1 (EC:1.4.1.3); K00261 glutamate dehydrogenase (NAD(P)+)
[EC:1.4.1.3]
Length=558
Score = 37.7 bits (86), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 57/208 (27%), Positives = 87/208 (41%), Gaps = 32/208 (15%)
Query 298 LDEAYKLAFTQQFKNK--DISEGGSKGIILLN-KTQTLAEAKR-----QAPLAFKAYIDN 349
+DE LA +K D+ GG+K + +N K T E ++ LA K +I
Sbjct 158 VDEVKALASLMTYKCAVVDVPFGGAKAGVKINPKNYTDNELEKITRRFTMELAKKGFIGP 217
Query 350 MLDLLLPHHDVDDGLGISEVWFLGPDENTGTGGLLDWAAQRAKERGSVWWKAFTTGKLIQ 409
+D+ P D G E+ ++ D T G D A A TGK I
Sbjct 218 GIDVPAP----DMSTGEREMSWIA-DTYASTIGHYDINAH-----------ACVTGKPIS 261
Query 410 HGGIPHDRFGMTTASV----EAYVKDAGIYNKLGLKE---EEMTRIQTGGPDGDLGCNAL 462
GGI H R T V E ++ +A + LG+ ++ +Q G G L
Sbjct 262 QGGI-HGRISATGRGVFHGIENFINEASYMSILGMTPGFGDKTFVVQGFGNVGLHSMRYL 320
Query 463 LQTKSKTIAVVDGSGVLYDPNGLDVGEL 490
+ +K IAV + G +++P+G+D EL
Sbjct 321 HRFGAKCIAVGESDGSIWNPDGIDPKEL 348
> cpv:cgd5_1320 actin-like protein
Length=367
Score = 33.9 bits (76), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 24/51 (47%), Gaps = 1/51 (1%)
Query 116 GSVGHSFPRIEEIVRQFGGKSLTMQEIYELRTRLKTCPFSEDLIFKVVGEN 166
S+ H FP VR+ K T +IY L CPFSE L+ V EN
Sbjct 8 NSLRHCFPNCIGKVRR-KDKIYTSDQIYSLNEYFSYCPFSEGLLIDVSMEN 57
> pfa:PF14_0164 NADP-specific glutamate dehydrogenase; K00262
glutamate dehydrogenase (NADP+) [EC:1.4.1.4]
Length=470
Score = 33.1 bits (74), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 28/99 (28%), Positives = 46/99 (46%), Gaps = 4/99 (4%)
Query 404 TGKLIQHGGIPHDRFGMTTASVEAYVKDAGIYNKLGLKEEEMTRIQTGGPDGDLGC-NAL 462
TGK ++ GG + R T + +V + + L + E+ T + +G + L C L
Sbjct 215 TGKNVKWGG-SNLRVEATGYGLVYFVLE--VLKSLNIPVEKQTAVVSGSGNVALYCVQKL 271
Query 463 LQTKSKTIAVVDGSGVLYDPNGLDVGELHRLCSLRFEGK 501
L K + + D +G +Y+PNG L L L+ E K
Sbjct 272 LHLNVKVLTLSDSNGYVYEPNGFTHENLEFLIDLKEEKK 310
> tgo:TGME49_021540 hypothetical protein
Length=765
Score = 33.1 bits (74), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 23/87 (26%), Positives = 36/87 (41%), Gaps = 25/87 (28%)
Query 447 RIQTGGPDGDLGCNALLQTKSKTIA-----------VVDGSG--------------VLYD 481
R+ G P G + +LL K K +A ++D S +LY+
Sbjct 577 RLYCGRPPGPITNKSLLDVKGKIVAGRQRGLNADYLILDASAWTFLHQRYGGGPPIILYN 636
Query 482 PNGLDVGELHRLCSLRFEGKPTNAMLY 508
PN + G+L+ C + FEG+ N Y
Sbjct 637 PNPCEFGDLYEGCFVTFEGEWQNGHPY 663
> tpv:TP01_1227 hypothetical protein; K03850 alpha-1,2-glucosyltransferase
[EC:2.4.1.-]
Length=780
Score = 33.1 bits (74), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 26/75 (34%), Positives = 38/75 (50%), Gaps = 6/75 (8%)
Query 123 PRIEEIVRQFGGKSLTMQEI--YELRTRLKTCPFSEDLIFKVVGENAN-IVKELYKEFAS 179
P+ EE+ + GK +EI Y+ + T P +ED +K+V +N V EL K A
Sbjct 661 PKPEEV--EGKGKRKICKEIKFYKKDEKGNTVPMTEDE-YKIVKDNDQRTVYELTKNIAE 717
Query 180 LHCPRVKRVLFKETN 194
LHC K + + TN
Sbjct 718 LHCDGEKVYIHRSTN 732
> tgo:TGME49_093660 hypothetical protein
Length=889
Score = 32.3 bits (72), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 28/95 (29%), Positives = 44/95 (46%), Gaps = 15/95 (15%)
Query 64 PIQERIDNLLSAVRLQFVMAKSKYADFARDRLLTLHEAAYAMNASHFALHFSGSVGHSFP 123
P+Q D +L+AV QF +A S ++ A LLT ++ LHF + +
Sbjct 524 PLQSIPDRVLTAVASQFEVASSSFSLSATVTLLTAFQSR--------GLHFPTCIAIAVA 575
Query 124 RIEEIVRQFGG----KSLTMQEIYELRTRLKTCPF 154
R+ + +R +LT+QEI L L+ C F
Sbjct 576 RVTQHLRVAANVRQRAALTIQEIGHL---LEACTF 607
> dre:559941 si:ch211-1o14.5
Length=1378
Score = 32.3 bits (72), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 33/119 (27%), Positives = 54/119 (45%), Gaps = 20/119 (16%)
Query 117 SVGHSFPRIEEIVRQFGGKSLTMQEIYELRTRLKTC---PFSEDLIFKVVG--------- 164
++G FP ++E++ Q LT +R R ++C S++ + VG
Sbjct 574 TIGRIFPCLDELL-QLHRDFLTA-----MRERRQSCVQNNSSKNFLIHRVGDIFLQQFSQ 627
Query 165 ENANIVKELYKEFASLHCPRVKRVLFKETNELKERILRLESSEAVAILLKFREFTKSIL 223
ENA +K++Y EF S H V FKE + +R ++ L+K RE + IL
Sbjct 628 ENAEKMKQVYGEFCSHHTEAVS--YFKELQQQNKRFQLFIKQQSSNSLVKRREIPECIL 684
> cpv:cgd7_1400 signal peptide containing protein
Length=730
Score = 32.0 bits (71), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 26/46 (56%), Gaps = 2/46 (4%)
Query 429 VKDAGIYNKLGLKEEEMTRIQTG--GPDGDLGCNALLQTKSKTIAV 472
VKD G + L + + E+T+I G GPDG++ C +S I++
Sbjct 680 VKDLGQFLLLSIDKMEVTKISKGGYGPDGNISCVQFFANESDDISI 725
> sce:YKL014C URB1, NPA1; Nucleolar protein required for the normal
accumulation of 25S and 5.8S rRNAs, associated with the
27SA2 pre-ribosomal particle; proposed to be involved in the
biogenesis of the 60S ribosomal subunit; K14861 nucleolar
pre-ribosomal-associated protein 1
Length=1764
Score = 31.6 bits (70), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 32/131 (24%), Positives = 56/131 (42%), Gaps = 21/131 (16%)
Query 86 KYADFARDRLLTLHEAAYAMNASHFALHFSGSV----GHSFPRIEEIVRQ--FGGKSL-- 137
+Y +FA +L A M S F + G+V GH EI+++ F G+++
Sbjct 844 RYINFALPKLAIEKRNALLMKKSRFMCNLIGAVCFETGHQLVEFREIIQKVVFSGENVEE 903
Query 138 --TMQEIYE----------LRTRLKTCPFSEDLIFKVVGENA-NIVKELYKEFASLHCPR 184
E+Y+ + L T + L+ E+ NI+++L+ E ++
Sbjct 904 YANYNELYQKEDVNAFLTSVSEYLSTSALTSLLMCSTKLESTRNILQKLFNEGKTIKISL 963
Query 185 VKRVLFKETNE 195
VK +L K NE
Sbjct 964 VKNILNKAANE 974
Lambda K H
0.322 0.139 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 25318618468
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40