bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0336_orf2
Length=153
Score E
Sequences producing significant alignments: (Bits) Value
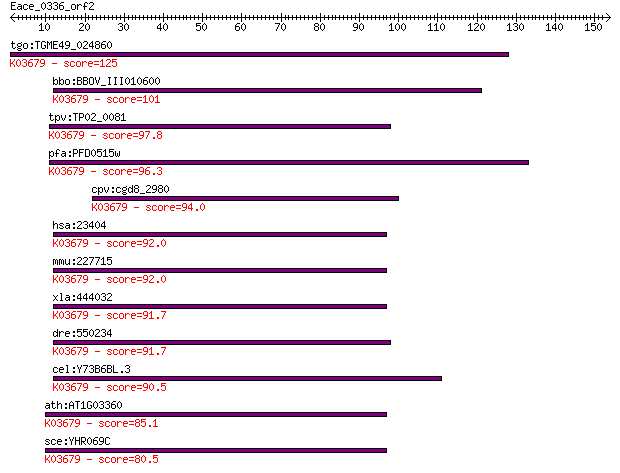
tgo:TGME49_024860 exosome complex exonuclease, putative ; K036... 125 4e-29
bbo:BBOV_III010600 17.m07916; hypothetical protein; K03679 exo... 101 1e-21
tpv:TP02_0081 hypothetical protein; K03679 exosome complex com... 97.8 1e-20
pfa:PFD0515w exosome complex exonuclease rrp4, putative; K0367... 96.3 3e-20
cpv:cgd8_2980 hypothetical protein ; K03679 exosome complex co... 94.0 2e-19
hsa:23404 EXOSC2, RRP4, Rrp4p, hRrp4p, p7; exosome component 2... 92.0 6e-19
mmu:227715 Exosc2, MGC30456, Rrp4; exosome component 2; K03679... 92.0 7e-19
xla:444032 exosc2, MGC82596; exosome component 2; K03679 exoso... 91.7 8e-19
dre:550234 exosc2, fa97b01, wu:fa97b01, zgc:110117; exosome co... 91.7 9e-19
cel:Y73B6BL.3 exos-2; EXOSome (multiexonuclease complex) compo... 90.5 2e-18
ath:AT1G03360 ATRRP4; ATRRP4 (ARABIDOPSIS THALIANA RIBOSOMAL R... 85.1 8e-17
sce:YHR069C RRP4; Exosome non-catalytic core component; involv... 80.5 2e-15
> tgo:TGME49_024860 exosome complex exonuclease, putative ; K03679
exosome complex component RRP4
Length=353
Score = 125 bits (315), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 60/127 (47%), Positives = 88/127 (69%), Gaps = 2/127 (1%)
Query 1 LSVAAVCLDVQRRRDDADVLIMRSMFQSADLLCCEVQKAQNDGTILLHTRSARYGRLQNG 60
L++AA+ L RRR D D+L M++ F D++CCEVQ+ + DG ILLHTRS RYGRL NG
Sbjct 165 LALAAISLPEHRRRLDEDMLEMQNFFVVGDVICCEVQRVRADGQILLHTRSTRYGRLMNG 224
Query 61 IGLRVSPQLIKRQSKHIGHLQCGLQLILGVNGFLWISVPGGAAGGSGDGVSTPSQQQQHE 120
+ L V+PQ I+RQS HI L CG+Q++LG+NG++WIS+P + + D ++ Q HE
Sbjct 225 VFLAVAPQQIQRQSHHIVQLSCGVQVVLGLNGYIWISLPMKTS--AKDTMNYAHVQTTHE 282
Query 121 EQQQQQQ 127
+ ++ +
Sbjct 283 KVSKEMR 289
> bbo:BBOV_III010600 17.m07916; hypothetical protein; K03679 exosome
complex component RRP4
Length=262
Score = 101 bits (251), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 50/109 (45%), Positives = 71/109 (65%), Gaps = 6/109 (5%)
Query 12 RRRDDADVLIMRSMFQSADLLCCEVQKAQNDGTILLHTRSARYGRLQNGIGLRVSPQLIK 71
RR+ D DV MR++F ++ CEVQ+ +GT+LL TR+++YGRL NGI ++V P L+
Sbjct 102 RRKLDEDVYEMRNLFGIGHVISCEVQRVSPNGTVLLQTRTSKYGRLNNGILVKVKPNLML 161
Query 72 RQSKHIGHLQCGLQLILGVNGFLWISVPGGAAGGSGDGVSTPSQQQQHE 120
RQSKH+ L CG+ +ILG NGF+W+ G SGD +T Q H+
Sbjct 162 RQSKHMQELSCGISMILGCNGFIWL----GPIQKSGD--ATIHAQVYHD 204
> tpv:TP02_0081 hypothetical protein; K03679 exosome complex component
RRP4
Length=263
Score = 97.8 bits (242), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 36/87 (41%), Positives = 64/87 (73%), Gaps = 0/87 (0%)
Query 11 QRRRDDADVLIMRSMFQSADLLCCEVQKAQNDGTILLHTRSARYGRLQNGIGLRVSPQLI 70
RR+ D D+ M ++F D++ CE+Q+ GTI+L TR+++YG+L+NGI +++ P L+
Sbjct 101 NRRKIDEDIYEMSNLFNVNDIVSCEIQRISPTGTIMLQTRTSKYGKLENGILVKIKPNLV 160
Query 71 KRQSKHIGHLQCGLQLILGVNGFLWIS 97
R+ KHI ++CG+++ILG NG++W++
Sbjct 161 IRKGKHIYDMECGVRIILGCNGYIWLT 187
> pfa:PFD0515w exosome complex exonuclease rrp4, putative; K03679
exosome complex component RRP4
Length=338
Score = 96.3 bits (238), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 45/122 (36%), Positives = 71/122 (58%), Gaps = 5/122 (4%)
Query 11 QRRRDDADVLIMRSMFQSADLLCCEVQKAQNDGTILLHTRSARYGRLQNGIGLRVSPQLI 70
QR R DV+ M +++ D++ CE+Q+ DGTI+LHTRS+ YG+L NG+ + V LI
Sbjct 148 QRIRLYNDVINMIHIYKPDDIIACEIQRISTDGTIILHTRSSIYGKLSNGVLIIVPQTLI 207
Query 71 KRQSKHIGHLQCGLQLILGVNGFLWISVPGGAAGGSGDGVSTPSQQQQHEEQQQQQQQQQ 130
Q KHI C +Q+ILG+NG++WIS P + + P+ Q+ E+ + +
Sbjct 208 HNQKKHIFVFPCNVQIILGMNGYIWISSPIKKSKDTN-----PNSVDQNIEENKFEDVDH 262
Query 131 QS 132
+
Sbjct 263 TT 264
> cpv:cgd8_2980 hypothetical protein ; K03679 exosome complex
component RRP4
Length=144
Score = 94.0 bits (232), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 40/78 (51%), Positives = 62/78 (79%), Gaps = 0/78 (0%)
Query 22 MRSMFQSADLLCCEVQKAQNDGTILLHTRSARYGRLQNGIGLRVSPQLIKRQSKHIGHLQ 81
M+S+ D++C EVQ+ Q++G LHTRSA+YG+L NG+ ++V +LI+RQ++HI L+
Sbjct 1 MKSILDEGDVICTEVQRVQSEGICHLHTRSAKYGKLANGMVVKVPNKLIQRQAQHIVKLK 60
Query 82 CGLQLILGVNGFLWISVP 99
G+QLILG+NG++WIS+P
Sbjct 61 YGIQLILGLNGYVWISLP 78
> hsa:23404 EXOSC2, RRP4, Rrp4p, hRrp4p, p7; exosome component
2; K03679 exosome complex component RRP4
Length=293
Score = 92.0 bits (227), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 43/85 (50%), Positives = 54/85 (63%), Gaps = 0/85 (0%)
Query 12 RRRDDADVLIMRSMFQSADLLCCEVQKAQNDGTILLHTRSARYGRLQNGIGLRVSPQLIK 71
RRR D L MR Q DL+ EVQ +DG + LHTRS +YG+L G+ ++VSP L+K
Sbjct 121 RRRSAEDELAMRGFLQEGDLISAEVQAVFSDGAVSLHTRSLKYGKLGQGVLVQVSPSLVK 180
Query 72 RQSKHIGHLQCGLQLILGVNGFLWI 96
RQ H L CG +ILG NGF+WI
Sbjct 181 RQKTHFHDLPCGASVILGNNGFIWI 205
> mmu:227715 Exosc2, MGC30456, Rrp4; exosome component 2; K03679
exosome complex component RRP4
Length=293
Score = 92.0 bits (227), Expect = 7e-19, Method: Compositional matrix adjust.
Identities = 43/85 (50%), Positives = 54/85 (63%), Gaps = 0/85 (0%)
Query 12 RRRDDADVLIMRSMFQSADLLCCEVQKAQNDGTILLHTRSARYGRLQNGIGLRVSPQLIK 71
RRR D L MR Q DL+ EVQ +DG + LHTRS +YG+L G+ ++VSP L+K
Sbjct 121 RRRSAEDELAMRGFLQEGDLISAEVQAVFSDGAVSLHTRSLKYGKLGQGVLVQVSPSLVK 180
Query 72 RQSKHIGHLQCGLQLILGVNGFLWI 96
RQ H L CG +ILG NGF+WI
Sbjct 181 RQKTHFHDLPCGASVILGNNGFIWI 205
> xla:444032 exosc2, MGC82596; exosome component 2; K03679 exosome
complex component RRP4
Length=293
Score = 91.7 bits (226), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 43/85 (50%), Positives = 56/85 (65%), Gaps = 0/85 (0%)
Query 12 RRRDDADVLIMRSMFQSADLLCCEVQKAQNDGTILLHTRSARYGRLQNGIGLRVSPQLIK 71
RRR D L MRS Q DL+ EVQ +DG + LHTRS +YG+L G+ ++VSP LIK
Sbjct 121 RRRSAEDELAMRSYLQEGDLISAEVQAVYSDGALSLHTRSLKYGKLGQGLLVQVSPSLIK 180
Query 72 RQSKHIGHLQCGLQLILGVNGFLWI 96
R+ H +L CG +ILG NGF+W+
Sbjct 181 RRKTHFHNLTCGASVILGNNGFIWL 205
> dre:550234 exosc2, fa97b01, wu:fa97b01, zgc:110117; exosome
component 2; K03679 exosome complex component RRP4
Length=254
Score = 91.7 bits (226), Expect = 9e-19, Method: Compositional matrix adjust.
Identities = 43/86 (50%), Positives = 55/86 (63%), Gaps = 0/86 (0%)
Query 12 RRRDDADVLIMRSMFQSADLLCCEVQKAQNDGTILLHTRSARYGRLQNGIGLRVSPQLIK 71
RRR D L MR Q DL+ EVQ +DG + LHTRS +YG+L G+ + VSP L+K
Sbjct 82 RRRSTQDELAMRDYLQEGDLISAEVQSIFSDGALSLHTRSLKYGKLGQGVLVYVSPSLVK 141
Query 72 RQSKHIGHLQCGLQLILGVNGFLWIS 97
RQ H +L CG LILG NG++W+S
Sbjct 142 RQKTHFHNLPCGASLILGNNGYVWLS 167
> cel:Y73B6BL.3 exos-2; EXOSome (multiexonuclease complex) component
family member (exos-2); K03679 exosome complex component
RRP4
Length=303
Score = 90.5 bits (223), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 42/101 (41%), Positives = 63/101 (62%), Gaps = 2/101 (1%)
Query 12 RRRDDADVLIMRSMFQSADLLCCEVQKAQNDGTILLHTRSARYGRLQNGIGLRVSPQLIK 71
RR+D D M ++ +L+C EVQ+ Q+DGT++LHTR+ +YG+LQ GI ++V P LIK
Sbjct 127 RRKDVEDEEKMSEFLKNGELICAEVQQVQHDGTLMLHTRNNKYGKLQQGILIKVPPHLIK 186
Query 72 RQSKHIGHLQCGLQLILGVNGFLWI--SVPGGAAGGSGDGV 110
+ KH L G+ +I+G NG +W+ S+P G V
Sbjct 187 KSKKHFHTLPYGMAVIIGCNGSVWVTPSLPETTLEEDGSHV 227
> ath:AT1G03360 ATRRP4; ATRRP4 (ARABIDOPSIS THALIANA RIBOSOMAL
RNA PROCESSING 4); RNA binding / exonuclease; K03679 exosome
complex component RRP4
Length=322
Score = 85.1 bits (209), Expect = 8e-17, Method: Compositional matrix adjust.
Identities = 38/88 (43%), Positives = 59/88 (67%), Gaps = 1/88 (1%)
Query 10 VQRRRDDADVLIMRSMFQSADLLCCEVQKAQNDGTILLHTRSARYGRLQNGIGLRVSPQL 69
+QRRR D L MR++F D++C EV+ Q+DG++ L RS +YG+L+ G L+V P L
Sbjct 132 IQRRRTSVDELNMRNIFVEHDVVCAEVRNFQHDGSLQLQARSQKYGKLEKGQLLKVDPYL 191
Query 70 IKRQSKHIGHLQC-GLQLILGVNGFLWI 96
+KR H +++ G+ LI+G NGF+W+
Sbjct 192 VKRSKHHFHYVESLGIDLIIGCNGFIWV 219
> sce:YHR069C RRP4; Exosome non-catalytic core component; involved
in 3'-5' RNA processing and degradation in both the nucleus
and the cytoplasm; predicted to contain RNA binding domains;
has similarity to human hRrp4p (EXOSC2) (EC:3.1.13.-);
K03679 exosome complex component RRP4
Length=359
Score = 80.5 bits (197), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 37/87 (42%), Positives = 55/87 (63%), Gaps = 0/87 (0%)
Query 10 VQRRRDDADVLIMRSMFQSADLLCCEVQKAQNDGTILLHTRSARYGRLQNGIGLRVSPQL 69
+ RR+ ++D L MRS + DLL EVQ DG+ LHTRS +YG+L+NG+ +V L
Sbjct 147 ILRRKSESDELQMRSFLKEGDLLNAEVQSLFQDGSASLHTRSLKYGKLRNGMFCQVPSSL 206
Query 70 IKRQSKHIGHLQCGLQLILGVNGFLWI 96
I R H +L + ++LGVNG++W+
Sbjct 207 IVRAKNHTHNLPGNITVVLGVNGYIWL 233
Lambda K H
0.322 0.135 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3256415000
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40