bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0374_orf1
Length=263
Score E
Sequences producing significant alignments: (Bits) Value
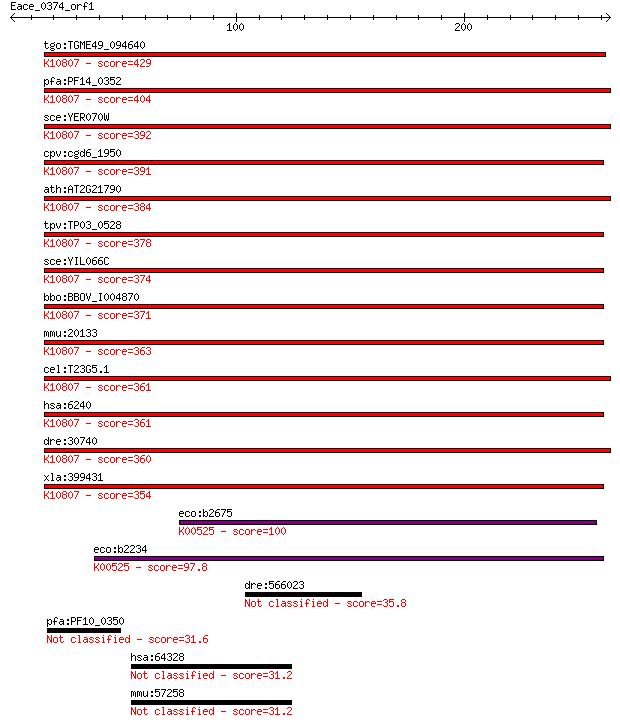
tgo:TGME49_094640 ribonucleoside-diphosphate reductase, large ... 429 6e-120
pfa:PF14_0352 ribonucleoside-diphosphate reductase, large subu... 404 1e-112
sce:YER070W RNR1, CRT7, RIR1, SDS12; One of two large regulato... 392 5e-109
cpv:cgd6_1950 ribonucleotide-diphosphate reductase large chain... 391 1e-108
ath:AT2G21790 RNR1; RNR1 (RIBONUCLEOTIDE REDUCTASE 1); ATP bin... 384 2e-106
tpv:TP03_0528 ribonucleotide-diphosphate reductase large subun... 378 2e-104
sce:YIL066C RNR3, DIN1, RIR3; One of two large regulatory subu... 374 2e-103
bbo:BBOV_I004870 19.m02176; ribonucleoside-diphosphate reducta... 371 1e-102
mmu:20133 Rrm1, RnrM1; ribonucleotide reductase M1 (EC:1.17.4.... 363 5e-100
cel:T23G5.1 rnr-1; RiboNucleotide Reductase family member (rnr... 361 1e-99
hsa:6240 RRM1, R1, RIR1, RR1; ribonucleotide reductase M1 (EC:... 361 1e-99
dre:30740 rrm1, CHUNP6866, cb396, cb838, r1, sb:cb548, wu:fb39... 360 2e-99
xla:399431 rrm1; ribonucleotide reductase M1 (EC:1.17.4.1); K1... 354 2e-97
eco:b2675 nrdE, ECK2669, JW2650; ribonucleoside-diphosphate re... 100 6e-21
eco:b2234 nrdA, dnaF, ECK2226, JW2228; ribonucleoside-diphosph... 97.8 3e-20
dre:566023 densin-180-like 35.8 0.17
pfa:PF10_0350 probable protein 31.6 3.1
hsa:64328 XPO4, FLJ13046, KIAA1721; exportin 4 31.2
mmu:57258 Xpo4, B430309A01Rik, mKIAA1721; exportin 4 31.2
> tgo:TGME49_094640 ribonucleoside-diphosphate reductase, large
subunit, putative (EC:5.1.3.4 1.17.4.1); K10807 ribonucleoside-diphosphate
reductase subunit M1 [EC:1.17.4.1]
Length=877
Score = 429 bits (1102), Expect = 6e-120, Method: Compositional matrix adjust.
Identities = 196/246 (79%), Positives = 220/246 (89%), Gaps = 0/246 (0%)
Query 16 LVHADVYAFVVKYKDEINKALVYDRDFDYDYFAFKTLERSYLLRAGGVIVERPQHTLMRV 75
LV +VY FV + + +N+AL Y RDFDYDYF FKTLERSYLL+ IVERPQH LMRV
Sbjct 172 LVSTEVYEFVRENEQALNEALNYSRDFDYDYFGFKTLERSYLLKIHDRIVERPQHMLMRV 231
Query 76 ACGIHCGDLQKTLETYELMSCKFFIHATPTLFNAGTPRPQMSSCFLLTMKEDSIDGIFST 135
ACGIHCGD++K +ETYELMS KFF HATPTLFNAGTPRPQMSSCFLLTM+EDSIDGIFST
Sbjct 232 ACGIHCGDVEKAIETYELMSQKFFTHATPTLFNAGTPRPQMSSCFLLTMQEDSIDGIFST 291
Query 136 LRQCALISKTAGGLGLSVTDIRATGSYIRGTNGYSNGLIPMLRVFNDASRYVDQGGGKRK 195
L+QCALISKTAGGLGL+VTDIRAT SYIRGTNGYSNGL+PMLRVFNDA+RYVDQGGGKRK
Sbjct 292 LKQCALISKTAGGLGLAVTDIRATNSYIRGTNGYSNGLLPMLRVFNDAARYVDQGGGKRK 351
Query 196 GSLAVYVEPWHADIFEFLEIRKNHGKEKMRARDLFPALWVPDLFMQRVYENGSWTLMCPC 255
GSLA+Y+EPWH D+F+FL+I+KNHGKE+ RARDLF ALW+PDLFM+RV +N WTLMCP
Sbjct 352 GSLAIYLEPWHFDVFDFLDIKKNHGKEERRARDLFCALWIPDLFMERVNDNAGWTLMCPN 411
Query 256 ECPGLT 261
ECPGLT
Sbjct 412 ECPGLT 417
> pfa:PF14_0352 ribonucleoside-diphosphate reductase, large subunit;
K10807 ribonucleoside-diphosphate reductase subunit M1
[EC:1.17.4.1]
Length=847
Score = 404 bits (1039), Expect = 1e-112, Method: Compositional matrix adjust.
Identities = 180/248 (72%), Positives = 211/248 (85%), Gaps = 0/248 (0%)
Query 16 LVHADVYAFVVKYKDEINKALVYDRDFDYDYFAFKTLERSYLLRAGGVIVERPQHTLMRV 75
L+ +VY F++ +KD +NK + Y RDF+YDYF FKTLERSYLLR I+ERPQH LMRV
Sbjct 154 LISKEVYDFILLHKDRLNKEIDYTRDFNYDYFGFKTLERSYLLRINNKIIERPQHLLMRV 213
Query 76 ACGIHCGDLQKTLETYELMSCKFFIHATPTLFNAGTPRPQMSSCFLLTMKEDSIDGIFST 135
+ GIH D+ K LETY LMS K+F HATPTLFN+GTPRPQMSSCFLL+MK DSI+GIF T
Sbjct 214 SIGIHIDDIDKALETYHLMSQKYFTHATPTLFNSGTPRPQMSSCFLLSMKADSIEGIFET 273
Query 136 LRQCALISKTAGGLGLSVTDIRATGSYIRGTNGYSNGLIPMLRVFNDASRYVDQGGGKRK 195
L+QCALISKTAGG+G++V DIR SYIRGTNG SNGL+PMLRVFND +RYVDQGGGKRK
Sbjct 274 LKQCALISKTAGGIGVAVQDIRGQNSYIRGTNGISNGLVPMLRVFNDTARYVDQGGGKRK 333
Query 196 GSLAVYVEPWHADIFEFLEIRKNHGKEKMRARDLFPALWVPDLFMQRVYENGSWTLMCPC 255
GS AVY+EPWH+DIFEFL++RKNHGKE++RARDLF A+WVPDLFM+RV EN +WTLMCP
Sbjct 334 GSFAVYIEPWHSDIFEFLDLRKNHGKEELRARDLFYAVWVPDLFMKRVKENKNWTLMCPN 393
Query 256 ECPGLTSC 263
ECPGL+
Sbjct 394 ECPGLSET 401
> sce:YER070W RNR1, CRT7, RIR1, SDS12; One of two large regulatory
subunits of ribonucleotide-diphosphate reductase; the RNR
complex catalyzes rate-limiting step in dNTP synthesis, regulated
by DNA replication and DNA damage checkpoint pathways
via localization of small subunits (EC:1.17.4.1); K10807
ribonucleoside-diphosphate reductase subunit M1 [EC:1.17.4.1]
Length=888
Score = 392 bits (1008), Expect = 5e-109, Method: Compositional matrix adjust.
Identities = 171/248 (68%), Positives = 209/248 (84%), Gaps = 0/248 (0%)
Query 16 LVHADVYAFVVKYKDEINKALVYDRDFDYDYFAFKTLERSYLLRAGGVIVERPQHTLMRV 75
++ DVY V++ KD++N A+VYDRDF Y YF FKTLERSYLLR G + ERPQH +MRV
Sbjct 115 MISDDVYNIVMENKDKLNSAIVYDRDFQYSYFGFKTLERSYLLRINGQVAERPQHLIMRV 174
Query 76 ACGIHCGDLQKTLETYELMSCKFFIHATPTLFNAGTPRPQMSSCFLLTMKEDSIDGIFST 135
A GIH D++ LETY LMS K+F HA+PTLFNAGTP+PQMSSCFL+ MKEDSI+GI+ T
Sbjct 175 ALGIHGRDIEAALETYNLMSLKYFTHASPTLFNAGTPKPQMSSCFLVAMKEDSIEGIYDT 234
Query 136 LRQCALISKTAGGLGLSVTDIRATGSYIRGTNGYSNGLIPMLRVFNDASRYVDQGGGKRK 195
L++CALISKTAGG+GL + +IR+TGSYI GTNG SNGLIPM+RVFN+ +RYVDQGG KR
Sbjct 235 LKECALISKTAGGIGLHIHNIRSTGSYIAGTNGTSNGLIPMIRVFNNTARYVDQGGNKRP 294
Query 196 GSLAVYVEPWHADIFEFLEIRKNHGKEKMRARDLFPALWVPDLFMQRVYENGSWTLMCPC 255
G+ A+Y+EPWHADIF+F++IRKNHGKE++RARDLFPALW+PDLFM+RV ENG+WTL P
Sbjct 295 GAFALYLEPWHADIFDFIDIRKNHGKEEIRARDLFPALWIPDLFMKRVEENGTWTLFSPT 354
Query 256 ECPGLTSC 263
PGL+ C
Sbjct 355 SAPGLSDC 362
> cpv:cgd6_1950 ribonucleotide-diphosphate reductase large chain;
RIR1; c-terminal PFL-like glycyl radical enzymes-like fold
; K10807 ribonucleoside-diphosphate reductase subunit M1
[EC:1.17.4.1]
Length=804
Score = 391 bits (1004), Expect = 1e-108, Method: Compositional matrix adjust.
Identities = 172/245 (70%), Positives = 210/245 (85%), Gaps = 0/245 (0%)
Query 16 LVHADVYAFVVKYKDEINKALVYDRDFDYDYFAFKTLERSYLLRAGGVIVERPQHTLMRV 75
L+ VY F+++ ++ IN + + +DF+YDYFAFKTLERSYLL+ +VERPQH LMRV
Sbjct 114 LISKPVYDFIMENRERINSKIDFSKDFEYDYFAFKTLERSYLLKIDNKVVERPQHLLMRV 173
Query 76 ACGIHCGDLQKTLETYELMSCKFFIHATPTLFNAGTPRPQMSSCFLLTMKEDSIDGIFST 135
+CGIHCGD++ LETYEL+S K+F HATPTLFN+GTPRPQMSSCFLL + EDSI+GIF T
Sbjct 174 SCGIHCGDIEAALETYELLSQKYFTHATPTLFNSGTPRPQMSSCFLLRIPEDSINGIFDT 233
Query 136 LRQCALISKTAGGLGLSVTDIRATGSYIRGTNGYSNGLIPMLRVFNDASRYVDQGGGKRK 195
L +CA ISKTAGGLG++V++IR TGSYIRGTNG SNGLIPMLRV+ND +RY+DQGGGKRK
Sbjct 234 LTKCANISKTAGGLGVAVSNIRGTGSYIRGTNGRSNGLIPMLRVYNDTARYIDQGGGKRK 293
Query 196 GSLAVYVEPWHADIFEFLEIRKNHGKEKMRARDLFPALWVPDLFMQRVYENGSWTLMCPC 255
G++A+Y+EPWH D+ EF+EIRKNHGKE+MR RDLFPALWVPDLFM+RV ++ WTLMCP
Sbjct 294 GAIAIYLEPWHVDVVEFIEIRKNHGKEEMRCRDLFPALWVPDLFMERVEKDQDWTLMCPD 353
Query 256 ECPGL 260
EC GL
Sbjct 354 ECRGL 358
> ath:AT2G21790 RNR1; RNR1 (RIBONUCLEOTIDE REDUCTASE 1); ATP binding
/ protein binding / ribonucleoside-diphosphate reductase
(EC:1.17.4.1); K10807 ribonucleoside-diphosphate reductase
subunit M1 [EC:1.17.4.1]
Length=816
Score = 384 bits (986), Expect = 2e-106, Method: Compositional matrix adjust.
Identities = 166/248 (66%), Positives = 208/248 (83%), Gaps = 0/248 (0%)
Query 16 LVHADVYAFVVKYKDEINKALVYDRDFDYDYFAFKTLERSYLLRAGGVIVERPQHTLMRV 75
L+ DV+ +++ ++ ++YDRDF+YDYF FKTLERSYLL+ G +VERPQH LMRV
Sbjct 115 LIADDVFEIIMQNAARLDSEIIYDRDFEYDYFGFKTLERSYLLKVQGTVVERPQHMLMRV 174
Query 76 ACGIHCGDLQKTLETYELMSCKFFIHATPTLFNAGTPRPQMSSCFLLTMKEDSIDGIFST 135
A GIH D+ ++TY LMS ++F HA+PTLFNAGTPRPQ+SSCFL+ MK+DSI+GI+ T
Sbjct 175 AVGIHKDDIDSVIQTYHLMSQRWFTHASPTLFNAGTPRPQLSSCFLVCMKDDSIEGIYET 234
Query 136 LRQCALISKTAGGLGLSVTDIRATGSYIRGTNGYSNGLIPMLRVFNDASRYVDQGGGKRK 195
L++CA+ISK+AGG+G+SV +IRATGSYIRGTNG SNG++PMLRVFND +RYVDQGGGKRK
Sbjct 235 LKECAVISKSAGGIGVSVHNIRATGSYIRGTNGTSNGIVPMLRVFNDTARYVDQGGGKRK 294
Query 196 GSLAVYVEPWHADIFEFLEIRKNHGKEKMRARDLFPALWVPDLFMQRVYENGSWTLMCPC 255
G+ AVY+EPWHAD++EFLE+RKNHGKE+ RARDLF ALW+PDLFM+RV NG W+L CP
Sbjct 295 GAFAVYLEPWHADVYEFLELRKNHGKEEHRARDLFYALWLPDLFMERVQNNGQWSLFCPN 354
Query 256 ECPGLTSC 263
E PGL C
Sbjct 355 EAPGLADC 362
> tpv:TP03_0528 ribonucleotide-diphosphate reductase large subunit
(EC:1.17.4.1); K10807 ribonucleoside-diphosphate reductase
subunit M1 [EC:1.17.4.1]
Length=898
Score = 378 bits (970), Expect = 2e-104, Method: Compositional matrix adjust.
Identities = 166/245 (67%), Positives = 203/245 (82%), Gaps = 0/245 (0%)
Query 16 LVHADVYAFVVKYKDEINKALVYDRDFDYDYFAFKTLERSYLLRAGGVIVERPQHTLMRV 75
L+ DV+ FV+ KD +N + Y RDF YDYF FKTLERSYLL+ G IVERPQH +MRV
Sbjct 172 LISDDVFEFVMANKDRLNAEIDYSRDFQYDYFGFKTLERSYLLKTNGKIVERPQHMIMRV 231
Query 76 ACGIHCGDLQKTLETYELMSCKFFIHATPTLFNAGTPRPQMSSCFLLTMKEDSIDGIFST 135
+ GIHCGDL++T++TY LMS ++F HATPTLFNAGT PQMSSCFLL M++DS+ GIF+T
Sbjct 232 SAGIHCGDLERTIQTYHLMSQRYFTHATPTLFNAGTRHPQMSSCFLLDMQDDSLAGIFNT 291
Query 136 LRQCALISKTAGGLGLSVTDIRATGSYIRGTNGYSNGLIPMLRVFNDASRYVDQGGGKRK 195
L QCA ISK+AGG+GL++ IRA+GSYIRGTNG SNG++PML++FN ++YVDQGGGKRK
Sbjct 292 LSQCAFISKSAGGIGLAIHKIRASGSYIRGTNGISNGIVPMLKIFNATAKYVDQGGGKRK 351
Query 196 GSLAVYVEPWHADIFEFLEIRKNHGKEKMRARDLFPALWVPDLFMQRVYENGSWTLMCPC 255
GS A+Y+EPWHADIF+ L++RKNHG E RARDLF ALW+PDLFM+RV N +WTLMCP
Sbjct 352 GSFAIYLEPWHADIFKLLDLRKNHGAEDQRARDLFYALWIPDLFMKRVEANKNWTLMCPD 411
Query 256 ECPGL 260
EC GL
Sbjct 412 ECRGL 416
> sce:YIL066C RNR3, DIN1, RIR3; One of two large regulatory subunits
of ribonucleotide-diphosphate reductase; the RNR complex
catalyzes rate-limiting step in dNTP synthesis, regulated
by DNA replication and DNA damage checkpoint pathways via
localization of small subunits (EC:1.17.4.1); K10807 ribonucleoside-diphosphate
reductase subunit M1 [EC:1.17.4.1]
Length=869
Score = 374 bits (961), Expect = 2e-103, Method: Compositional matrix adjust.
Identities = 161/245 (65%), Positives = 203/245 (82%), Gaps = 0/245 (0%)
Query 16 LVHADVYAFVVKYKDEINKALVYDRDFDYDYFAFKTLERSYLLRAGGVIVERPQHTLMRV 75
++ ++Y V++ KD +N A+VYDRDF Y YF FKTLERSYLLR G + ERPQH +MRV
Sbjct 115 MISDEIYNIVMENKDTLNSAIVYDRDFQYTYFGFKTLERSYLLRLNGEVAERPQHLVMRV 174
Query 76 ACGIHCGDLQKTLETYELMSCKFFIHATPTLFNAGTPRPQMSSCFLLTMKEDSIDGIFST 135
A GIH D++ L+TY LMS ++F HA+PTLFNAGTP PQMSSCFL+ MK+DSI+GI+ T
Sbjct 175 ALGIHGSDIESVLKTYNLMSLRYFTHASPTLFNAGTPHPQMSSCFLIAMKDDSIEGIYDT 234
Query 136 LRQCALISKTAGGLGLSVTDIRATGSYIRGTNGYSNGLIPMLRVFNDASRYVDQGGGKRK 195
L++CA+ISKTAGG+GL + +IR+TGSYI GTNG SNGLIPM+RVFN+ +RYVDQGG KR
Sbjct 235 LKECAMISKTAGGVGLHINNIRSTGSYIAGTNGTSNGLIPMIRVFNNTARYVDQGGNKRP 294
Query 196 GSLAVYVEPWHADIFEFLEIRKNHGKEKMRARDLFPALWVPDLFMQRVYENGSWTLMCPC 255
G+ A+++EPWHADIF+F++IRK HGKE++RARDLFPALW+PDLFM+RV E+G WTL P
Sbjct 295 GAFALFLEPWHADIFDFVDIRKTHGKEEIRARDLFPALWIPDLFMKRVQEDGPWTLFSPS 354
Query 256 ECPGL 260
PGL
Sbjct 355 AAPGL 359
> bbo:BBOV_I004870 19.m02176; ribonucleoside-diphosphate reductase
large chain (EC:1.17.4.1); K10807 ribonucleoside-diphosphate
reductase subunit M1 [EC:1.17.4.1]
Length=838
Score = 371 bits (953), Expect = 1e-102, Method: Compositional matrix adjust.
Identities = 168/245 (68%), Positives = 198/245 (80%), Gaps = 0/245 (0%)
Query 16 LVHADVYAFVVKYKDEINKALVYDRDFDYDYFAFKTLERSYLLRAGGVIVERPQHTLMRV 75
L+ +VY F++ D IN + Y RDF YDYF KTLERSYLLR IVERPQH LMRV
Sbjct 140 LISEEVYDFIMANIDRINAEIDYSRDFQYDYFGLKTLERSYLLRINDKIVERPQHMLMRV 199
Query 76 ACGIHCGDLQKTLETYELMSCKFFIHATPTLFNAGTPRPQMSSCFLLTMKEDSIDGIFST 135
+ GIH GD+++T++TY LMS K+F HATPTLF +GTPRPQMSSCFLL MK+DS+ GIF T
Sbjct 200 SAGIHTGDIERTIQTYHLMSQKYFTHATPTLFFSGTPRPQMSSCFLLDMKDDSLAGIFET 259
Query 136 LRQCALISKTAGGLGLSVTDIRATGSYIRGTNGYSNGLIPMLRVFNDASRYVDQGGGKRK 195
L QCA ISK AGG+GL+ IRA+G+YIRGTNG SNGL+PMLR+FN +RYVDQGGGKRK
Sbjct 260 LTQCAFISKCAGGIGLACHKIRASGAYIRGTNGKSNGLVPMLRIFNSTARYVDQGGGKRK 319
Query 196 GSLAVYVEPWHADIFEFLEIRKNHGKEKMRARDLFPALWVPDLFMQRVYENGSWTLMCPC 255
GS A+Y+EPWHADI +FL++RKNHG E+ RARDLF ALW+PDLFM+RV NG WTLMCP
Sbjct 320 GSFAIYLEPWHADIMDFLDLRKNHGAEEARARDLFYALWIPDLFMKRVESNGDWTLMCPD 379
Query 256 ECPGL 260
EC GL
Sbjct 380 ECRGL 384
> mmu:20133 Rrm1, RnrM1; ribonucleotide reductase M1 (EC:1.17.4.1);
K10807 ribonucleoside-diphosphate reductase subunit M1
[EC:1.17.4.1]
Length=792
Score = 363 bits (931), Expect = 5e-100, Method: Compositional matrix adjust.
Identities = 161/245 (65%), Positives = 201/245 (82%), Gaps = 0/245 (0%)
Query 16 LVHADVYAFVVKYKDEINKALVYDRDFDYDYFAFKTLERSYLLRAGGVIVERPQHTLMRV 75
+V + V+ KD +N A++YDRDF Y+YF FKTLERSYLL+ G + ERPQH LMRV
Sbjct 115 MVASSTLDIVMANKDRLNSAIIYDRDFSYNYFGFKTLERSYLLKINGKVAERPQHMLMRV 174
Query 76 ACGIHCGDLQKTLETYELMSCKFFIHATPTLFNAGTPRPQMSSCFLLTMKEDSIDGIFST 135
+ GIH D+ +ETY L+S K+F HA+PTLFNAGT RPQ+SSCFLL+MK+DSI+GI+ T
Sbjct 175 SVGIHKEDIDAAIETYNLLSEKWFTHASPTLFNAGTNRPQLSSCFLLSMKDDSIEGIYDT 234
Query 136 LRQCALISKTAGGLGLSVTDIRATGSYIRGTNGYSNGLIPMLRVFNDASRYVDQGGGKRK 195
L+QCALISK+AGG+G++V+ IRATGSYI GTNG SNGL+PMLRV+N+ +RYVDQGG KR
Sbjct 235 LKQCALISKSAGGIGVAVSCIRATGSYIAGTNGNSNGLVPMLRVYNNTARYVDQGGNKRP 294
Query 196 GSLAVYVEPWHADIFEFLEIRKNHGKEKMRARDLFPALWVPDLFMQRVYENGSWTLMCPC 255
G+ A+Y+EPWH DIFEFL+++KN GKE+ RARDLF ALW+PDLFM+RV N W+LMCP
Sbjct 295 GAFAIYLEPWHLDIFEFLDLKKNTGKEEQRARDLFFALWIPDLFMKRVETNQDWSLMCPN 354
Query 256 ECPGL 260
ECPGL
Sbjct 355 ECPGL 359
> cel:T23G5.1 rnr-1; RiboNucleotide Reductase family member (rnr-1);
K10807 ribonucleoside-diphosphate reductase subunit M1
[EC:1.17.4.1]
Length=788
Score = 361 bits (927), Expect = 1e-99, Method: Compositional matrix adjust.
Identities = 162/248 (65%), Positives = 198/248 (79%), Gaps = 0/248 (0%)
Query 16 LVHADVYAFVVKYKDEINKALVYDRDFDYDYFAFKTLERSYLLRAGGVIVERPQHTLMRV 75
++ + +A + K D++N A+VYDRD+ Y YF FKTLERSYLL+ IVERPQ LMRV
Sbjct 121 MISDETWAIIEKNADKLNSAIVYDRDYSYTYFGFKTLERSYLLKINKEIVERPQQMLMRV 180
Query 76 ACGIHCGDLQKTLETYELMSCKFFIHATPTLFNAGTPRPQMSSCFLLTMKEDSIDGIFST 135
+ GIH D+ +ETY LMS ++ HA+PTLFN+GT RPQMSSCFLLTM EDSI GI+ T
Sbjct 181 SIGIHGDDITSAIETYNLMSERYMTHASPTLFNSGTCRPQMSSCFLLTMSEDSILGIYDT 240
Query 136 LRQCALISKTAGGLGLSVTDIRATGSYIRGTNGYSNGLIPMLRVFNDASRYVDQGGGKRK 195
L+QCALISK+AGG+GL+V IRATGS I GTNG SNGLIPMLRV+N+ +RYVDQGG KR
Sbjct 241 LKQCALISKSAGGIGLNVHKIRATGSVIAGTNGTSNGLIPMLRVYNNTARYVDQGGNKRP 300
Query 196 GSLAVYVEPWHADIFEFLEIRKNHGKEKMRARDLFPALWVPDLFMQRVYENGSWTLMCPC 255
G+ A+Y+EPWHADIFEF+ +RKN G E+ RARDLF ALW+PDLFM+RV ++ W+LMCPC
Sbjct 301 GAFAIYLEPWHADIFEFVSLRKNTGPEEERARDLFLALWIPDLFMKRVEKDQEWSLMCPC 360
Query 256 ECPGLTSC 263
ECPGL C
Sbjct 361 ECPGLDDC 368
> hsa:6240 RRM1, R1, RIR1, RR1; ribonucleotide reductase M1 (EC:1.17.4.1);
K10807 ribonucleoside-diphosphate reductase subunit
M1 [EC:1.17.4.1]
Length=792
Score = 361 bits (927), Expect = 1e-99, Method: Compositional matrix adjust.
Identities = 160/245 (65%), Positives = 200/245 (81%), Gaps = 0/245 (0%)
Query 16 LVHADVYAFVVKYKDEINKALVYDRDFDYDYFAFKTLERSYLLRAGGVIVERPQHTLMRV 75
+V V+ KD +N A++YDRDF Y+YF FKTLERSYLL+ G + ERPQH LMRV
Sbjct 115 MVAKSTLDIVLANKDRLNSAIIYDRDFSYNYFGFKTLERSYLLKINGKVAERPQHMLMRV 174
Query 76 ACGIHCGDLQKTLETYELMSCKFFIHATPTLFNAGTPRPQMSSCFLLTMKEDSIDGIFST 135
+ GIH D+ +ETY L+S ++F HA+PTLFNAGT RPQ+SSCFLL+MK+DSI+GI+ T
Sbjct 175 SVGIHKEDIDAAIETYNLLSERWFTHASPTLFNAGTNRPQLSSCFLLSMKDDSIEGIYDT 234
Query 136 LRQCALISKTAGGLGLSVTDIRATGSYIRGTNGYSNGLIPMLRVFNDASRYVDQGGGKRK 195
L+QCALISK+AGG+G++V+ IRATGSYI GTNG SNGL+PMLRV+N+ +RYVDQGG KR
Sbjct 235 LKQCALISKSAGGIGVAVSCIRATGSYIAGTNGNSNGLVPMLRVYNNTARYVDQGGNKRP 294
Query 196 GSLAVYVEPWHADIFEFLEIRKNHGKEKMRARDLFPALWVPDLFMQRVYENGSWTLMCPC 255
G+ A+Y+EPWH DIFEFL+++KN GKE+ RARDLF ALW+PDLFM+RV N W+LMCP
Sbjct 295 GAFAIYLEPWHLDIFEFLDLKKNTGKEEQRARDLFFALWIPDLFMKRVETNQDWSLMCPN 354
Query 256 ECPGL 260
ECPGL
Sbjct 355 ECPGL 359
> dre:30740 rrm1, CHUNP6866, cb396, cb838, r1, sb:cb548, wu:fb39b07,
wu:fi14b02, wu:fk95f07; ribonucleotide reductase M1 polypeptide
(EC:1.17.4.1); K10807 ribonucleoside-diphosphate
reductase subunit M1 [EC:1.17.4.1]
Length=794
Score = 360 bits (925), Expect = 2e-99, Method: Compositional matrix adjust.
Identities = 159/248 (64%), Positives = 200/248 (80%), Gaps = 0/248 (0%)
Query 16 LVHADVYAFVVKYKDEINKALVYDRDFDYDYFAFKTLERSYLLRAGGVIVERPQHTLMRV 75
++ + V+ KD +N A++YDRDF Y++F FKTLERSYLL+ G + ERPQH LMRV
Sbjct 115 MISKETLDIVLANKDRLNSAIIYDRDFSYNFFGFKTLERSYLLKINGKVAERPQHMLMRV 174
Query 76 ACGIHCGDLQKTLETYELMSCKFFIHATPTLFNAGTPRPQMSSCFLLTMKEDSIDGIFST 135
+ GIH D+ +ETY L+S K+F HA+PTLFNAGT RPQ+SSCFLL MK+DSI+GI+ T
Sbjct 175 SVGIHKEDIAAAIETYNLLSEKWFTHASPTLFNAGTNRPQLSSCFLLAMKDDSIEGIYDT 234
Query 136 LRQCALISKTAGGLGLSVTDIRATGSYIRGTNGYSNGLIPMLRVFNDASRYVDQGGGKRK 195
L+QCALISK+AGG+G++V+ IRATG YI GTNG SNGL+PMLRV N+ +RYVDQGG KR
Sbjct 235 LKQCALISKSAGGIGVAVSCIRATGRYIAGTNGNSNGLVPMLRVNNNTARYVDQGGNKRP 294
Query 196 GSLAVYVEPWHADIFEFLEIRKNHGKEKMRARDLFPALWVPDLFMQRVYENGSWTLMCPC 255
G+ A+Y+EPWH DIF+FLE++KN GKE+ RARDLF ALW+PDLFM+RV NG W+LMCP
Sbjct 295 GAFAMYLEPWHFDIFDFLELKKNTGKEEQRARDLFYALWIPDLFMKRVETNGDWSLMCPN 354
Query 256 ECPGLTSC 263
+CPGL C
Sbjct 355 DCPGLDEC 362
> xla:399431 rrm1; ribonucleotide reductase M1 (EC:1.17.4.1);
K10807 ribonucleoside-diphosphate reductase subunit M1 [EC:1.17.4.1]
Length=797
Score = 354 bits (909), Expect = 2e-97, Method: Compositional matrix adjust.
Identities = 155/245 (63%), Positives = 199/245 (81%), Gaps = 0/245 (0%)
Query 16 LVHADVYAFVVKYKDEINKALVYDRDFDYDYFAFKTLERSYLLRAGGVIVERPQHTLMRV 75
+V + V+ KD +N +++YDRDF Y++F FKTLERSYLL+ G + ERPQH LMRV
Sbjct 115 MVSRETLDIVLANKDRLNSSIIYDRDFSYNFFGFKTLERSYLLKINGKVAERPQHMLMRV 174
Query 76 ACGIHCGDLQKTLETYELMSCKFFIHATPTLFNAGTPRPQMSSCFLLTMKEDSIDGIFST 135
+ GIH D+ +ETY L+S K+F HA+PTLFNAGT RPQ+SSCFLL MK+DSI+GI+ T
Sbjct 175 SVGIHKTDIDAAIETYNLLSEKWFTHASPTLFNAGTNRPQLSSCFLLCMKDDSIEGIYDT 234
Query 136 LRQCALISKTAGGLGLSVTDIRATGSYIRGTNGYSNGLIPMLRVFNDASRYVDQGGGKRK 195
L+QCALISK+AGG+G++V+ IRATGSYI GTNG SNGL+PMLRV+N+ +RYVDQGG KR
Sbjct 235 LKQCALISKSAGGIGVAVSCIRATGSYIAGTNGNSNGLVPMLRVYNNTARYVDQGGNKRP 294
Query 196 GSLAVYVEPWHADIFEFLEIRKNHGKEKMRARDLFPALWVPDLFMQRVYENGSWTLMCPC 255
G+ A+Y+EPWH D+F+FL+++KN GKE+ RARDLF A+W+PDLFM+R N W+LMCP
Sbjct 295 GAFAIYLEPWHYDVFDFLDLKKNTGKEEQRARDLFYAMWIPDLFMKRAENNLDWSLMCPH 354
Query 256 ECPGL 260
ECPGL
Sbjct 355 ECPGL 359
> eco:b2675 nrdE, ECK2669, JW2650; ribonucleoside-diphosphate
reductase 2, alpha subunit (EC:1.17.4.1); K00525 ribonucleoside-diphosphate
reductase alpha chain [EC:1.17.4.1]
Length=714
Score = 100 bits (249), Expect = 6e-21, Method: Compositional matrix adjust.
Identities = 58/184 (31%), Positives = 96/184 (52%), Gaps = 3/184 (1%)
Query 75 VACGIHCGDLQKTLETYELMSCKFFIHATPTLFNAG-TPRPQMSSCFLLTMKEDSIDGIF 133
VA + GD L+ + M F ATPT N G R ++ SCFLL + ED+++ I
Sbjct 133 VALTLAQGDETLALQLTDEMLSGRFQPATPTFLNCGKQQRGELVSCFLLRI-EDNMESIG 191
Query 134 STLRQCALISKTAGGLGLSVTDIRATGSYIRGTNGYSNGLIPMLRVFNDASRYVDQGGGK 193
+ +SK GG+ ++++R G+ I+ S+G+IP++++ DA Y +Q G
Sbjct 192 RAVNSALQLSKRGGGVAFLLSNLREAGAPIKRIENQSSGVIPVMKMLEDAFSYANQLGA- 250
Query 194 RKGSLAVYVEPWHADIFEFLEIRKNHGKEKMRARDLFPALWVPDLFMQRVYENGSWTLMC 253
R+G+ AVY+ H DI FL+ ++ + EK+R + L + +PD+ EN L
Sbjct 251 RQGAGAVYLHAHHPDILRFLDTKRENADEKIRIKTLSLGVVIPDITFHLAKENAQMALFS 310
Query 254 PCEC 257
P +
Sbjct 311 PYDV 314
> eco:b2234 nrdA, dnaF, ECK2226, JW2228; ribonucleoside-diphosphate
reductase 1, alpha subunit (EC:1.17.4.1); K00525 ribonucleoside-diphosphate
reductase alpha chain [EC:1.17.4.1]
Length=761
Score = 97.8 bits (242), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 71/230 (30%), Positives = 101/230 (43%), Gaps = 9/230 (3%)
Query 38 YDRDFDYDYFAFKTLERSYLL--RAGGVIVERPQHTLMRVACGIHCG-----DLQKTLET 90
+DRD + Y A K LE YL+ R G I E Q + VA + LQ
Sbjct 137 HDRDMTFSYAAVKQLEGKYLVQNRVTGEIYESAQFLYILVAACLFSNYPRETRLQYVKRF 196
Query 91 YELMSCKFFIHATPTLFNAGTPRPQMSSCFLLTMKEDSIDGIFSTLRQCALISKTAGGLG 150
Y+ +S TP + TP Q SSC L+ DS+D I +T G+G
Sbjct 197 YDAVSTFKISLPTPIMSGVRTPTRQFSSCVLIECG-DSLDSINATSSAIVKYVSQRAGIG 255
Query 151 LSVTDIRATGSYIRGTNGYSNGLIPMLRVFNDASRYVDQGGGKRKGSLAVYVEPWHADIF 210
++ IRA GS IRG + G IP + F A + Q GG R G+ ++ WH ++
Sbjct 256 INAGRIRALGSPIRGGEAFHTGCIPFYKHFQTAVKSCSQ-GGVRGGAATLFYPMWHLEVE 314
Query 211 EFLEIRKNHGKEKMRARDLFPALWVPDLFMQRVYENGSWTLMCPCECPGL 260
L ++ N G E R R + + + L R+ + TL P + PGL
Sbjct 315 SLLVLKNNRGVEGNRVRHMDYGVQINKLMYTRLLKGEDITLFSPSDVPGL 364
> dre:566023 densin-180-like
Length=1330
Score = 35.8 bits (81), Expect = 0.17, Method: Composition-based stats.
Identities = 19/51 (37%), Positives = 26/51 (50%), Gaps = 3/51 (5%)
Query 104 PTLFNAGTPRPQMSSCFLLTMKEDSIDGIFSTLRQCALISKTAGGLGLSVT 154
P +PRPQ + CF+ T + S+DG L C I K GLG S++
Sbjct 1208 PMPITTTSPRPQSARCFIQTKGQKSMDGFPEQL--CVRIEKNP-GLGFSIS 1255
> pfa:PF10_0350 probable protein
Length=712
Score = 31.6 bits (70), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 12/32 (37%), Positives = 17/32 (53%), Gaps = 0/32 (0%)
Query 17 VHADVYAFVVKYKDEINKALVYDRDFDYDYFA 48
H + + F DE N L Y+ D+DY+YF
Sbjct 637 THEESHNFYTPTHDEFNVPLNYNHDYDYNYFE 668
> hsa:64328 XPO4, FLJ13046, KIAA1721; exportin 4
Length=1151
Score = 31.2 bits (69), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 21/77 (27%), Positives = 36/77 (46%), Gaps = 16/77 (20%)
Query 54 RSYLLRAGGVIVERP-------QHTLMRVACGIHCGDLQKTLETYELMSCKFFIHATPTL 106
R++LL +++RP + L+ VA + G L K+++ CK H L
Sbjct 94 RTFLL---TYVLQRPNLQKYVREQILLAVAVIVKRGSLDKSID------CKSIFHEVSQL 144
Query 107 FNAGTPRPQMSSCFLLT 123
++G P Q +C +LT
Sbjct 145 ISSGNPTVQTLACSILT 161
> mmu:57258 Xpo4, B430309A01Rik, mKIAA1721; exportin 4
Length=1151
Score = 31.2 bits (69), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 21/77 (27%), Positives = 36/77 (46%), Gaps = 16/77 (20%)
Query 54 RSYLLRAGGVIVERP-------QHTLMRVACGIHCGDLQKTLETYELMSCKFFIHATPTL 106
R++LL +++RP + L+ VA + G L K+++ CK H L
Sbjct 94 RTFLL---TYVLQRPNLQKYVREQILLAVAVIVKRGSLDKSID------CKSIFHEVSQL 144
Query 107 FNAGTPRPQMSSCFLLT 123
++G P Q +C +LT
Sbjct 145 ISSGNPTVQTLACSILT 161
Lambda K H
0.326 0.141 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 9788454684
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40