bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0419_orf1
Length=153
Score E
Sequences producing significant alignments: (Bits) Value
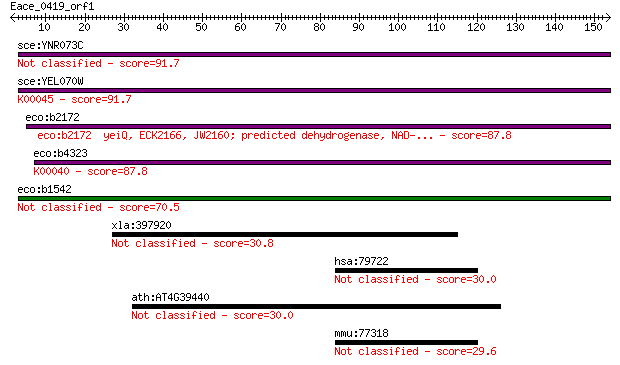
sce:YNR073C Putative mannitol dehydrogenase (EC:1.1.-.-) 91.7 8e-19
sce:YEL070W DSF1; Deletion suppressor of mpt5 mutation (EC:1.1... 91.7 8e-19
eco:b2172 yeiQ, ECK2166, JW2160; predicted dehydrogenase, NAD-... 87.8 1e-17
eco:b4323 uxuB, ECK4314, JW4286; D-mannonate oxidoreductase, N... 87.8 1e-17
eco:b1542 ydfI, ECK1535, JW1535; predicted mannonate dehydroge... 70.5 2e-12
xla:397920 numa; nuclear/mitotic apparatus protein 30.8 1.6
hsa:79722 ANKRD55, FLJ11795, MGC126013, MGC126014; ankyrin rep... 30.0 2.7
ath:AT4G39440 hypothetical protein 30.0 3.0
mmu:77318 Ankrd55, C030011J08Rik; ankyrin repeat domain 55 29.6
> sce:YNR073C Putative mannitol dehydrogenase (EC:1.1.-.-)
Length=502
Score = 91.7 bits (226), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 48/153 (31%), Positives = 82/153 (53%), Gaps = 13/153 (8%)
Query 3 HKGPWAVCGIGILPGDKRMSNVLHEQEFLYTVLTRSQRVCEARVIGALMDFVYAPEEPLR 62
H W++CG+G++ D M + + Q+ LYT++ R + A ++G++ ++YAP++P
Sbjct 63 HLKDWSICGVGLMKADALMRDAMKAQDCLYTLVERGIKDTNAYIVGSITAYMYAPDDPRA 122
Query 63 LKGLLLDIRTKLLSLTITEKGY--CMATNGDLDKSLAAVKQDLALMKCKDIRKQESCTPQ 120
+ + + T ++SLT+TE GY ATN + + + D+ E P
Sbjct 123 VIEKMANPDTHIVSLTVTENGYYHSEATNSLMTDAPEII---------NDLNHPEK--PD 171
Query 121 TALGLIYLGLKLRRDAKIRPFTVLSCDNIPNNG 153
T G +Y L LR + PFT++SCDN+P NG
Sbjct 172 TLYGYLYEALLLRYKRGLTPFTIMSCDNMPQNG 204
> sce:YEL070W DSF1; Deletion suppressor of mpt5 mutation (EC:1.1.-.-);
K00045 mannitol 2-dehydrogenase [EC:1.1.1.67]
Length=502
Score = 91.7 bits (226), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 48/153 (31%), Positives = 82/153 (53%), Gaps = 13/153 (8%)
Query 3 HKGPWAVCGIGILPGDKRMSNVLHEQEFLYTVLTRSQRVCEARVIGALMDFVYAPEEPLR 62
H W++CG+G++ D M + + Q+ LYT++ R + A ++G++ ++YAP++P
Sbjct 63 HLKDWSICGVGLMKADALMRDAMKAQDCLYTLVERGIKDTNAYIVGSITAYMYAPDDPRA 122
Query 63 LKGLLLDIRTKLLSLTITEKGY--CMATNGDLDKSLAAVKQDLALMKCKDIRKQESCTPQ 120
+ + + T ++SLT+TE GY ATN + + + D+ E P
Sbjct 123 VIEKMANPDTHIVSLTVTENGYYHSEATNSLMTDAPEII---------NDLNHPEK--PD 171
Query 121 TALGLIYLGLKLRRDAKIRPFTVLSCDNIPNNG 153
T G +Y L LR + PFT++SCDN+P NG
Sbjct 172 TLYGYLYEALLLRYKRGLTPFTIMSCDNMPQNG 204
> eco:b2172 yeiQ, ECK2166, JW2160; predicted dehydrogenase, NAD-dependent;
K00540 [EC:1.-.-.-]
Length=488
Score = 87.8 bits (216), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 56/151 (37%), Positives = 80/151 (52%), Gaps = 13/151 (8%)
Query 5 GPWAVCGIGILPGDKRMSNVLHEQEFLYTVLTRSQRVCEARVIGALMDFVYAPEEPLR-L 63
G W +C I + GD+ MS L Q LYTVL + + ++GA+ + + A + L +
Sbjct 53 GDWGICEISLFSGDQLMSQ-LRAQNHLYTVLEKGADGNQVIIVGAVHECLNAKLDSLAAI 111
Query 64 KGLLLDIRTKLLSLTITEKGYCM-ATNGDLDKSLAAVKQDLALMKCKDIRKQESCTPQTA 122
+ + ++SLTITEKGYC+ G LD S + DL Q P +A
Sbjct 112 IEKFCEPQVAIVSLTITEKGYCIDPATGALDTSNPRIIHDL----------QTPEEPHSA 161
Query 123 LGLIYLGLKLRRDAKIRPFTVLSCDNIPNNG 153
G++ LK RR+ + PFTVLSCDNIP+NG
Sbjct 162 PGILVEALKRRRERGLTPFTVLSCDNIPDNG 192
> eco:b4323 uxuB, ECK4314, JW4286; D-mannonate oxidoreductase,
NAD-binding (EC:1.1.1.57); K00040 fructuronate reductase [EC:1.1.1.57]
Length=486
Score = 87.8 bits (216), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 51/152 (33%), Positives = 87/152 (57%), Gaps = 17/152 (11%)
Query 7 WAVCGIGILPGDKR-MSNVLHEQEFLYTVLTRSQRVCEARVIGALMDFVYAPEEPLRLKG 65
W +C + ++PG+ R + L +Q+ LYTV + E ++IG++ + ++ E +G
Sbjct 53 WGICEVNLMPGNDRVLIENLKKQQLLYTVAEKGAESTELKIIGSMKEALHP--EIDGCEG 110
Query 66 LLLDI---RTKLLSLTITEKGYCM-ATNGDLDKSLAAVKQDLALMKCKDIRKQESCTPQT 121
+L + +T ++SLT+TEKGYC A +G LD + +K DL + P++
Sbjct 111 ILNAMARPQTAIVSLTVTEKGYCADAASGQLDLNNPLIKHDL----------ENPTAPKS 160
Query 122 ALGLIYLGLKLRRDAKIRPFTVLSCDNIPNNG 153
A+G I L+LRR+ ++ FTV+SCDN+ NG
Sbjct 161 AIGYIVEALRLRREKGLKAFTVMSCDNVRENG 192
> eco:b1542 ydfI, ECK1535, JW1535; predicted mannonate dehydrogenase
Length=486
Score = 70.5 bits (171), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 48/153 (31%), Positives = 78/153 (50%), Gaps = 13/153 (8%)
Query 3 HKGPWAVCGIGILPGDKRMSNVLHEQEFLYTVLTRSQRVCEARVIGALMDFVYAPEEPLR 62
H W + ++ G++++++ L +Q+ LYTV S V ARV+G + ++ + L
Sbjct 48 HFSDWGYYEVNLIGGEQQIAD-LQQQDNLYTVAEMSADVWTARVVGVVKKALHVQIDGLE 106
Query 63 -LKGLLLDIRTKLLSLTITEKGYCMA-TNGDLDKSLAAVKQDLALMKCKDIRKQESCTPQ 120
+ + + + ++SLTITEKGY + G L V D+ Q P+
Sbjct 107 TVLAAMCEPQIAIVSLTITEKGYFHSPATGQLMLDHPMVAADV----------QNPHQPK 156
Query 121 TALGLIYLGLKLRRDAKIRPFTVLSCDNIPNNG 153
TA G+I L R+ A + FTV+SCDN+P NG
Sbjct 157 TATGVIVEALARRKAAGLPAFTVMSCDNMPENG 189
> xla:397920 numa; nuclear/mitotic apparatus protein
Length=2253
Score = 30.8 bits (68), Expect = 1.6, Method: Composition-based stats.
Identities = 28/93 (30%), Positives = 49/93 (52%), Gaps = 9/93 (9%)
Query 27 EQEFLYTVLTRSQRVCEARVIGALMDFVYAPEEPLRLKG---LLLDIRTKLLSLTITEKG 83
EQE + + Q E+ + GA+ + Y E L L+G +L D ++ + + E G
Sbjct 353 EQELSLSNWQQKQNQLESELSGAVGEKKYLEEHNLILQGKISMLEDQLKEMGEIDMPETG 412
Query 84 YCMATNGDLDKSLAAVKQDLALM--KCKDIRKQ 114
CM GD+ K L +KQ+LA++ +C +++Q
Sbjct 413 DCM---GDILK-LDDLKQELAVLNTQCLSLKEQ 441
> hsa:79722 ANKRD55, FLJ11795, MGC126013, MGC126014; ankyrin repeat
domain 55
Length=614
Score = 30.0 bits (66), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 13/36 (36%), Positives = 23/36 (63%), Gaps = 3/36 (8%)
Query 84 YCMATNGDLDKSLAAVKQDLALMKCKDIRKQESCTP 119
Y A+NGD++ A +++D ++++C D E CTP
Sbjct 32 YQAASNGDVNALTAVIREDPSILECCD---SEGCTP 64
> ath:AT4G39440 hypothetical protein
Length=443
Score = 30.0 bits (66), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 30/102 (29%), Positives = 43/102 (42%), Gaps = 13/102 (12%)
Query 32 YTVLTRSQRVCEARVIGA-LMDFVYAPEEPLRLKGLLLDIRTKLLSLTITEKGYCMA--- 87
Y+V +RS+ +C + A LMD Y + RL G DIR L T+ E+
Sbjct 72 YSVSSRSEYICSIEEVDAVLMDVPYI--KIFRLPG---DIRCSLWLTTLMEQELARKLIF 126
Query 88 ----TNGDLDKSLAAVKQDLALMKCKDIRKQESCTPQTALGL 125
LD + + L C+ K+E+CTP L L
Sbjct 127 LKEYWENALDVVYLLARAGVILGNCEVSFKEETCTPSLDLCL 168
> mmu:77318 Ankrd55, C030011J08Rik; ankyrin repeat domain 55
Length=598
Score = 29.6 bits (65), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 12/36 (33%), Positives = 23/36 (63%), Gaps = 3/36 (8%)
Query 84 YCMATNGDLDKSLAAVKQDLALMKCKDIRKQESCTP 119
Y A+NGD++ + +++D ++++C D E CTP
Sbjct 3 YQAASNGDVNSLTSVIREDPSILECCD---SEGCTP 35
Lambda K H
0.323 0.140 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3256415000
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40