bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0433_orf6
Length=1096
Score E
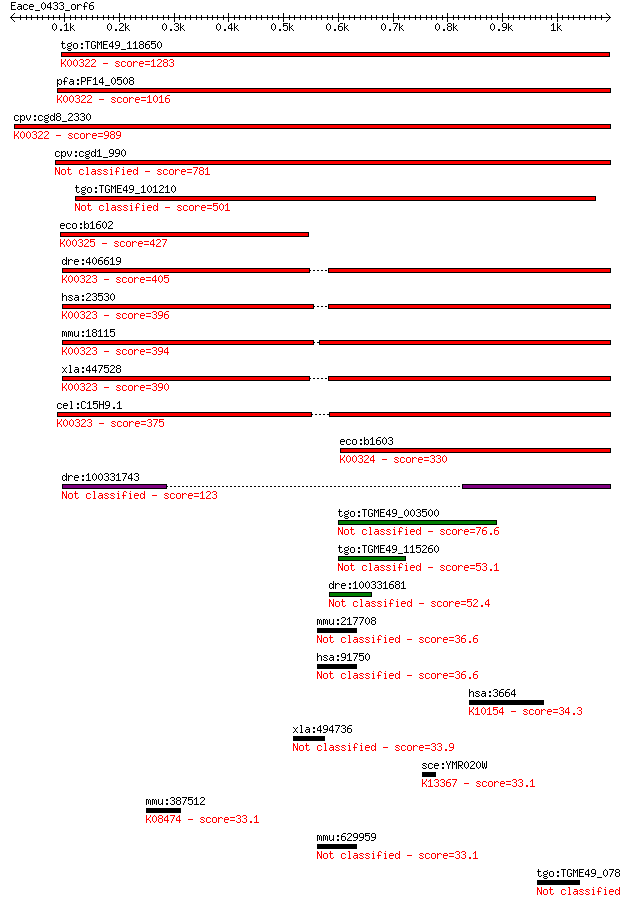
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_118650 transhydrogenase, putative (EC:1.6.1.2); K00... 1283 0.0
pfa:PF14_0508 pyridine nucleotide transhydrogenase, putative (... 1016 0.0
cpv:cgd8_2330 pyridine nucleotide/ NAD(P) transhydrogenase alp... 989 0.0
cpv:cgd1_990 pyridine nucleotide/ NAD(P) transhydrogenase alph... 781 0.0
tgo:TGME49_101210 NAD(P) transhydrogenase, alpha subunit, puta... 501 9e-141
eco:b1602 pntB, ECK1597, JW1594; pyridine nucleotide transhydr... 427 1e-118
dre:406619 nnt, wu:fa20d10, wu:fc86a04, zgc:76979; nicotinamid... 405 4e-112
hsa:23530 NNT, MGC126502, MGC126503; nicotinamide nucleotide t... 396 3e-109
mmu:18115 Nnt, 4930423F13Rik, AI323702, BB168308; nicotinamide... 394 1e-108
xla:447528 nnt, MGC83563; nicotinamide nucleotide transhydroge... 390 2e-107
cel:C15H9.1 nnt-1; Nicotinamide Nucleotide Transhydrogenase fa... 375 4e-103
eco:b1603 pntA, ECK1598, JW1595; pyridine nucleotide transhydr... 330 2e-89
dre:100331743 Nicotinamide Nucleotide Transhydrogenase family ... 123 4e-27
tgo:TGME49_003500 alanine dehydrogenase, putative (EC:1.4.1.1) 76.6 5e-13
tgo:TGME49_115260 alanine dehydrogenase, putative (EC:1.4.1.1) 53.1 6e-06
dre:100331681 MGC83563 protein-like 52.4 1e-05
mmu:217708 Lin52, 5830457H20Rik; lin-52 homolog (C. elegans) 36.6 0.60
hsa:91750 LIN52, C14orf46, c14_5549; lin-52 homolog (C. elegans) 36.6 0.60
hsa:3664 IRF6, LPS, OFC6, PIT, PPS, VWS, VWS1; interferon regu... 34.3 3.2
xla:494736 ankrd13a; ankyrin repeat domain 13A 33.9 3.6
sce:YMR020W FMS1; Fms1p (EC:1.5.3.11); K13367 non-specific pol... 33.1 6.5
mmu:387512 Tas2r135, Tas2r35, mt2r38; taste receptor, type 2, ... 33.1 6.6
mmu:629959 Gm7020, EG629959; predicted gene 7020 33.1
tgo:TGME49_078850 zinc finger DHHC domain-containing protein (... 32.7 8.5
> tgo:TGME49_118650 transhydrogenase, putative (EC:1.6.1.2); K00322
NAD(P) transhydrogenase [EC:1.6.1.1]
Length=1013
Score = 1283 bits (3319), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 649/1005 (64%), Positives = 804/1005 (80%), Gaps = 6/1005 (0%)
Query 94 GAVYLFSAVCFILCLRGLSTPQTAKRGNILGLVGMVAAVVVTFTEAGFGQHYLLFFATAA 153
G VYLFSA+CFIL LRGLSTP+TAKRGNILG+ GM AA+++TF+ GFG HY FF T A
Sbjct 3 GFVYLFSAICFILSLRGLSTPETAKRGNILGMTGMGAAILITFSTEGFGGHYAAFFLTVA 62
Query 154 PAVGLGLYIAQSVNMTEMPQLVALFHSFVGLAAVMVGFANFHSPAGVERASSLLRLLEVY 213
P +G+YIAQSV+M +MPQLVALFHSFVG AAV+VG ++FH+ G E ++ R +E
Sbjct 63 PPAIVGVYIAQSVSMVQMPQLVALFHSFVGFAAVLVGISSFHAGLG-ESGTNATRAVETA 121
Query 214 VGVFVAGITFTGSVVAAAKLHGSMESRSLRVPGRHALNSATIAAIGVLGALFCVSSGHFT 273
VG FVA +TFTGS+VAAAKLH M+ +SL++PGRHA+N+ T+AA+ V+G FC ++ T
Sbjct 122 VGEFVAAVTFTGSLVAAAKLHELMDPKSLKIPGRHAINAMTVAAVAVVGITFCATADTTT 181
Query 274 RMLCLYVNAGLSMWLGFHLVAAIGGADMPVVISLLNSYSGVALAASGFMLDNNLLIIAGA 333
+++CLY LS+WLGFHLVA+IGGADMPVVIS+LNSYSG A AASGFMLDNNLLIIAG+
Sbjct 182 KIICLYTLITLSLWLGFHLVASIGGADMPVVISMLNSYSGFATAASGFMLDNNLLIIAGS 241
Query 334 LIASSGAILSYIMCKGMNRSLWNVVLGGFEEAEGVGAAAPQGAVQQANADEVADELLAAR 393
LI SSGAILSYIMC+GMNR LW+VVLGG+E+A G +G V++ + D VA+E+L A+
Sbjct 242 LIGSSGAILSYIMCRGMNRDLWSVVLGGWEDAPAEGVPGVEGVVREISPDSVAEEVLLAK 301
Query 394 KVLIVPGYGMAVARCQSELADIAKNLMNCGITVDFGIHPVAGRMPGHMNVLLAEADVPYK 453
KVLIVPGYGMAV+RCQS++ADIA+ L + G+ V+FGIHPVAGRMPGHMNVLLAEA+V Y+
Sbjct 302 KVLIVPGYGMAVSRCQSDVADIAQILASRGVEVEFGIHPVAGRMPGHMNVLLAEANVSYR 361
Query 454 IVKEMSEVNPEMSSYDVVLVVGANDTVNPAALEPGSKISGMPVIEAWKARRVFVLKRSMA 513
VKEMSEVN +M YDVV+VVGANDTVNPA+LEPG+KI GMPVIE WKA+RV VLKRS+A
Sbjct 362 SVKEMSEVNKQMLEYDVVIVVGANDTVNPASLEPGTKIYGMPVIEVWKAKRVIVLKRSLA 421
Query 514 AGYASIENPLFHLENTRMLFGNAKNTTSAVFARVNARAEQMPPSAARDDLEAGLLEFERD 573
GYA+I+NPLF L+NTRMLFGNAK+TT+A+FA ++ RA + SA DLE G E
Sbjct 422 PGYAAIDNPLFFLDNTRMLFGNAKDTTTAIFACLSQRAAFVGKSAVFLDLEGGARALEEG 481
Query 574 ERADP-SSWPYPRLAVGVLKDSNG--SVMVPIAPKFVPRLRKMAFRVIVESGAGANAGFS 630
R P ++WP P+ VGVLKDSNG + +V +AP+FV RLRK AFRVIVESGAGA A FS
Sbjct 482 RRETPPTTWPSPKRTVGVLKDSNGRGTPLVSLAPRFVKRLRKQAFRVIVESGAGAEANFS 541
Query 631 DEEYRRAGAEIASNADAVINGAEVLLRVSAPTPEMVSRMPRDKVLISYLFPSVNTQALDM 690
D++Y +AGAEI NAD VI+ ++V+L+VS P+ E++ R+PR K+LIS++FP N L++
Sbjct 542 DDDYVKAGAEIMPNADTVISRSDVILKVSVPSEELIRRIPRGKILISHVFPGQNAPLLEL 601
Query 691 LARQGVTALAVDEVPRVTRAQKLDVKSAMQGLQGYRAVIEAFNALPKLSKASISAAGRVE 750
+A QG+TALAVDEVPR+TRAQ +DVKS+MQGLQGYRAV+EAFNALP+LSK SI+AAGRV+
Sbjct 602 MASQGLTALAVDEVPRITRAQNVDVKSSMQGLQGYRAVLEAFNALPRLSKTSITAAGRVD 661
Query 751 AAKVFVIGAGVAGLQAISTAHGLGAQVFGHDVRSATREEVESCGGKFIGLRMGEEAEVLG 810
AA+V V+GAGVAGLQAISTAHGL A+VF +DVR+ATREEVESCGG F+ + + EE EV G
Sbjct 662 AARVLVLGAGVAGLQAISTAHGLQAEVFAYDVRAATREEVESCGGTFLSVELEEEGEVEG 721
Query 811 GYAREMGDAYQRAQRELIANTIKHCDVVICTAAIHGKPSPKLISRDMLRSMKPGSVIVDI 870
GYAREMG+AY+ AQ+++++ + + DV+ICTAAIHGKPSPKLIS DML +M+PGSV++D+
Sbjct 722 GYAREMGEAYEMAQKQMLSRVVPNVDVIICTAAIHGKPSPKLISNDMLATMRPGSVVIDL 781
Query 871 ATEFGDTRSGWGGNVEVSPKDDQVVVDGITVIGRKRIETRMPVQASELFSMNICNLLEDL 930
ATEFGD RS WGGNVE SP D + + G+T+IGR RIET+MP QASELFSMN+ NLLE+L
Sbjct 782 ATEFGDRRSNWGGNVEGSPTDGETQIHGVTIIGRSRIETQMPTQASELFSMNMSNLLEEL 841
Query 931 GGGSNFRVNMDDEVIRGLVAVYQGRNVWQPPQPTPVSRTPPRVPMPSAPSAAPAKAGGAF 990
GGG NFR+++ +EVI+GL V++GR W P P+ R PP S+P P K
Sbjct 842 GGGENFRIDLTNEVIKGLCCVHEGRVSWA--PPEPLPRRPPTPSPRSSPRLVPPKEVSLL 899
Query 991 AQALASDAFFAMCLVVAAAVVGLLGIVLDPMELKHLTLLALSLIVGYYCVWAVTPALHTP 1050
Q + SDAFFAM LV AA GLLG+ L +EL+ TLLALSLIVGYY VW+VTPALHTP
Sbjct 900 DQLVTSDAFFAMSLVCTAAFAGLLGVTLKALELQQFTLLALSLIVGYYSVWSVTPALHTP 959
Query 1051 LMSVTNALSGVIVIGCMLEYGTALISGFTLLALVGTFLASVNIAG 1095
LMSVTNALSGVI+IG MLEYG + S + AL TF +S+NIAG
Sbjct 960 LMSVTNALSGVIIIGSMLEYGPSATSASAICALCATFFSSLNIAG 1004
> pfa:PF14_0508 pyridine nucleotide transhydrogenase, putative
(EC:1.6.1.2); K00322 NAD(P) transhydrogenase [EC:1.6.1.1]
Length=1176
Score = 1016 bits (2626), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 500/1076 (46%), Positives = 709/1076 (65%), Gaps = 66/1076 (6%)
Query 86 FSNMPSLLGAVYLFSAVCFILCLRGLSTPQTAKRGNILGLVGMVAAVVVTFTEAGFGQHY 145
+S +PSLL AVYLFS++CFILCL GL+ +T+KRGNILG +G+VAA+++TF++ GFG Y
Sbjct 89 YSVLPSLLNAVYLFSSICFILCLTGLNAHRTSKRGNILGFIGIVAAIMITFSQVGFGFRY 148
Query 146 LLFFATAAPAVGLGLYIAQSVNMTEMPQLVALFHSFVGLAAVMVGFANFHSPAGVERASS 205
LF PA+ +G+YIA +V+M +MPQLVALFHSFVGLAA+ VGF+ +HS + S
Sbjct 149 KLFLLIVIPAITIGIYIAHNVSMVQMPQLVALFHSFVGLAALFVGFSKYHSESFENYEIS 208
Query 206 LLRLLEVYVGVFVAGITFTGSVVAAAKLHGSMESRSLRVPGRHALNSATIAAIGVLGALF 265
+ LLE+YVG F+A I F GS+VAA KL G ++S+SL++ + +N I I +LG F
Sbjct 209 TIHLLELYVGTFIASIAFIGSLVAAGKLSGILDSKSLKLQIKKIINILCIVLIIILGYYF 268
Query 266 CVSSGHFTRMLCLYVNAGLSMWLGFHLVAAIGGADMPVVISLLNSYSGVALAASGFMLDN 325
+ + +CLY++ + +LGFHL+A+IG ADMPV+IS+LNSYSG A A SGF+L N
Sbjct 269 VTLKLLYLKSICLYISLIIDSFLGFHLIASIGAADMPVIISVLNSYSGFATAISGFLLHN 328
Query 326 NLLIIAGALIASSGAILSYIMCKGMNRSLWNVVLGGFEEAEGVGAA-APQGAVQQANADE 384
NLLII+GALI SSGAILSYIMC GMNR ++N++LGG+++ E +G + Q +++ N
Sbjct 329 NLLIISGALIGSSGAILSYIMCIGMNRDIFNIILGGWDDYENMGESIYDQNFIEKKNKQT 388
Query 385 --------VADELLAARKVLIVPGYGMAVARCQSELADIAKNLMNCGITVDFGIHPVAGR 436
VA+ L+ A+ ++IVPGYG A+++CQ ELA+I L + I V F IHPVAGR
Sbjct 389 INSTTNKYVAENLINAKNIIIVPGYGTALSKCQRELAEICSILTSRNINVTFAIHPVAGR 448
Query 437 MPGHMNVLLAEADVPYKIVKEMSEVNPEMSSYDVVLVVGANDTVNPAALEPGSKISGMPV 496
MPGH+NVLLAEA++PY IVKEM+E+NP +S D+VLVVGAND VNP++L+P SKI GMPV
Sbjct 449 MPGHLNVLLAEANIPYNIVKEMNEINPIISEADIVLVVGANDIVNPSSLDPSSKIYGMPV 508
Query 497 IEAWKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVFARVNARAEQMPP 556
IE WK+++V V KR++ GY++I+NPLF+ NT +LFG+AK+TT+ + +N P
Sbjct 509 IEVWKSKQVIVFKRTLNTGYSAIDNPLFYFSNTFLLFGDAKHTTNQILTILNDYVNNKYP 568
Query 557 SAARDD--LEAGLLEF------------ERDERADPSSWPYPRLAVGVLKDSN------- 595
+ D + +F D + ++P PR +G++KD N
Sbjct 569 DISDQDRHINHDKTQFRYSYSLTDSSHSNDDSTSKEQNYPKPRRVIGLIKDDNVQGGNDL 628
Query 596 ----------------------GSVMVPIAPKFVPRLRKMAFRVIVESGAGANAGFSDEE 633
+VPI+PKF+P+LR M FR++VE G N ++E
Sbjct 629 LIEHIQSKVENAPNIKDQKRDVNLSIVPISPKFIPKLRLMGFRILVERDIGTNILMQNDE 688
Query 634 YRRAGAEIASNADAVINGAEVLLRVSAPTPEMVSRMPRDKVLISYLFPSVNTQALDML-- 691
Y + GAE+ S + ++ + ++L+V PT + + + +LISYL+PS+N LD +
Sbjct 689 YTKYGAEVVS-RNVILQQSNIILKVDPPTVNFIEEIQNNTILISYLWPSINYHLLDKMIQ 747
Query 692 --ARQGVTALAVDEVPRVTRAQKLDVKSAMQGLQGYRAVIEAFNALPKLSKASISAAGRV 749
+ +T LA+DEVPR TRAQKLDV+S+M LQGYRAV+EAF LP+ SK+SI+AAG++
Sbjct 748 DEEKHNITYLAIDEVPRSTRAQKLDVRSSMSNLQGYRAVLEAFFILPRFSKSSITAAGKI 807
Query 750 EAAKVFVIGAGVAGLQAISTAHGLGAQVFGHDVRSATREEVESCGGKFIGLRMGEEAEVL 809
AKVFVIGAGVAGLQAI TA LGA V+ HD R AT EEV+SCGG FI + E ++L
Sbjct 808 NPAKVFVIGAGVAGLQAIITAKSLGAIVYSHDSRLATEEEVKSCGGIFIRIPTSERGDIL 867
Query 810 GGYAREMGDAYQRAQRELIANTIKHCDVVICTAAIHGKPSPKLISRDMLRSMKPGSVIVD 869
+ Y + Q L IK CD++IC+A+I GK SPKL++ +M++ MKPGSV VD
Sbjct 868 NMSTDMNNEEYIKVQSNLFKKIIKKCDILICSASIPGKTSPKLVTTEMIKLMKPGSVAVD 927
Query 870 IATEFGDTRSGWGGNVEVSPKDDQVVVDGITVIGRKRIETRMPVQASELFSMNICNLLED 929
++TEFGD ++ WGGN+E S ++ ++++G+ V+GR +IE MP+QAS+LFSMN+ NLLE+
Sbjct 928 LSTEFGDKKNNWGGNIECSQSNENILINGVHVLGRDKIERNMPMQASDLFSMNMINLLEE 987
Query 930 LGGGSNFRVNMDDEVIRGLVAVYQGRNVWQPPQPTP--------VSRTPPRVPMPSAPSA 981
+GGG +F ++M++++I+ LV + G ++ P + ++ P +
Sbjct 988 MGGGVHFNIDMNNDIIKSLVVIKDGNILYSPDKSVEKLIKSESVFINEKNQLIEPISEQK 1047
Query 982 APAKAGGAFAQA-LASDAFFAMCLVVAAAVVGLLGIVLDPMELKHLTLLALSLIVGYYCV 1040
G + + SD FF + L+ + L L +L+ L L LS IVGYYCV
Sbjct 1048 IKYPTGLRLTEKFIESDTFFYISLLFVIILTFLAATYLSQSDLQSLFLFTLSTIVGYYCV 1107
Query 1041 WAVTPALHTPLMSVTNALSGVIVIGCMLEYGTALISGFTLLALVGTFLASVNIAGG 1096
W+VTPALHTPLMS+TNALSGVI+IG M+EYG ++L+++ TFL+SVN++GG
Sbjct 1108 WSVTPALHTPLMSMTNALSGVIIIGSMIEYGNGYKDLSSILSMLATFLSSVNLSGG 1163
> cpv:cgd8_2330 pyridine nucleotide/ NAD(P) transhydrogenase alpha
plus beta subunits, duplicated gene, possible signal peptide
plus 12 transmembrane regions ; K00322 NAD(P) transhydrogenase
[EC:1.6.1.1]
Length=1143
Score = 989 bits (2558), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 530/1124 (47%), Positives = 738/1124 (65%), Gaps = 56/1124 (4%)
Query 8 WRPSGLSTVFLATCLGVALLSTTTNAEFDFPAANLRSIQRHLVRMGEFFPVNRPGEGDEG 67
W +++ LA CL + L+ + E D P +R + +R G P E
Sbjct 8 WMLKSSTSLILAICLVLFLVLENVSCE-DVP---IRLKTSNNLRGG-------PSTNRES 56
Query 68 FDGARPENPEGGAISKFAFSNMPSLLGAV-YL---------FSAVCFILCLRGLSTPQTA 117
P + K+ FS + S L V YL FS++CF+LCLRGLST +TA
Sbjct 57 NILVPAFEPSKEYVDKY-FSYVESFLKNVNYLSTLSVSLEVFSSICFVLCLRGLSTQETA 115
Query 118 KRGNILGLVGMVAAVVVTFTEAGFGQHYLLFFATAAPAVGLGLYIAQSVNMTEMPQLVAL 177
KRGN LG+VG++ A+ TF F ++++F A A+ +G+ I+ V+M +PQLVAL
Sbjct 116 KRGNSLGIVGIICAIAATFLAPTFTMNWIMFIVPFALAIIIGIPISHCVSMVNIPQLVAL 175
Query 178 FHSFVGLAAVMVGFANFHSP-AGVERASSLLRLLEVYVGVFVAGITFTGSVVAAAKLHGS 236
FHSFVGLAA+++ FAN +P E S + +E+++G ++ ITFTGS+VAA KLH
Sbjct 176 FHSFVGLAAMLISFANLWTPFQSSEEEFSAVHAIEMFIGEAISAITFTGSIVAAGKLHEI 235
Query 237 MESRSLRVPGRHALNSATIAAIGVLGALFCVSSGHFTRMLCLYVNAGLSMWLGFHLVAAI 296
S +L++PGRH LN+ +A + LG++F + + + R +Y N+ LSM LG HLVA+I
Sbjct 236 FPSGALKLPGRHFLNALMVAGLIALGSVFIIITDYANRTYLMYGNSLLSMLLGIHLVASI 295
Query 297 GGADMPVVISLLNSYSGVALAASGFMLDNNLLIIAGALIASSGAILSYIMCKGMNRSLWN 356
GGADMPV IS+LNSYSG A + +GF++++ +LII GALI SSGAILS+IMC GMNRSL N
Sbjct 296 GGADMPVAISMLNSYSGWATSFTGFLINSKMLIICGALIGSSGAILSHIMCLGMNRSLLN 355
Query 357 VVLGGFE------EAEGVGAAAPQGAVQQANADEVADELLAARKVLIVPGYGMAVARCQS 410
V+LGG+E EA+ +G Q V + NA +VA +LL+A+KVLIVPGYGMAV+R Q
Sbjct 356 VMLGGWESNGDSNEAQALG---DQSDVNKTNAMKVARDLLSAKKVLIVPGYGMAVSRSQQ 412
Query 411 ELADIAKNLMNCGITVDFGIHPVAGRMPGHMNVLLAEADVPYKIVKEMSEVNPEMSSYDV 470
++A I L I V+F IHPVAGRMPGHMNVLLAEADVPY IVKEM +VNP M S+DV
Sbjct 413 DVASIVNALRLRDIYVEFAIHPVAGRMPGHMNVLLAEADVPYSIVKEMEDVNPHMESFDV 472
Query 471 VLVVGANDTVNPAALEPGSKISGMPVIEAWKARRVFVLKRSMAAGYASIENPLFHLENTR 530
VLV+GANDTVNP ALE SKI+GMPVIE W+A +V V KRS+ GYA+I+NPLF ++N
Sbjct 473 VLVIGANDTVNPLALEKDSKINGMPVIEVWRASKVIVSKRSLGKGYAAIDNPLFFMKNVE 532
Query 531 MLFGNAKNTTSAVFARVNARAEQMPPSAARDDLEAGLLEFERDERADPSSWPYPRLAVGV 590
M+FGNAK++ + + + + + + DD E ++ + D +P P + +GV
Sbjct 533 MIFGNAKDSMINILQNIISISPEQKKANLNDDQET--IDGTTASQEDNEEYPTPTMTIGV 590
Query 591 LK-DSNGSVMVPIAPKFVPRLRKMAFRVIVESGAGANAGFSDEEYRRAGAEIASNADAVI 649
LK D +V IAP FV +LRK+ FRV+VESGAG + F D++Y A I + V+
Sbjct 591 LKEDLPSEKLVAIAPNFVKKLRKLGFRVLVESGAGTKSQFDDQKYENASCTIMATRQDVV 650
Query 650 NGAEVLLRVSAPTPEMVSRMPRDKVLISYLFPSVNTQALDMLARQGVTALAVDEVPRVTR 709
+ ++V+++V PT E +S+M + L+SY++P+ N L+ LA++GVT +A+DEVPR TR
Sbjct 651 SRSDVIVKVQKPTDEEISQMKSGQTLVSYIWPAQNPSLLESLAKKGVTTIALDEVPRTTR 710
Query 710 AQKLDVKSAMQGLQGYRAVIEAFNALPKLSKASISAAGRVEAAKVFVIGAGVAGLQAIST 769
AQKLD++S+M L GYRAVIEAF LPKLSK+SI+AAGRV+AA+VFVIGAGVAGLQAI+T
Sbjct 711 AQKLDIRSSMSNLAGYRAVIEAFVQLPKLSKSSITAAGRVDAARVFVIGAGVAGLQAIAT 770
Query 770 AHGLGAQVFGHDVRSATREEVESCGGKFIGLRMGEEAEVLGGYAREMGDAYQRAQRELIA 829
A LGA V+ D R+ATREEVES G KF+ + + E+ + GYA+ M Y +AQ +L +
Sbjct 771 AKNLGADVYASDTRTATREEVESLGAKFVTVDIKEDGDSGSGYAKVMSPEYLKAQSKLYS 830
Query 830 NTIKHCDVVICTAAIHGKPSPKLISRDMLRSMKPGSVIVDIATEFGDTRSGWGGNVEVSP 889
I+ CDVVI TA I GKPSPK+I+R+M+ SMKPGSVIVD+A E DT SGWGGN E++
Sbjct 831 KMIRSCDVVITTALIPGKPSPKIITREMVNSMKPGSVIVDMAAEMADTASGWGGNCEIT- 889
Query 890 KDDQVVVD---GITVIGRKRIETRMPVQASELFSMNICNLLEDLGGGSNFRVNMDDEVIR 946
K DQ+ +D G+T+IG + + MP QASELFSMN+ ++LE+LGG +F V+M D++++
Sbjct 890 KKDQIYLDEKSGVTIIGLTNLPSTMPSQASELFSMNVVHMLEELGGAEHFSVDMKDDLLK 949
Query 947 GLVAVYQGRNVWQPPQPTPVSRTPPRVPMPSAPSAAPAKA----------GGAFAQALAS 996
+V + + P P PP+ ++ S++ + + + + S
Sbjct 950 EMVVTINEKVTYVPVDKRP---PPPKSESSNSSSSSSSSSSSTETSHRSFSSIMERVIYS 1006
Query 997 DAFFAMCLVVAAAVVGLLGIVLDPMELKHLTLLALSLIVGYYCVWAVTPALHTPLMSVTN 1056
+ F + ++ V LG ++D L ++ + +LS+IVGYYC+W VTP+LHTPLMSVTN
Sbjct 1007 NVSFGFFVFISVLVSIGLGYIIDHDTLGNILVFSLSVIVGYYCIWNVTPSLHTPLMSVTN 1066
Query 1057 ALSGVIVIGCMLEYGTALI----SGFTLLALVGTFLASVNIAGG 1096
ALSG+I+IG MLE G ++ ++ L + L+S+NI GG
Sbjct 1067 ALSGIIIIGAMLECGPVILFTDFQVYSFLLFLAMLLSSINIIGG 1110
> cpv:cgd1_990 pyridine nucleotide/ NAD(P) transhydrogenase alpha
plus beta subunits, duplicated gene, 12 transmembrane domain
(EC:1.6.1.2)
Length=1147
Score = 781 bits (2017), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 434/1101 (39%), Positives = 648/1101 (58%), Gaps = 104/1101 (9%)
Query 84 FAFSNMPSLLGAVYLFSAVCFILCLRGLSTPQTAKRGNILGLVGMVAAVVVTFTEAGFGQ 143
+ SN SL A F+A+CFI C++GLS+ +TA R NI+G VGM+ A++V+ +E GFG
Sbjct 50 YQTSNSGSLTMAGQFFAALCFIYCIKGLSSEKTAYRSNIVGFVGMMVAILVSLSEEGFGN 109
Query 144 HYLLFFATAAPAVGLGLYIAQSVNMTEMPQLVALFHSFVGLAAVMVGFANFHSPAGVERA 203
HY +FF + +G+YIA + +MPQLVA+FHS VGLAA+ V ++ F+S G++
Sbjct 110 HYFVFFLATIVSGAVGVYIADKTQLIKMPQLVAIFHSLVGLAALFVSYSYFYSSIGMKEG 169
Query 204 SSLLRLLEVYVGVFVAGITFTGSVVAAAKLHGSMESRSLRVPGRHALNSATIAAIGVLGA 263
+LR +E++VG + ITF GSV+AA KL + S+S+++P ++ S + +I + G
Sbjct 170 IYVLRRMELFVGGVMGMITFVGSVIAALKLDDIIPSKSVKIPLKNVSLSILLFSICMTGL 229
Query 264 LFCVSSGHFTRMLCLYVNAGLSMWLGFHLVAAIGGADMPVVISLLNSYSGVALAASGFML 323
FCVS+ L+ LS G ++ +IGGADMPV+IS+LNSYSG + A +GF+L
Sbjct 230 SFCVSNNELVITTNLHCGMILSALFGIFMIISIGGADMPVIISMLNSYSGWSTAITGFLL 289
Query 324 DNNLLIIAGALIASSGAILSYIMCKGMNRSLWNVVLGGFEEAEGVGAAAPQGAVQQANAD 383
DN+LLII+GALI SSGAILSYIMCKGMNR +V+LGGF++ E ++
Sbjct 290 DNSLLIISGALIGSSGAILSYIMCKGMNRDFVSVLLGGFDQEE-TEKMVGDTKCYTTSSK 348
Query 384 EVADELLAARKVLIVPGYGMAVARCQSELADIAKNLMNCGITVDFGIHPVAGRMPGHMNV 443
E A+ L + K+LIVPGYGMAV++CQ ++DI K L + V+ GIHPVAGRMPGHMNV
Sbjct 349 ETAEILAESSKILIVPGYGMAVSKCQDIISDIIKELESRSTIVEIGIHPVAGRMPGHMNV 408
Query 444 LLAEADVPYKIVKEMSEVNPEMSSYDVVLVVGANDTVNPAALEPGSKISGMPVIEAWKAR 503
LLAE+ VP +IVKEM VN M YD+VLVVGAND VNPAAL+P SKISGMPVI WKA+
Sbjct 409 LLAESGVPSRIVKEMDAVNNCMHEYDLVLVVGANDIVNPAALDPQSKISGMPVINVWKAK 468
Query 504 RVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVFA------------RVNAR- 550
+V V KRS+A GYA IEN LF E TRML G+++ T V+ +++R
Sbjct 469 QVVVSKRSLAYGYACIENELFTCERTRMLLGDSRETLQQVYKILKGCAGFVPRQYIDSRV 528
Query 551 AEQMPPSAARDDLEAGLLEFE-------------RDERADPSSWPYPRLAVG-------- 589
E++ D++ LL + R ++A + ++++
Sbjct 529 TEEIEEVEDTDEVTTALLSSKQRKSSKTSGNVSSRSKKAGGAETETEKVSMSANKGSKKI 588
Query 590 ----VLKDSNGSVMVPIAPKFVPRLRKMAFRVIVE----SGAGANAGFSDEEYRRAGAEI 641
++ + + +++ PI P V + R+ + V + + + + FS+E Y R+GA +
Sbjct 589 ALLPIVSEQDKTLLFPIPPSKVYKFREKGYEVAISRDVLTSSFQSKYFSEEIYMRSGALV 648
Query 642 ASNADAVINGAEVLLRVSAPTPEMVSRMPRDKVLISYLFPS-------VNTQALDMLARQ 694
N + +I +V+++++ P+ + M + + LI + S Q L L +
Sbjct 649 YKNIEELIKECDVVIKMARPSEKEAKCMKQGQFLICNMHISQYQDKENSQDQLLASLTKN 708
Query 695 GVTALAVDEVPRVTRAQKLDVKSAMQGLQGYRAVIEAFNALPKLSKASISAAGRVEAAKV 754
GVT +A+DEVPR +RAQ +D+++ + GYRAV++AFN LP++SK+ SAAG VE + V
Sbjct 709 GVTVIALDEVPRTSRAQTMDIRTTTSTISGYRAVVDAFNYLPRISKSISSAAGNVEESTV 768
Query 755 FVIGAGVAGLQAISTAHGLGAQVFGHDVRSATREEVESCGGKFIGLRMGEEAEVLGGYAR 814
VIGAGVAGLQAI+TA +G +VF D RS ++EE ESCG +FI ++ E E L
Sbjct 769 LVIGAGVAGLQAIATAKSMGTKVFAMDSRSTSKEEAESCGARFI--QVPSEGESL----- 821
Query 815 EMGDAYQRAQRELIANTIKHCDVVICTAAIHGKPSPKLISRDMLRSMKPGSVIVDIATEF 874
+ + QR+LI + D+VI +A G+ P LI+++ +R MK GSVIVD+ +EF
Sbjct 822 -RKEEILKKQRDLIEKYLCISDIVITSACKPGEECPILITKEAVRKMKSGSVIVDLCSEF 880
Query 875 GDTRSGWGGNVEVSPKDDQV--VVDGITVIGRKRIETRMPVQASELFSMNICNLLEDLGG 932
GGN E++ KD V G T+IG+ MP+Q+SELFS N+ +L+ +LG
Sbjct 881 -------GGNCELTQKDRTFSDVQSGTTIIGKCNYVFSMPLQSSELFSGNLLSLISELGP 933
Query 933 GSN-FRVNMDDEVIRGLVAVYQGRNVWQP-------------------------PQPTPV 966
S+ F+ ++++E+I + ++ + +W+P Q T +
Sbjct 934 TSDRFKCDLNNEIISKMCVAHENKMLWRPFTEQNQEKHENQEMLEKKSLLENISKQSTLI 993
Query 967 S-----RTPPRVPMPSAPSAAPA------KAGGAFAQALASDAFFAMCLVVAAAVVGLLG 1015
S T + + + + K F + L F +++A +LG
Sbjct 994 SIKDEDTTTSKTSVFCSKTEIDGYLQKVMKEFKNFDKQLNGGVNFYAGVILATLFFTVLG 1053
Query 1016 IVLDPMELKHLTLLALSLIVGYYCVWAVTPALHTPLMSVTNALSGVIVIGCMLEYGTALI 1075
+ + ++++++ +S ++GYYCVW V P LHTPLMSVTNALSGVI+IG M++YG +
Sbjct 1054 LTMTTIQIQNIFSFIISTMLGYYCVWDVDPKLHTPLMSVTNALSGVIIIGSMMQYGNQTV 1113
Query 1076 SGFTLLALVGTFLASVNIAGG 1096
+ TL+++ TFLAS+N GG
Sbjct 1114 TYTTLMSMFSTFLASINTFGG 1134
> tgo:TGME49_101210 NAD(P) transhydrogenase, alpha subunit, putative
(EC:1.6.1.2)
Length=1165
Score = 501 bits (1289), Expect = 9e-141, Method: Compositional matrix adjust.
Identities = 373/1109 (33%), Positives = 548/1109 (49%), Gaps = 176/1109 (15%)
Query 120 GNILGLVGMVAAVVVTFTEAG-FGQHYLLFFATAAPAVGLGLYIAQSVNMTEMPQLVALF 178
GNI G++GM+ AV+ T + + +FF P L + +A V MT +PQLV
Sbjct 2 GNIYGVIGMLVAVIGTLVSPFVYDRALWIFFVVCVPPAVLAVIVAMLVKMTSIPQLVGAL 61
Query 179 HSFVGLAAVMVGFANFHSPAGVERASSLLRL-------LEVYVGVF------VAGITFTG 225
++F GLAA + FA + SP V + ++ + +VY VF + ITFTG
Sbjct 62 NAFGGLAATLESFALYFSPYEVAKQEAMQSVGETRYLQFQVYQTVFYLIGASIGMITFTG 121
Query 226 SVVAAAKLHGSMESRSLRVPGR------HALNSATIAAIGVLGALFCVSSGHFTRMLCLY 279
S+VA KL G + S+ +P R A+ + L V G LC +
Sbjct 122 SLVACGKLSGWIASKPRVMPLRCFWMPVVLALLLAAGAVAGVLGLDNVPVGTVCLSLCTF 181
Query 280 VNAGLSMWLGFHLVAAIGGADMPVVISLLNSYSGVALAASGFMLDNNLLIIAGALIASSG 339
LS G V AIGGADMPVVIS+LNS SG A +G L N+++IIAGA + +SG
Sbjct 182 ----LSAVYGVCFVLAIGGADMPVVISVLNSGSGWAGVFAGLTLQNSIIIIAGAFVGASG 237
Query 340 AILSYIMCKGMNRSLWNVVLGGF-EEAEGVGAAAPQGAVQQANADEVADELLAARKVLIV 398
ILSY+MC MNRSL NV+LGGF + + A A +G + A +++VA LLA++ V+IV
Sbjct 238 IILSYVMCTAMNRSLCNVLLGGFGDTGKESNAQAFEGEAKIATSEQVAAYLLASQSVIIV 297
Query 399 PGYGMAVARCQSELADIAKNLMNCGITVDFGIHPVAGRMPGHMNVLLAEADVPYKIVKEM 458
PGYGMAV+ Q + ++ + L G V F IHPVAGR+PGHMNVLLAEA+VPY IV M
Sbjct 298 PGYGMAVSHAQFAVHELTQQLRERGTQVRFAIHPVAGRLPGHMNVLLAEANVPYDIVLSM 357
Query 459 SEVNPEMSSYDVVLVVGANDTVNPAALE-PGSKISGMPVIEAWKARRVFVLKRSMAAGYA 517
E+N + D V+++GANDTVNPAA PG I GMPV+E WKA++V VLKRSM GYA
Sbjct 358 DEINEDFPQTDTVIIIGANDTVNPAAQNAPGCPIYGMPVLEVWKAKKVIVLKRSMRVGYA 417
Query 518 SIENPLFHLENTRMLFGNAKNTTSAVFARVNARAEQMPPSAARDDLEAGLLEFER----- 572
++NPLF N+ ML G+AK + + + + + P A+R E + R
Sbjct 418 GVDNPLFFYPNSEMLLGDAKASLQTLLTDMQDKLDAHPIEASRLSSETQTNKSARIAVNV 477
Query 573 -DERADPSSWPY-------------------------------------PRLAVGVLKDS 594
+++ P+ P VGVLK+
Sbjct 478 AGDKSAKQEKPHGAAADKPDATETAGTVGVTVTGSGAPGEVPPVAESAPPLFFVGVLKER 537
Query 595 NG-SVMVPIAPKFVPRLRKMAFRVIVESGAGANAGFSDEEYRRAGAEIASNADAVINGAE 653
V + P V +LR++ I+E AG +AGF DE+Y +GA IA A+ V+ +
Sbjct 538 RPLERRVAMTPSVVKKLRELQLGCIIEKQAGVSAGFLDEQYMESGALIADTAEEVVRLSR 597
Query 654 VLLRVSA------------PTPEMVSRMPRDKVLISYLFPSVNTQALDMLARQGVTA--- 698
V+++V+ E SR + L + P+ N + ++ + +G
Sbjct 598 VVVKVTTFECDEVALAKPQNDDEQSSRESTEPPLPALATPA-NKEDTELYSVEGNGKGHR 656
Query 699 -LAVDEV-------PRVTRAQKLDVKSAMQGLQGYRAVIEAF----NALPKLSKASISAA 746
L+ ++V P T ++ SA +G +G + ++A A+P+++ S+
Sbjct 657 LLSSEKVFVAGFVGPN-TATSEVSADSAPEGEKGKLSPVDALLRSALAIPQVTLISLDVL 715
Query 747 GRVEAAK---VFVIGAGVAGLQAISTAHGLGAQVFGHDVRSA------------------ 785
R+ ++ V A +AG +A+ A ++ G ++ SA
Sbjct 716 PRLTISQKMDVLSSTAKLAGYRAVVEAISHFGRLMGPEITSAGKYPPAKVLVIGAGVAGL 775
Query 786 ---------------------TREEVESCGGKFIGLRM---GEEAEVL-GGYAREMGDAY 820
+E+VES GG+F+ + GE GGYA+ M + +
Sbjct 776 QAIGDAHRLGADVRGFDVRLECKEQVESMGGEFLAMTFEGPGESGRATAGGYAKPMSEEF 835
Query 821 QRAQRELIANTIKHCDVVICTAAIHGKPSPKLISRDMLRSMKPGSVIVDIATEFGDTRSG 880
R + EL A + D++I TA++ G+P+PKLI ++ + +MK GSVIVD+A +
Sbjct 836 LRKEMELFAEQCREIDILITTASLPGRPAPKLIKKEHVDTMKAGSVIVDLA-------AP 888
Query 881 WGGNVEVSPKDDQVVVDG-ITVIGRKRIETRMPVQASELFSMNICNLLEDLGGGSNFRVN 939
GGNVEV+ + + +G +TV+G + +RM QASE+F+ NI NLLE +G G +F VN
Sbjct 889 TGGNVEVTRPGETYLYNGKVTVVGWTDLPSRMAPQASEMFARNIFNLLEHMGAGRDFSVN 948
Query 940 MDDEVIRGLVAVYQGRNVWQPPQPTPVSRTPPRVPMPSA------PSAAPAKAGGAFAQA 993
DDE+IR + GR + PP+ P SA AAP + ++
Sbjct 949 FDDEIIRAITVARHGRKTYPPPRK---ESDPENTNAGSALLTGSNTDAAPVEESAQRSEN 1005
Query 994 LAS--------------DAFFAMCLVVAAAVVGLLGIVLDPMELKHLTLLALSLIVGYYC 1039
L + ++V AA L P L + L+ VGY
Sbjct 1006 LWHLVQKRMRNRWRGIVAPLDILTILVIAAFTALFAATAPPSFPPLLFIFFLACFVGYLL 1065
Query 1040 VWAVTPALHTPLMSVTNALSGVIVIGCML 1068
VW V PALHTPLMSVTNA+SG +++G ML
Sbjct 1066 VWNVAPALHTPLMSVTNAISGTVLVGGML 1094
> eco:b1602 pntB, ECK1597, JW1594; pyridine nucleotide transhydrogenase,
beta subunit (EC:1.6.1.2); K00325 NAD(P) transhydrogenase
subunit beta [EC:1.6.1.2]
Length=462
Score = 427 bits (1098), Expect = 1e-118, Method: Compositional matrix adjust.
Identities = 228/456 (50%), Positives = 305/456 (66%), Gaps = 4/456 (0%)
Query 92 LLGAVYLFSAVCFILCLRGLSTPQTAKRGNILGLVGMVAAVVVTFTEAGFGQHYLLFFAT 151
L+ A Y+ +A+ FI L GLS +T+++GN G+ GM A++ T G + A
Sbjct 5 LVTAAYIVAAILFIFSLAGLSKHETSRQGNNFGIAGMAIALIATIFGPDTGNVGWILLAM 64
Query 152 AAPAVGLGLYIAQSVNMTEMPQLVALFHSFVGLAAVMVGFANF-HSPAGVERASSLLRLL 210
+G+ +A+ V MTEMP+LVA+ HSFVGLAAV+VGF ++ H AG+ + L
Sbjct 65 VIGG-AIGIRLAKKVEMTEMPELVAILHSFVGLAAVLVGFNSYLHHDAGMAPILVNIHLT 123
Query 211 EVYVGVFVAGITFTGSVVAAAKLHGSMESRSLRVPGRHALNSATIAAIGVLGALFCVSSG 270
EV++G+F+ +TFTGSVVA KL G + S+ L +P RH +N A + +L +F +
Sbjct 124 EVFLGIFIGAVTFTGSVVAFGKLCGKISSKPLMLPNRHKMNLAALVVSFLLLIVFVRTDS 183
Query 271 HFTRMLCLYVNAGLSMWLGFHLVAAIGGADMPVVISLLNSYSGVALAASGFMLDNNLLII 330
++L L + +++ G+HLVA+IGGADMPVV+S+LNSYSG A AA+GFML N+LLI+
Sbjct 184 VGLQVLALLIMTAIALVFGWHLVASIGGADMPVVVSMLNSYSGWAAAAAGFMLSNDLLIV 243
Query 331 AGALIASSGAILSYIMCKGMNRSLWNVVLGGF-EEAEGVGAAAPQGAVQQANADEVADEL 389
GAL+ SSGAILSYIMCK MNRS +V+ GGF + G G ++ A+E A+ L
Sbjct 244 TGALVGSSGAILSYIMCKAMNRSFISVIAGGFGTDGSSTGDDQEVGEHREITAEETAELL 303
Query 390 LAARKVLIVPGYGMAVARCQSELADIAKNLMNCGITVDFGIHPVAGRMPGHMNVLLAEAD 449
+ V+I PGYGMAVA+ Q +A+I + L GI V FGIHPVAGR+PGHMNVLLAEA
Sbjct 304 KNSHSVIITPGYGMAVAQAQYPVAEITEKLRARGINVRFGIHPVAGRLPGHMNVLLAEAK 363
Query 450 VPYKIVKEMSEVNPEMSSYDVVLVVGANDTVNPAAL-EPGSKISGMPVIEAWKARRVFVL 508
VPY IV EM E+N + + D VLV+GANDTVNPAA +P S I+GMPV+E WKA+ V V
Sbjct 364 VPYDIVLEMDEINDDFADTDTVLVIGANDTVNPAAQDDPKSPIAGMPVLEVWKAQNVIVF 423
Query 509 KRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVF 544
KRSM GYA ++NPLF ENT MLFG+AK + A+
Sbjct 424 KRSMNTGYAGVQNPLFFKENTHMLFGDAKASVDAIL 459
> dre:406619 nnt, wu:fa20d10, wu:fc86a04, zgc:76979; nicotinamide
nucleotide transhydrogenase (EC:1.6.1.2); K00323 NAD(P)
transhydrogenase [EC:1.6.1.2]
Length=1079
Score = 405 bits (1041), Expect = 4e-112, Method: Compositional matrix adjust.
Identities = 226/462 (48%), Positives = 306/462 (66%), Gaps = 14/462 (3%)
Query 96 VYLFSAVCFILCLRGLSTPQTAKRGNILGLVGMVAAVVVTFTEAGFGQHYLLFFATAAPA 155
+YL S +C + L GLST TA+ GN LG++G+ + TF L+ +AA A
Sbjct 619 MYLGSGLCCVGALGGLSTQSTARLGNALGMIGVAGGIAATFGVLKPSPE-LMAQMSAAMA 677
Query 156 VG--LGLYIAQSVNMTEMPQLVALFHSFVGLAAVMVGFANF---HSPAGVERASSLLRLL 210
VG GL IA+ + ++++PQLVA FHS VGLAAV+ A + + + A++L +++
Sbjct 678 VGGTAGLTIAKKIQISDLPQLVAAFHSLVGLAAVLTCVAEYMVEYPHFATDPAANLTKIV 737
Query 211 EVYVGVFVAGITFTGSVVAAAKLHGSMESRSLRVPGRHALNSATIAAIGVLGAL-FCVSS 269
Y+G ++ G+TF+GS+VA KL G + S L +PGRHALN AT+ A V G + + +
Sbjct 738 -AYLGTYIGGVTFSGSLVAYGKLQGLLNSAPLMLPGRHALN-ATLMAASVGGMIPYMLDP 795
Query 270 GHFTRMLCLYVNAGLSMWLGFHLVAAIGGADMPVVISLLNSYSGVALAASGFMLDNNLLI 329
+ T + CL + LS +G L AAIGGADMPVVI++LNSYSG AL A GF+L+NNLL
Sbjct 796 SYTTGITCLGSVSALSAVMGLTLTAAIGGADMPVVITVLNSYSGWALCAEGFLLNNNLLT 855
Query 330 IAGALIASSGAILSYIMCKGMNRSLWNVVLGGFEEAEGVGAAAP---QGAVQQANADEVA 386
I GALI SSGAILSYIMC MNRSL NV+LGG+ + G P G + N D+
Sbjct 856 IVGALIGSSGAILSYIMCVAMNRSLANVILGGYGTSS-TGTGKPMEITGTHTEVNVDQTV 914
Query 387 DELLAARKVLIVPGYGMAVARCQSELADIAKNLMNCGITVDFGIHPVAGRMPGHMNVLLA 446
D + A ++IVPGYG+ A+ Q +AD+ K+L + G V FGIHPVAGRMPG +NVLLA
Sbjct 915 DLIKEAHNIIIVPGYGLCAAKAQYPIADLVKSLTDQGKKVRFGIHPVAGRMPGQLNVLLA 974
Query 447 EADVPYKIVKEMSEVNPEMSSYDVVLVVGANDTVNPAALE-PGSKISGMPVIEAWKARRV 505
EA VPY +V EM E+N + D+VLV+GANDTVN AA E P S I+GMPV+E WK+++V
Sbjct 975 EAGVPYDVVLEMDEINEDFPETDLVLVIGANDTVNSAAQEDPNSIIAGMPVLEVWKSKQV 1034
Query 506 FVLKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVFARV 547
V+KRS+ GYA+++NP+F+ NT ML G+AK T A+ A+V
Sbjct 1035 VVMKRSLGVGYAAVDNPIFYKPNTSMLLGDAKKTCDALSAKV 1076
Score = 286 bits (732), Expect = 3e-76, Method: Compositional matrix adjust.
Identities = 201/535 (37%), Positives = 292/535 (54%), Gaps = 33/535 (6%)
Query 582 PYPRLAVGVLKDS-NGSVMVPIAPKFVPRLRKMAFRVIVESGAGANAGFSDEEYRRAGAE 640
PY +L VGV K+ V I+P V L K F V+VESGAG +A FSD+ Y +AGA
Sbjct 49 PYKQLTVGVPKEIFQNERRVAISPAGVEALIKQGFNVVVESGAGESAKFSDDMYTKAGAT 108
Query 641 IASNADAVINGAEVLLRVSAP--TPEM----VSRMPRDKVLISYLFPSVNTQALDMLARQ 694
I D + ++VLL+V AP P + S M L+S+++P+ N + +D L+++
Sbjct 109 IRDVKD--VFSSDVLLKVRAPMLNPTLGVHEASLMSEGATLVSFIYPAQNPELMDTLSQR 166
Query 695 GVTALAVDEVPRVTRAQKLDVKSAMQGLQGYRAVIEAFNALPKLSKASISAAGRVEAAKV 754
T LA+D+VPRVT AQ D S+M + GY+AV+ A N + I+AAG+V AKV
Sbjct 167 KATVLAMDQVPRVTIAQGYDALSSMANIAGYKAVVLAANNFGRFFTGQITAAGKVPPAKV 226
Query 755 FVIGAGVAGLQAISTAHGLGAQVFGHDVRSATREEVESCGGKFIGLRMGEEAEVLGGYAR 814
+IG GVAGL A +A +GA V G D R+A E+ +S G + + + + E E GGYA+
Sbjct 227 LIIGGGVAGLAAAGSARAMGAIVRGFDTRAAALEQFKSLGAEPLEVDIKESGEGQGGYAK 286
Query 815 EMGDAYQRAQRELIANTIKHCDVVICTAAIHGKPSPKLISRDMLRSMKPGSVIVDIATEF 874
EM + A+ +L A D++I TA I G+ +P LI+++M+ +MK GSV+VD+A E
Sbjct 287 EMSKEFIEAEMKLFAKQCLDVDIIITTALIPGRKAPVLITKEMVETMKDGSVVVDLAAE- 345
Query 875 GDTRSGWGGNVEVSPKDDQVVVDGITVIGRKRIETRMPVQASELFSMNICNLLEDLGGG- 933
GGN+E + + V G+ +G I +R+P QAS L+S NI L+ +
Sbjct 346 ------AGGNIETTVPGELSVHKGVIHVGYTDIPSRLPTQASTLYSNNITKLIRAISPDK 399
Query 934 --------SNFRVNMDDEVIRGLVAVYQGRNVWQPPQP--TPVSRTPPRVPMPSAPSAAP 983
+ F D VIRG V + G+ ++ PQP PV+ PP+
Sbjct 400 ETFYFDVKNEFDFGTMDHVIRGSVVMQDGKVLFPAPQPQNVPVA-APPKQKTVQELQKEK 458
Query 984 AKAGGAFAQALASDAFFAMCLVVAAAVVGLLGIVLDPMELKHLTLLALSLIVGYYCVWAV 1043
A A F L + + L A +GL + + +T L+ IVGY+ VW V
Sbjct 459 ASAVSPFRATLTTAGVYTGGLGTA---IGLGLCAPNAAFTQMVTTFGLAGIVGYHTVWGV 515
Query 1044 TPALHTPLMSVTNALSGVIVIGCMLEYGTALISGFT--LLALVGTFLASVNIAGG 1096
TPALH+PLMSVTNA+SG+ +G + G + T LA++ F++SVNIAGG
Sbjct 516 TPALHSPLMSVTNAISGLTAVGGLSLMGGGYLPSSTAETLAVLAAFISSVNIAGG 570
> hsa:23530 NNT, MGC126502, MGC126503; nicotinamide nucleotide
transhydrogenase (EC:1.6.1.2); K00323 NAD(P) transhydrogenase
[EC:1.6.1.2]
Length=1086
Score = 396 bits (1018), Expect = 3e-109, Method: Compositional matrix adjust.
Identities = 224/466 (48%), Positives = 299/466 (64%), Gaps = 10/466 (2%)
Query 96 VYLFSAVCFILCLRGLSTPQTAKRGNILGLVGMVAAVVVTFTEAGFGQHYLLFFATAAPA 155
+YL S +C + L GLST TA+ GN LG++G+ + T G LL + A A
Sbjct 623 MYLGSGLCCVGALAGLSTQGTARLGNALGMIGVAGGLAATLGVLKPGPE-LLAQMSGAMA 681
Query 156 VG--LGLYIAQSVNMTEMPQLVALFHSFVGLAAVMVGFANF--HSPAGVERASSLLRLLE 211
+G +GL IA+ + ++++PQLVA FHS VGLAAV+ A + P A++ L +
Sbjct 682 LGGTIGLTIAKRIQISDLPQLVAAFHSLVGLAAVLTCIAEYIIEYPHFATDAAANLTKIV 741
Query 212 VYVGVFVAGITFTGSVVAAAKLHGSMESRSLRVPGRHALNSATIAAIGVLGAL-FCVSSG 270
Y+G ++ G+TF+GS++A KL G ++S L +PGRH LN+ +AA V G + F V
Sbjct 742 AYLGTYIGGVTFSGSLIAYGKLQGLLKSAPLLLPGRHLLNAGLLAA-SVGGIIPFMVDPS 800
Query 271 HFTRMLCLYVNAGLSMWLGFHLVAAIGGADMPVVISLLNSYSGVALAASGFMLDNNLLII 330
T + CL + LS +G L AAIGGADMPVVI++LNSYSG AL A GF+L+NNLL I
Sbjct 801 FTTGITCLGSVSALSAVMGVTLTAAIGGADMPVVITVLNSYSGWALCAEGFLLNNNLLTI 860
Query 331 AGALIASSGAILSYIMCKGMNRSLWNVVLGGFEEAEGVGAAAPQ--GAVQQANADEVADE 388
GALI SSGAILSYIMC MNRSL NV+LGG+ G + G + N D D
Sbjct 861 VGALIGSSGAILSYIMCVAMNRSLANVILGGYGTTSTAGGKPMEISGTHTEINLDNAIDM 920
Query 389 LLAARKVLIVPGYGMAVARCQSELADIAKNLMNCGITVDFGIHPVAGRMPGHMNVLLAEA 448
+ A ++I PGYG+ A+ Q +AD+ K L G V FGIHPVAGRMPG +NVLLAEA
Sbjct 921 IREANSIIITPGYGLCAAKAQYPIADLVKMLTEQGKKVRFGIHPVAGRMPGQLNVLLAEA 980
Query 449 DVPYKIVKEMSEVNPEMSSYDVVLVVGANDTVNPAALE-PGSKISGMPVIEAWKARRVFV 507
VPY IV EM E+N + D+VLV+GANDTVN AA E P S I+GMPV+E WK+++V V
Sbjct 981 GVPYDIVLEMDEINHDFPDTDLVLVIGANDTVNSAAQEDPNSIIAGMPVLEVWKSKQVIV 1040
Query 508 LKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVFARVNARAEQ 553
+KRS+ GYA+++NP+F+ NT ML G+AK T A+ A+V ++
Sbjct 1041 MKRSLGVGYAAVDNPIFYKPNTAMLLGDAKKTCDALQAKVRESYQK 1086
Score = 299 bits (765), Expect = 5e-80, Method: Compositional matrix adjust.
Identities = 194/537 (36%), Positives = 296/537 (55%), Gaps = 37/537 (6%)
Query 582 PYPRLAVGVLKD-SNGSVMVPIAPKFVPRLRKMAFRVIVESGAGANAGFSDEEYRRAGAE 640
PY +L VGV K+ V ++P V L K F V+VESGAG + FSD+ YR AGA+
Sbjct 53 PYKQLTVGVPKEIFQNEKRVALSPAGVQNLVKQGFNVVVESGAGEASKFSDDHYRVAGAQ 112
Query 641 IASNADAVINGAEVLLRVSAP--TPEM----VSRMPRDKVLISYLFPSVNTQALDMLARQ 694
I + + ++++++V AP P + + LIS+++P+ N + L+ L+++
Sbjct 113 IQGAKEVL--ASDLVVKVRAPMVNPTLGVHEADLLKTSGTLISFIYPAQNPELLNKLSQR 170
Query 695 GVTALAVDEVPRVTRAQKLDVKSAMQGLQGYRAVIEAFNALPKLSKASISAAGRVEAAKV 754
T LA+D+VPRVT AQ D S+M + GY+AV+ A N + I+AAG+V AK+
Sbjct 171 KTTVLAMDQVPRVTIAQGYDALSSMANIAGYKAVVLAANHFGRFFTGQITAAGKVPPAKI 230
Query 755 FVIGAGVAGLQAISTAHGLGAQVFGHDVRSATREEVESCGGKFIGLRMGEEAEVLGGYAR 814
++G GVAGL + A +GA V G D R+A E+ +S G + + + + E E GGYA+
Sbjct 231 LIVGGGVAGLASAGAAKSMGAIVRGFDTRAAALEQFKSLGAEPLEVDLKESGEGQGGYAK 290
Query 815 EMGDAYQRAQRELIANTIKHCDVVICTAAIHGKPSPKLISRDMLRSMKPGSVIVDIATEF 874
EM + A+ +L A K D++I TA I GK +P L +++M+ SMK GSV+VD+A E
Sbjct 291 EMSKEFIEAEMKLFAQQCKEVDILISTALIPGKKAPVLFNKEMIESMKEGSVVVDLAAE- 349
Query 875 GDTRSGWGGNVEVSPKDDQVVVDGITVIGRKRIETRMPVQASELFSMNICNLLEDLG-GG 933
GGN E + + + GIT IG + +RM QAS L+S NI LL+ +
Sbjct 350 ------AGGNFETTKPGELYIHKGITHIGYTDLPSRMATQASTLYSNNITKLLKAISPDK 403
Query 934 SNFRVNMDDE--------VIRGLVAVYQGRNVWQPPQPTPVSRTPP-RVPMPSAPSAAPA 984
NF ++ D+ VIRG V + G+ ++ P P + + P + + A A
Sbjct 404 DNFYFDVKDDFDFGTMGHVIRGTVVMKDGKVIFPAPTPKNIPQGAPVKQKTVAELEAEKA 463
Query 985 KAGGAFAQALASDAFFAMCLVVAAAVVGLLGIVLDPMEL---KHLTLLALSLIVGYYCVW 1041
F + +++ + + A + G+LG+ + L + +T L+ IVGY+ VW
Sbjct 464 ATITPFRKTMSTASAY------TAGLTGILGLGIAAPNLAFSQMVTTFGLAGIVGYHTVW 517
Query 1042 AVTPALHTPLMSVTNALSGVIVIGCMLEYGTALISGFTL--LALVGTFLASVNIAGG 1096
VTPALH+PLMSVTNA+SG+ +G + G L T LA + F++SVNIAGG
Sbjct 518 GVTPALHSPLMSVTNAISGLTAVGGLALMGGHLYPSTTSQGLAALAAFISSVNIAGG 574
> mmu:18115 Nnt, 4930423F13Rik, AI323702, BB168308; nicotinamide
nucleotide transhydrogenase (EC:1.6.1.2); K00323 NAD(P) transhydrogenase
[EC:1.6.1.2]
Length=1086
Score = 394 bits (1011), Expect = 1e-108, Method: Compositional matrix adjust.
Identities = 224/467 (47%), Positives = 298/467 (63%), Gaps = 12/467 (2%)
Query 96 VYLFSAVCFILCLRGLSTPQTAKRGNILGLVGMVAAVVVTFTEAGFGQH-YLLFFATAAP 154
+YL S +C + L GLST TA+ GN LG++G+ + T G LL + A
Sbjct 623 MYLGSGLCCVGALGGLSTQGTARLGNALGMIGVAGGLAATL--GGLKPDPQLLAQMSGAM 680
Query 155 AVG--LGLYIAQSVNMTEMPQLVALFHSFVGLAAVMVGFANF--HSPAGVERASSLLRLL 210
A+G +GL IA+ + ++++PQLVA FHS VGLAAV+ A + P A+S +
Sbjct 681 AMGGTIGLTIAKRIQISDLPQLVAAFHSLVGLAAVLTCMAEYIVEYPHFAMDATSNFTKI 740
Query 211 EVYVGVFVAGITFTGSVVAAAKLHGSMESRSLRVPGRHALNSATIAAIGVLGAL-FCVSS 269
Y+G ++ G+TF+GS+VA KL G ++S L +PGRHALN+ +AA V G + F
Sbjct 741 VAYLGTYIGGVTFSGSLVAYGKLQGILKSAPLLLPGRHALNAGLLAA-SVGGIIPFMADP 799
Query 270 GHFTRMLCLYVNAGLSMWLGFHLVAAIGGADMPVVISLLNSYSGVALAASGFMLDNNLLI 329
T + CL + LS +G L AAIGGADMPVVI++LNSYSG AL A GF+L+NNLL
Sbjct 800 SFTTGITCLGSVSALSTLMGVTLTAAIGGADMPVVITVLNSYSGWALCAEGFLLNNNLLT 859
Query 330 IAGALIASSGAILSYIMCKGMNRSLWNVVLGGFEEAEGVGAAAPQ--GAVQQANADEVAD 387
I GALI SSGAILSYIMC MNRSL NV+LGG+ G + G + N D +
Sbjct 860 IVGALIGSSGAILSYIMCVAMNRSLANVILGGYGTTSTAGGKPMEISGTHTEINLDNAVE 919
Query 388 ELLAARKVLIVPGYGMAVARCQSELADIAKNLMNCGITVDFGIHPVAGRMPGHMNVLLAE 447
+ A ++I PGYG+ A+ Q +AD+ K L G V FGIHPVAGRMPG +NVLLAE
Sbjct 920 MIREANSIVITPGYGLCAAKAQYPIADLVKMLTEQGKKVRFGIHPVAGRMPGQLNVLLAE 979
Query 448 ADVPYKIVKEMSEVNPEMSSYDVVLVVGANDTVNPAALE-PGSKISGMPVIEAWKARRVF 506
A VPY IV EM E+N + D+VLV+GANDTVN AA E P S I+GMPV+E WK+++V
Sbjct 980 AGVPYDIVLEMDEINSDFPDTDLVLVIGANDTVNSAAQEDPNSIIAGMPVLEVWKSKQVI 1039
Query 507 VLKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVFARVNARAEQ 553
V+KRS+ GYA+++NP+F+ NT ML G+AK T A+ A+V ++
Sbjct 1040 VMKRSLGVGYAAVDNPIFYKPNTAMLLGDAKKTCDALQAKVRESYQK 1086
Score = 300 bits (767), Expect = 3e-80, Method: Compositional matrix adjust.
Identities = 204/563 (36%), Positives = 300/563 (53%), Gaps = 48/563 (8%)
Query 566 GLLEFERDERADPSSW---------PYPRLAVGVLKD-SNGSVMVPIAPKFVPRLRKMAF 615
G +F R R + W PY +L VGV K+ V ++P V L K F
Sbjct 28 GKRDFVRMLRTHQALWCKSPVKPGIPYKQLTVGVPKEIFQNEKRVALSPAGVQALVKQGF 87
Query 616 RVIVESGAGANAGFSDEEYRRAGAEIASNADAVINGAEVLLRVSAP--TPEMVSR----M 669
V+VESGAG + F D+ YR AGA+I + + ++++++V AP P + + +
Sbjct 88 NVVVESGAGEASKFPDDLYRAAGAQIQGMKEVL--ASDLVVKVRAPMVNPTLGAHEADFL 145
Query 670 PRDKVLISYLFPSVNTQALDMLARQGVTALAVDEVPRVTRAQKLDVKSAMQGLQGYRAVI 729
LIS+++P+ N L+ L+ + T LA+D+VPRVT AQ D S+M + GY+AV+
Sbjct 146 KPSGTLISFIYPAQNPDLLNKLSERKTTVLAMDQVPRVTIAQGYDALSSMANISGYKAVV 205
Query 730 EAFNALPKLSKASISAAGRVEAAKVFVIGAGVAGLQAISTAHGLGAQVFGHDVRSATREE 789
A N + I+AAG+V AK+ ++G GVAGL + A +GA V G D R+A E+
Sbjct 206 LAANHFGRFFTGQITAAGKVPPAKILIVGGGVAGLASAGAAKSMGAVVRGFDTRAAALEQ 265
Query 790 VESCGGKFIGLRMGEEAEVLGGYAREMGDAYQRAQRELIANTIKHCDVVICTAAIHGKPS 849
+S G + + + + E E GGYA+EM + A+ +L A K D++I TA I GK +
Sbjct 266 FKSLGAEPLEVDLKESGEGQGGYAKEMSKEFIEAEMKLFAQQCKEVDILISTALIPGKKA 325
Query 850 PKLISRDMLRSMKPGSVIVDIATEFGDTRSGWGGNVEVSPKDDQVVVDGITVIGRKRIET 909
P L S++M+ SMK GSV+VD+A E GGN E + + V GIT IG + +
Sbjct 326 PVLFSKEMIESMKEGSVVVDLAAE-------AGGNFETTKPGELYVHKGITHIGYTDLPS 378
Query 910 RMPVQASELFSMNICNLLEDLG-GGSNFRVNMDDE--------VIRGLVAVYQGRNVWQP 960
RM QAS L+S NI LL+ + NF + D+ VIRG V + G+ ++
Sbjct 379 RMATQASTLYSNNITKLLKAISPDKDNFHFEVKDDFDFGTMSHVIRGTVVMKDGKVIFPA 438
Query 961 PQPTPVSRTPPRVPMPSAPSAAPAKAG--GAFAQALASDAFFAMCLVVAAAVVGL--LGI 1016
P P + P P A A KAG + + L + + + +A + G+ LGI
Sbjct 439 PTPKNIPEEAPVKPKTVAELEA-EKAGTVSMYTKTLTTASVY------SAGLTGMLGLGI 491
Query 1017 VLDPMELKHL-TLLALSLIVGYYCVWAVTPALHTPLMSVTNALSGVIVIGCMLEYGTALI 1075
V + + T L+ I+GY+ VW VTPALH+PLMSVTNA+SG+ +G + G
Sbjct 492 VAPNVAFSQMVTTFGLAGIIGYHTVWGVTPALHSPLMSVTNAISGLTAVGGLALMGGHFY 551
Query 1076 SGFT--LLALVGTFLASVNIAGG 1096
T LA + TF++SVNIAGG
Sbjct 552 PSTTSQSLAALATFISSVNIAGG 574
> xla:447528 nnt, MGC83563; nicotinamide nucleotide transhydrogenase
(EC:1.6.1.2); K00323 NAD(P) transhydrogenase [EC:1.6.1.2]
Length=1086
Score = 390 bits (1002), Expect = 2e-107, Method: Compositional matrix adjust.
Identities = 219/461 (47%), Positives = 298/461 (64%), Gaps = 12/461 (2%)
Query 96 VYLFSAVCFILCLRGLSTPQTAKRGNILGLVGMVAAVVVTFTEAGFGQHYLLFFATAAPA 155
VYL S +C + L GLST TA+ GN LG++G+ + T LL + A A
Sbjct 623 VYLGSGLCCVGALAGLSTQGTARLGNSLGMMGVAGGIAATLGALKPSPE-LLAQMSGAMA 681
Query 156 VG--LGLYIAQSVNMTEMPQLVALFHSFVGLAAVMVGFANF---HSPAGVERASSLLRLL 210
+G +GL IA+ + ++++PQLVA FHS VGLAAV+ A + + + A++L +++
Sbjct 682 LGGTIGLTIAKRIQISDLPQLVAAFHSLVGLAAVLTCVAEYMIEYPHFATDPAANLTKIV 741
Query 211 EVYVGVFVAGITFTGSVVAAAKLHGSMESRSLRVPGRHALNSATIAAIGVLGAL-FCVSS 269
Y+G ++ G+TF+GS+VA KL G + S L +PGRH LN+ + A V G + + +
Sbjct 742 -AYLGTYIGGVTFSGSLVAYGKLQGVLNSAPLLLPGRHMLNAGLLTA-SVGGIIPYMLDP 799
Query 270 GHFTRMLCLYVNAGLSMWLGFHLVAAIGGADMPVVISLLNSYSGVALAASGFMLDNNLLI 329
+ T + CL + LS +G L AAIGGADMPVVI++LNSYSG AL A GF+L+NNLL
Sbjct 800 SYTTGITCLGSVSALSAVMGVTLTAAIGGADMPVVITVLNSYSGWALCAEGFLLNNNLLT 859
Query 330 IAGALIASSGAILSYIMCKGMNRSLWNVVLGGFEEAEGVGAAAPQ--GAVQQANADEVAD 387
I GALI SSGAILSYIMC MNRSL NV+LGG+ G + G + N D +
Sbjct 860 IVGALIGSSGAILSYIMCVAMNRSLTNVILGGYGTTSTAGGKPMEITGTHTEINLDNAVE 919
Query 388 ELLAARKVLIVPGYGMAVARCQSELADIAKNLMNCGITVDFGIHPVAGRMPGHMNVLLAE 447
+ A ++I PGYG+ A+ Q +AD+ K L G V FGIHPVAGRMPG +NVLLAE
Sbjct 920 YIREANNIIITPGYGLCAAKAQYPIADLVKILKEAGKNVRFGIHPVAGRMPGQLNVLLAE 979
Query 448 ADVPYKIVKEMSEVNPEMSSYDVVLVVGANDTVNPAALE-PGSKISGMPVIEAWKARRVF 506
A VPY IV EM E+N + D+VLV+GANDTVN AA E P S I+GMPV+E WK+++V
Sbjct 980 AGVPYDIVLEMDEINEDFPETDLVLVIGANDTVNSAAQEDPNSIIAGMPVLEVWKSKQVI 1039
Query 507 VLKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVFARV 547
V+KRS+ GYA+++NP+F+ NT ML G+AK T ++ A+V
Sbjct 1040 VMKRSLGVGYAAVDNPIFYKPNTSMLLGDAKKTCDSLQAKV 1080
Score = 285 bits (728), Expect = 9e-76, Method: Compositional matrix adjust.
Identities = 201/543 (37%), Positives = 291/543 (53%), Gaps = 49/543 (9%)
Query 582 PYPRLAVGVLKDS-NGSVMVPIAPKFVPRLRKMAFRVIVESGAGANAGFSDEEYRRAGAE 640
PY ++ VGV K+ V ++P V L K F V+VE+GAG + FSD+ Y+ AGA+
Sbjct 53 PYKQITVGVPKEIFQNEKRVALSPAGVQALVKQGFNVVVETGAGEASKFSDDHYKEAGAK 112
Query 641 IASNADAVINGAEVLLRVSAP--TPEM----VSRMPRDKVLISYLFPSVNTQALDMLARQ 694
I D + ++++L+V AP P + L+S+++P+ N L L+ +
Sbjct 113 IQGTKDVL--ASDLVLKVRAPMLNPALGVHEADMFKPSSTLVSFVYPAQNPDLLSKLSEK 170
Query 695 GVTALAVDEVPRVTRAQKLDVKSAMQGLQGYRAVIEAFNALPKLSKASISAAGRVEAAKV 754
+T LA+D+VPRVT AQ D S+M + GY+AV+ A N + I+AAG+V AKV
Sbjct 171 NMTVLAMDQVPRVTIAQGYDALSSMANISGYKAVVMAANNFGRFFTGQITAAGKVPPAKV 230
Query 755 FVIGAGVAGLQAISTAHGLGAQVFGHDVRSATREEVESCGGKFIGLRMGEEAEVLGGYAR 814
+IG GVAGL A A +GA V G D R+A E+ +S G + + + + E E GGYA+
Sbjct 231 LIIGGGVAGLAAAGAAKSMGAIVRGFDTRAAALEQFKSLGAEPLEVDLKESGEGQGGYAK 290
Query 815 EMGDAYQRAQRELIANTIKHCDVVICTAAIHGKPSPKLISRDMLRSMKPGSVIVDIATEF 874
EM + A+ +L A + D+++ TA I GK +P L +DM+ MK GSV+VD+A E
Sbjct 291 EMSKEFIEAEMKLFAKQCQDVDIIVTTALIPGKTAPILFRKDMIELMKEGSVVVDLAAE- 349
Query 875 GDTRSGWGGNVEVSPKDDQVVVDGITVIGRKRIETRMPVQASELFSMNICNLLEDLG-GG 933
GGN+E + D V G+T IG I +RM QAS L+S NI LL+ +
Sbjct 350 ------AGGNIETTKPGDIYVHKGVTHIGYTDIPSRMASQASTLYSNNITKLLKAISPDK 403
Query 934 SNFRVNMDDE--------VIRGLVAVYQGRNVWQPPQPTPVSRTPPRVPMPSAPSAAPAK 985
NF ++ D+ VIRG V + G ++ P P + P AAP K
Sbjct 404 DNFYYDIKDDFDYGTMDHVIRGTVVMKDGNVIFPAPPPNKI------------PQAAPVK 451
Query 986 AGGAF---AQALASDAFFAMCLVVAAAVVGLLGIVL-----DPME--LKHLTLLALSLIV 1035
A+ AS + F + AAA LG +L P + +T L+ IV
Sbjct 452 QKSVAQIEAEKAASISPFRKTMNGAAAYTAGLGTLLSLGIASPHSAFTQMVTTFGLAGIV 511
Query 1036 GYYCVWAVTPALHTPLMSVTNALSGVIVIGCMLEYGTALISGFT--LLALVGTFLASVNI 1093
GY+ VW VTPALH+PLMSVTNA+SG+ +G + G + T LLA++ F++S+NI
Sbjct 512 GYHTVWGVTPALHSPLMSVTNAISGLTAVGGLALMGGGYLPTNTHELLAVLAAFVSSINI 571
Query 1094 AGG 1096
AGG
Sbjct 572 AGG 574
> cel:C15H9.1 nnt-1; Nicotinamide Nucleotide Transhydrogenase
family member (nnt-1); K00323 NAD(P) transhydrogenase [EC:1.6.1.2]
Length=1041
Score = 375 bits (964), Expect = 4e-103, Method: Compositional matrix adjust.
Identities = 216/474 (45%), Positives = 300/474 (63%), Gaps = 11/474 (2%)
Query 86 FSNMPSLLGAVYLFSAVCFILCLRGLSTPQTAKRGNILGLVGMVAAVVVTF--TEAGFGQ 143
++ P + YL S++C + L GLS+ TA+ GN LG++G+ + T + F
Sbjct 568 YTAAPLIHSYAYLGSSLCCVGALAGLSSQSTARVGNALGIIGVTGGIGATLGLLQPDFNT 627
Query 144 HYLLFFATAAPAVGLGLYIAQSVNMTEMPQLVALFHSFVGLAAVMVGFANF--HSPAGVE 201
+ + A ++ +GL IA + +T++PQLVA FHSFVGLAA + ANF P +E
Sbjct 628 LCQMGGSVAMGSL-IGLGIANRIKVTDLPQLVAAFHSFVGLAATLTCLANFIQEHPHFLE 686
Query 202 RASSLLRLLEVYV-GVFVAGITFTGSVVAAAKLHGSMESRSLRVPGRHALNSATIAA-IG 259
S+ G ++ G+TFTGS++A KL G + S +P RH LN A +A +G
Sbjct 687 DPSNAAAAKLALFLGTYIGGVTFTGSLMAYGKLQGILASAPTYLPARHVLNGALLAGNVG 746
Query 260 VLGALFCVSSGHFTRMLCLYVNAGLSMWLGFHLVAAIGGADMPVVISLLNSYSGVALAAS 319
LG + S+ T M L GLS +G L AIGGADMP+VI++LNSYSG AL A
Sbjct 747 ALGT-YMYSTDFGTGMSMLGGTVGLSSLMGVTLTMAIGGADMPIVITVLNSYSGWALCAE 805
Query 320 GFMLDNNLLIIAGALIASSGAILSYIMCKGMNRSLWNVVLGGF-EEAEGVG-AAAPQGAV 377
GFMLDN+LL + GALI SSGAILS+IMCK MNRSL NV+LGG +++G G A A +G
Sbjct 806 GFMLDNSLLTVLGALIGSSGAILSHIMCKAMNRSLLNVILGGVGTKSKGTGEAKAIEGTA 865
Query 378 QQANADEVADELLAARKVLIVPGYGMAVARCQSELADIAKNLMNCGITVDFGIHPVAGRM 437
++ E AD LL AR V+I+PGYG+ A+ Q +A + K L G+ V F IHPVAGRM
Sbjct 866 KEIAPVETADMLLNARSVIIIPGYGLCAAQAQYPIAQLVKELQQRGVRVRFAIHPVAGRM 925
Query 438 PGHMNVLLAEADVPYKIVKEMSEVNPEMSSYDVVLVVGANDTVNPAAL-EPGSKISGMPV 496
PG +NVLLAEA VPY IV+EM E+N + DV LV+G+NDT+N AA +P S I+GMPV
Sbjct 926 PGQLNVLLAEAGVPYDIVEEMEEINEDFKETDVALVIGSNDTINSAAEDDPNSSIAGMPV 985
Query 497 IEAWKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVFARVNAR 550
+ W +++V ++KR++ GYA+++NP+F ENT+ML G+AK + + V ++
Sbjct 986 LRVWNSKQVIIVKRTLGTGYAAVDNPVFFNENTQMLLGDAKKMSEKLLEEVKSK 1039
Score = 339 bits (870), Expect = 4e-92, Method: Compositional matrix adjust.
Identities = 211/519 (40%), Positives = 293/519 (56%), Gaps = 21/519 (4%)
Query 583 YPRLAVGVLKD-SNGSVMVPIAPKFVPRLRKMAFRVIVESGAGANAGFSDEEYRRAGAEI 641
Y +L V V K+ G V ++P V L+K V++E AG AG+S+EEY R+GA++
Sbjct 26 YSKLKVAVPKEIFPGEKRVSLSPNGVALLKKNGISVLIEENAGVLAGYSNEEYVRSGADV 85
Query 642 ASNADAVINGAEVLLRVSAPTPEMVSRMPRDKVLISYLFPSVNTQALDMLARQGVTALAV 701
+ + V N +++L+V PT VS++ LIS++ P N LD L + T A+
Sbjct 86 GKH-NEVFN-TDIMLKVRPPTENEVSKLKSGCTLISFIHPGQNQALLDSLTKTDKTVFAM 143
Query 702 DEVPRVTRAQKLDVKSAMQGLQGYRAVIEAFNALPKLSKASISAAGRVEAAKVFVIGAGV 761
D VPR++RAQ D S+M + GYRAVIEA N + I+AAG+V AKV VIG GV
Sbjct 144 DCVPRISRAQVFDALSSMANIAGYRAVIEAANHFGRFFTGQITAAGKVPPAKVLVIGGGV 203
Query 762 AGLQAISTAHGLGAQVFGHDVRSATREEVESCGGKFIGLRMGEEAEVLGGYAREMGDAYQ 821
AGL AI T+ G+GA V G D R+A +E VES G +F+ + + E+ E GGYA+EM +
Sbjct 204 AGLSAIGTSRGMGAVVRGFDTRAAVKEHVESLGAQFLTVNVKEDGEGGGGYAKEMSKEFI 263
Query 822 RAQRELIANTIKHCDVVICTAAIHGKPSPKLISRDMLRSMKPGSVIVDIATEFGDTRSGW 881
A+ +L A+ K D++I TA I GK +P LI+ +M++SMKPGSV+VD+A E
Sbjct 264 DAEMKLFADQCKDVDIIITTALIPGKKAPILITEEMIKSMKPGSVVVDLAAE-------S 316
Query 882 GGNVEVSPKDDQVVVDGITVIGRKRIETRMPVQASELFSMNICNLLEDLGGGSNFRVNMD 941
GGN+ + + V G+T IG + +R+P Q+SEL+S NI L LG F VN +
Sbjct 317 GGNIATTRPGEVYVKHGVTHIGFTDLPSRLPTQSSELYSNNIAKFLLHLGKDKTFFVNEE 376
Query 942 DEVIRGLVAVYQGRNVWQPPQPTPVSRTPPRVPMPSAPSA-APAKAGGAFAQALASDAFF 1000
DEV RG + V G+ W PP P+ PS +A P A
Sbjct 377 DEVARGALVVRDGQMKWPPPPINFPPPAAPKSDKPSENTALVP-------LTPFRKTANQ 429
Query 1001 AMCLVVAAAVVGLLGIV-LDPMELKHLTLLALSLIVGYYCVWAVTPALHTPLMSVTNALS 1059
+ L V LLGI +P T AL+ +VGY+ VW VTPALH+PLMSVTNA+S
Sbjct 430 TLLLTSGLGSVSLLGIAGTNPQISSMSTTFALAGLVGYHTVWGVTPALHSPLMSVTNAIS 489
Query 1060 GVIVIGCMLEYGTALI--SGFTLLALVGTFLASVNIAGG 1096
G G + G L+ + +AL+ TF++SVNI GG
Sbjct 490 GTTAAGALCLMGGGLMPQNSAQTMALLATFISSVNIGGG 528
> eco:b1603 pntA, ECK1598, JW1595; pyridine nucleotide transhydrogenase,
alpha subunit (EC:1.6.1.2); K00324 NAD(P) transhydrogenase
subunit alpha [EC:1.6.1.2]
Length=510
Score = 330 bits (847), Expect = 2e-89, Method: Compositional matrix adjust.
Identities = 198/496 (39%), Positives = 287/496 (57%), Gaps = 22/496 (4%)
Query 604 PKFVPRLRKMAFRVIVESGAGANAGFSDEEYRRAGAEIASNADAVINGAEVLLRVSAPTP 663
PK V +L K+ F V VESGAG A F D+ + +AGAEI + +E++L+V+AP
Sbjct 20 PKTVEQLLKLGFTVAVESGAGQLASFDDKAFVQAGAEIVEGNS--VWQSEIILKVNAPLD 77
Query 664 EMVSRMPRDKVLISYLFPSVNTQALDMLARQGVTALAVDEVPRVTRAQKLDVKSAMQGLQ 723
+ ++ + L+S+++P+ N + + LA + VT +A+D VPR++RAQ LD S+M +
Sbjct 78 DEIALLNPGTTLVSFIWPAQNPELMQKLAERNVTVMAMDSVPRISRAQSLDALSSMANIA 137
Query 724 GYRAVIEAFNALPKLSKASISAAGRVEAAKVFVIGAGVAGLQAISTAHGLGAQVFGHDVR 783
GYRA++EA + + I+AAG+V AKV VIGAGVAGL AI A+ LGA V D R
Sbjct 138 GYRAIVEAAHEFGRFFTGQITAAGKVPPAKVMVIGAGVAGLAAIGAANSLGAIVRAFDTR 197
Query 784 SATREEVESCGGKFIGLRMGEEAEVLGGYAREMGDAYQRAQRELIANTIKHCDVVICTAA 843
+E+V+S G +F+ L EEA GYA+ M DA+ +A+ EL A K D+++ TA
Sbjct 198 PEVKEQVQSMGAEFLELDFKEEAGSGDGYAKVMSDAFIKAEMELFAAQAKEVDIIVTTAL 257
Query 844 IHGKPSPKLISRDMLRSMKPGSVIVDIATEFGDTRSGWGGNVEVS-PKDDQVVVDGITVI 902
I GKP+PKLI+R+M+ SMK GSVIVD+A + GGN E + P + +G+ VI
Sbjct 258 IPGKPAPKLITREMVDSMKAGSVIVDLAAQN-------GGNCEYTVPGEIFTTENGVKVI 310
Query 903 GRKRIETRMPVQASELFSMNICNLLEDL--GGGSNFRVNMDDEVIRGLVAVYQGRNVWQP 960
G + R+P Q+S+L+ N+ NLL+ L N V+ DD VIRG+ + G W P
Sbjct 311 GYTDLPGRLPTQSSQLYGTNLVNLLKLLCKEKDGNITVDFDDVVIRGVTVIRAGEITW-P 369
Query 961 PQPTPVSRTPPRVPMPSAPSAAPAKAGGAFAQALASDAFFAMCLVVAAAVVGLLGIVLDP 1020
P VS P + K + + A + +A + G + V
Sbjct 370 APPIQVSAQPQAAQKAAPEVKTEEKCTCSPWRKYA-------LMALAIILFGWMASVAPK 422
Query 1021 MELKHLTLLALSLIVGYYCVWAVTPALHTPLMSVTNALSGVIVIGCMLEYGTALISGFTL 1080
L H T+ AL+ +VGYY VW V+ ALHTPLMSVTNA+SG+IV+G +L+ G +
Sbjct 423 EFLGHFTVFALACVVGYYVVWNVSHALHTPLMSVTNAISGIIVVGALLQIGQG--GWVSF 480
Query 1081 LALVGTFLASVNIAGG 1096
L+ + +AS+NI GG
Sbjct 481 LSFIAVLIASINIFGG 496
> dre:100331743 Nicotinamide Nucleotide Transhydrogenase family
member (nnt-1)-like
Length=518
Score = 123 bits (309), Expect = 4e-27, Method: Compositional matrix adjust.
Identities = 103/283 (36%), Positives = 148/283 (52%), Gaps = 24/283 (8%)
Query 827 LIANTIKHCDVVICTAAIHGKPSPKLISRDMLRSMKPGSVIVDIATEFGDTRSGWGGNVE 886
L A + D++I TA GK +P LI R+M+ SM+ GSV+VD+A E GGN+E
Sbjct 3 LFAKQCRDVDIIISTALNPGKRAPVLIKREMVESMRDGSVVVDLAAE-------AGGNIE 55
Query 887 VSPKDDQVVVDGITVIGRKRIETRMPVQASELFSMNICNLLEDLGGGSNFRVNMD----- 941
+ + V G+T IG + +RM QAS L+S NI LL+ + F N +
Sbjct 56 TTKPGELYVHQGVTHIGYTDLPSRMATQASSLYSNNIIKLLKAISPDKEF-FNFEPTDEF 114
Query 942 -----DEVIRGLVAVYQGRNVWQPPQPTPVSRTPPRVPMPSAPSAAPAKAG-GAFAQALA 995
D VIRG + + QG+N++ P P PP A A +A F + +
Sbjct 115 DYGTLDHVIRGTMVMKQGQNLFPAPLPKTTPPPPPAKQKSVAELEAEKRAVISPFRRTMT 174
Query 996 SDAFFAMCLVVAAAVVGLLGIVLDPMELKHLTLLALSLIVGYYCVWAVTPALHTPLMSVT 1055
S + L + V+GL + + +T L+ IVGY+ VW VTPALH+PLMSVT
Sbjct 175 SAGVYTAGL---STVLGLGIASPNAAFTQMVTTFGLAGIVGYHTVWGVTPALHSPLMSVT 231
Query 1056 NALSGVIVIGCMLEYGTALI--SGFTLLALVGTFLASVNIAGG 1096
NA+SG+ +G ++ G L S LAL+ F++S+NIAGG
Sbjct 232 NAISGLTAVGGLVLMGGGLTPSSLPESLALLAAFVSSINIAGG 274
Score = 103 bits (256), Expect = 6e-21, Method: Compositional matrix adjust.
Identities = 71/195 (36%), Positives = 112/195 (57%), Gaps = 7/195 (3%)
Query 96 VYLFSAVCFILCLRGLSTPQTAKRGNILGLVGMVAAVVVTFTEAGFGQHYLLFFATAAPA 155
+YL S +C + L GLS T++ N LG++G+ + T L + A +
Sbjct 323 MYLGSGMCCVGALAGLSAQGTSRLRNALGMIGVAGGIAATLGALKPSPELLSQMSIAMAS 382
Query 156 VG-LGLYIAQSVNMTEMPQLVALFHSFVGLAAVMVGFANF---HSPAGVERASSLLRLLE 211
G LGL IA+ + ++++PQLVA FHS VGLAAV+ A + + V A+ +L+++
Sbjct 383 GGTLGLTIAKRIEISDLPQLVAAFHSLVGLAAVLTCVAEYMIEYPHLDVHPAAGVLKIV- 441
Query 212 VYVGVFVAGITFTGSVVAAAKLHGSMESRSLRVPGRHALNSA-TIAAIGVLGALFCVSSG 270
Y+G ++ G+TF+GS+VA KL G ++S L +PGRH LN+ +A++G + F +S
Sbjct 442 AYLGTYIGGVTFSGSLVAYGKLQGLLDSAPLLLPGRHMLNAGLMLASVGGM-VPFMLSDS 500
Query 271 HFTRMLCLYVNAGLS 285
T M CL +GLS
Sbjct 501 FHTGMGCLLGVSGLS 515
> tgo:TGME49_003500 alanine dehydrogenase, putative (EC:1.4.1.1)
Length=462
Score = 76.6 bits (187), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 81/303 (26%), Positives = 139/303 (45%), Gaps = 33/303 (10%)
Query 600 VPIAPKFVPRLRKMAFRVIVESGAGANAGFSDEEYRRAGAEIASNADAVINGAEVLLRVS 659
V + P V L +V+V++GAG +GF+DE Y AGA + + V ++++++V
Sbjct 43 VALTPATVAELVNNGHKVVVQTGAGVASGFTDESYTAAGATMVQTTEEVYKTSQMIVKVQ 102
Query 660 APTPEMVSRMPRDKVLISYL-FPSVNTQALDMLARQGVTALAVDEVPRVTRAQKLDVKSA 718
AP P+ S + +V+ ++ F + T M+ R+ V + +++ T + A
Sbjct 103 APQPQEYSFIQPGQVIFAFFSFENNKTLFETMMQREAV-CFSYEQMK--TETGTTPILQA 159
Query 719 MQGLQGYRAVIEAFNALPKLSKASISAAGRV---EAAKVFVIGAGVAGLQAISTAHGLGA 775
M + G A+ +A L K S G V + V V+G GVA +QA A GA
Sbjct 160 MCEISGQLAISQAMKYLEKTMGGSGCMLGTVTGMTSGYVLVLGGGVAAMQAARVAASTGA 219
Query 776 QVFGHDV-----RSATREEVESCGGKFIG----LRMGEEAEVLGGYAREMGDAYQRAQRE 826
QV DV R+ T ++C + ++ ++A+V+ +G + Q
Sbjct 220 QVCIMDVCMSNMRTMTEMLPKNCTTMYFCTENLVQQIQKADVV------IGAVFGSFQ-- 271
Query 827 LIANTIKHCDVV-ICTAAIHGKPSPKLISRDMLRSMKPGSVIVDIATEFGDTRSGWGGNV 885
+ T+ + V+ T +P L+ ++ + MKPGSVIVD+A GGN
Sbjct 272 -MPATVTNTTVMNQKTTMTQVTKAPMLVKKEHVAMMKPGSVIVDLAV-------SRGGNF 323
Query 886 EVS 888
E +
Sbjct 324 ETT 326
> tgo:TGME49_115260 alanine dehydrogenase, putative (EC:1.4.1.1)
Length=390
Score = 53.1 bits (126), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 31/122 (25%), Positives = 59/122 (48%), Gaps = 0/122 (0%)
Query 600 VPIAPKFVPRLRKMAFRVIVESGAGANAGFSDEEYRRAGAEIASNADAVINGAEVLLRVS 659
V +AP L K V++E AG A F+DE+Y + GAEI ++A+ + +E++++V
Sbjct 16 VGLAPAGALELVKAGHTVLIEKDAGKKAHFTDEDYVQQGAEIVASAEELYGRSEMIVKVK 75
Query 660 APTPEMVSRMPRDKVLISYLFPSVNTQALDMLARQGVTALAVDEVPRVTRAQKLDVKSAM 719
P P+ + ++L Y + + + G ++ + V + R L+ S +
Sbjct 76 EPQPDEWKLVKSGQILFCYFHFCASQSLTEAMLNSGAICISYETVQKDGRLPLLEPMSEV 135
Query 720 QG 721
G
Sbjct 136 AG 137
> dre:100331681 MGC83563 protein-like
Length=142
Score = 52.4 bits (124), Expect = 1e-05, Method: Composition-based stats.
Identities = 35/78 (44%), Positives = 46/78 (58%), Gaps = 3/78 (3%)
Query 583 YPRLAVGVLKDSNGSVM-VPIAPKFVPRLRKMAFRVIVESGAGANAGFSDEEYRRAGAEI 641
Y + VGV K+ S V ++P V L K F V VESGAGA A FSDE+YR AGA+I
Sbjct 38 YKDVVVGVPKEIFPSERRVSVSPAGVELLVKQGFSVCVESGAGAAAQFSDEQYRAAGAKI 97
Query 642 ASNADAVINGAEVLLRVS 659
A+ + ++L+VS
Sbjct 98 TDTHTAL--ASHLVLKVS 113
> mmu:217708 Lin52, 5830457H20Rik; lin-52 homolog (C. elegans)
Length=116
Score = 36.6 bits (83), Expect = 0.60, Method: Composition-based stats.
Identities = 21/71 (29%), Positives = 33/71 (46%), Gaps = 0/71 (0%)
Query 562 DLEAGLLEFERDERADPSSWPYPRLAVGVLKDSNGSVMVPIAPKFVPRLRKMAFRVIVES 621
DLEA LL FE+ +RA P WP V S S + PK++ + + ++ E
Sbjct 13 DLEASLLSFEKLDRASPDLWPEQLPGVAEFAASFKSPITSSPPKWMAEIERDDIDMLKEL 72
Query 622 GAGANAGFSDE 632
G+ A ++
Sbjct 73 GSLTTANLMEK 83
> hsa:91750 LIN52, C14orf46, c14_5549; lin-52 homolog (C. elegans)
Length=116
Score = 36.6 bits (83), Expect = 0.60, Method: Composition-based stats.
Identities = 21/71 (29%), Positives = 33/71 (46%), Gaps = 0/71 (0%)
Query 562 DLEAGLLEFERDERADPSSWPYPRLAVGVLKDSNGSVMVPIAPKFVPRLRKMAFRVIVES 621
DLEA LL FE+ +RA P WP V S S + PK++ + + ++ E
Sbjct 13 DLEASLLSFEKLDRASPDLWPEQLPGVAEFAASFKSPITSSPPKWMAEIERDDIDMLKEL 72
Query 622 GAGANAGFSDE 632
G+ A ++
Sbjct 73 GSLTTANLMEK 83
> hsa:3664 IRF6, LPS, OFC6, PIT, PPS, VWS, VWS1; interferon regulatory
factor 6; K10154 interferon regulatory factor 6
Length=467
Score = 34.3 bits (77), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 33/148 (22%), Positives = 61/148 (41%), Gaps = 14/148 (9%)
Query 840 CTAAIHGKPSPKLISRDMLRSMKPGSVIVDIATEFGDTRSGWGGNVEVSPKDDQVVVDGI 899
C G +P L++ +++ K + + T D + G +E P + + G
Sbjct 319 CKVYWSGPCAPSLVAPNLIERQKKVKLFC-LETFLSDLIAHQKGQIEKQPPFEIYLCFGE 377
Query 900 TVIGRKRIETRM------PVQASELFSMNICNLLEDLGGGS-NFRV---NMDDEVIRGLV 949
K +E ++ PV A ++ M + GS ++ ++ D ++ L
Sbjct 378 EWPDGKPLERKLILVQVIPVVARMIYEMFSGDFTRSFDSGSVRLQISTPDIKDNIVAQLK 437
Query 950 AVY---QGRNVWQPPQPTPVSRTPPRVP 974
+Y Q + WQP QPTP + PP +P
Sbjct 438 QLYRILQTQESWQPMQPTPSMQLPPALP 465
> xla:494736 ankrd13a; ankyrin repeat domain 13A
Length=593
Score = 33.9 bits (76), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 25/59 (42%), Positives = 32/59 (54%), Gaps = 11/59 (18%)
Query 518 SIENPLFHLENTRMLFGNAKNTTSAVFARVNARAEQMPPSAARD--DLEAGLLEFERDE 574
IE PLFH+ N R+ FGN NT S RAE P S R+ D ++ L +FE D+
Sbjct 410 KIEIPLFHVLNARITFGNV-NTCS--------RAEDSPASTPRESQDEDSLLPKFEVDQ 459
> sce:YMR020W FMS1; Fms1p (EC:1.5.3.11); K13367 non-specific polyamine
oxidase [EC:1.5.3.17]
Length=508
Score = 33.1 bits (74), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 15/24 (62%), Positives = 18/24 (75%), Gaps = 0/24 (0%)
Query 753 KVFVIGAGVAGLQAISTAHGLGAQ 776
KV +IGAG+AGL+A ST H G Q
Sbjct 10 KVIIIGAGIAGLKAASTLHQNGIQ 33
> mmu:387512 Tas2r135, Tas2r35, mt2r38; taste receptor, type 2,
member 135; K08474 taste receptor type 2
Length=321
Score = 33.1 bits (74), Expect = 6.6, Method: Composition-based stats.
Identities = 15/62 (24%), Positives = 33/62 (53%), Gaps = 0/62 (0%)
Query 250 LNSATIAAIGVLGALFCVSSGHFTRMLCLYVNAGLSMWLGFHLVAAIGGADMPVVISLLN 309
L +ATI +LG +CV T + +++ L W+ + L++A+G + + ++ +
Sbjct 112 LTAATIWLCSLLGFFYCVKIATLTHPVFVWLKYRLPGWVPWMLLSAVGMSSLTSILCFIG 171
Query 310 SY 311
+Y
Sbjct 172 NY 173
> mmu:629959 Gm7020, EG629959; predicted gene 7020
Length=116
Score = 33.1 bits (74), Expect = 7.2, Method: Composition-based stats.
Identities = 20/71 (28%), Positives = 32/71 (45%), Gaps = 0/71 (0%)
Query 562 DLEAGLLEFERDERADPSSWPYPRLAVGVLKDSNGSVMVPIAPKFVPRLRKMAFRVIVES 621
DLEA LL FE+ + A P WP V S S + PK++ + + ++ E
Sbjct 13 DLEASLLSFEKLDCASPDLWPEQLPGVAEFAASFKSPITSSPPKWMAEIERDDIDMLKEL 72
Query 622 GAGANAGFSDE 632
G+ A ++
Sbjct 73 GSLTTANLMEK 83
> tgo:TGME49_078850 zinc finger DHHC domain-containing protein
(EC:3.4.24.35)
Length=361
Score = 32.7 bits (73), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 21/77 (27%), Positives = 37/77 (48%), Gaps = 8/77 (10%)
Query 963 PTPVSRTPPRVPMPSAPSAAPAKAGGAFAQALASDAFFAMCLVVAAAVVGLLGIVLDPME 1022
P+P +R P RVP+ A + G F FA+ L+++ +G + I+L P+
Sbjct 60 PSPTNRAPTRVPVSGAAGTSRRSGNGCF--------LFAVVLLLSFLYLGYVFILLAPLL 111
Query 1023 LKHLTLLALSLIVGYYC 1039
++L L V ++C
Sbjct 112 WPIPSMLGSVLFVAFHC 128
Lambda K H
0.321 0.136 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 60168743740
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40