bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0434_orf5
Length=218
Score E
Sequences producing significant alignments: (Bits) Value
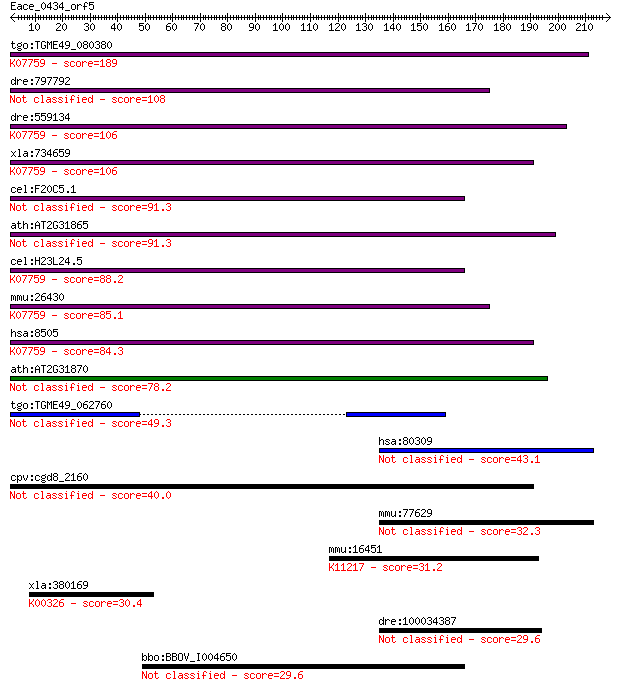
tgo:TGME49_080380 non-transmembrane antigen (EC:3.2.1.143); K0... 189 6e-48
dre:797792 si:dkey-259k14.2 108 2e-23
dre:559134 poly(ADP-ribose) glycohydrolase 63 kDa-like; K07759... 106 7e-23
xla:734659 parg, MGC115697; poly (ADP-ribose) glycohydrolase; ... 106 8e-23
cel:F20C5.1 pme-3; Poly(ADP-ribose) Metabolism Enzyme family m... 91.3 2e-18
ath:AT2G31865 poly (ADP-ribose) glycohydrolase (PARG) family p... 91.3 2e-18
cel:H23L24.5 pme-4; Poly(ADP-ribose) Metabolism Enzyme family ... 88.2 2e-17
mmu:26430 Parg, AI413217; poly (ADP-ribose) glycohydrolase (EC... 85.1 2e-16
hsa:8505 PARG, FLJ54459, FLJ60257, FLJ60456, PARG99; poly (ADP... 84.3 3e-16
ath:AT2G31870 TEJ; TEJ (Sanskrit for 'bright'); poly(ADP-ribos... 78.2 2e-14
tgo:TGME49_062760 poly(ADP-ribose) glycohydrolase, putative 49.3 1e-05
hsa:80309 SPHKAP, DKFZp781H143, DKFZp781J171, KIAA1678, MGC132... 43.1 8e-04
cpv:cgd8_2160 shares a domain with poly(ADP) ribose glycohydro... 40.0 0.006
mmu:77629 Sphkap, 4930544G21Rik, A930009L15Rik, AI852220, mKIA... 32.3 1.1
mmu:16451 Jak1, AA960307, C130039L05Rik, MGC37919; Janus kinas... 31.2 2.7
xla:380169 cyb5r3, MGC52778, dia1; cytochrome b5 reductase 3 (... 30.4 4.7
dre:100034387 sphkap, si:dkey-223n17.6, si:dkey-99o15.1; SPHK1... 29.6 7.4
bbo:BBOV_I004650 19.m02247; tRNA nucleotidyltransferase/poly(A... 29.6 8.6
> tgo:TGME49_080380 non-transmembrane antigen (EC:3.2.1.143);
K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=553
Score = 189 bits (480), Expect = 6e-48, Method: Compositional matrix adjust.
Identities = 90/215 (41%), Positives = 134/215 (62%), Gaps = 7/215 (3%)
Query 1 ELIFALRPELHVIMLFMSAMKDDEAVVVKGAESFVLYSGYEGTFQVFPPV-----VPVGS 55
ELIF + PE+ +MLF MK +EAVVVKG E FV GYE TFQ+ +P
Sbjct 338 ELIFIIYPEMLCVMLFSDIMKPNEAVVVKGVERFVFSQGYEWTFQITAAAGDWTELPSNK 397
Query 56 HWSIHGYTPLDSMGRRETTVVAIDAIIVKNEREQYSTPSMKREILKAALGFQGDPFEDAI 115
H+S+ G PLDS+GRR +V IDA+ +QYS + RE++KAA+GF+GDP+E +
Sbjct 398 HFSVQGVVPLDSLGRRNVAIVGIDAVQFHEPNKQYSPVMVNRELMKAAVGFKGDPYELIV 457
Query 116 HATRAGVATGKWGCVIFGGDNELKTLMQWISAAAAGRELHYKAFGDTRLSHMQVTLHAIR 175
+R +ATG WGC +F GD +LKTL+QW++A+ AGR + + F + + + + + +R
Sbjct 458 SGSRQPIATGLWGCGVFNGDAQLKTLIQWLAASYAGRSMKFYTFSNKSVDGLGLVISKLR 517
Query 176 EKFPTTRDLFEAIVQSVQQREERPNGNWSLWQSLL 210
+ + T +L++AI ++ +R R WSLW+ LL
Sbjct 518 QSYATVGNLYKAIQNALSRRSGRL--GWSLWKELL 550
> dre:797792 si:dkey-259k14.2
Length=609
Score = 108 bits (270), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 63/174 (36%), Positives = 91/174 (52%), Gaps = 17/174 (9%)
Query 1 ELIFALRPELHVIMLFMSAMKDDEAVVVKGAESFVLYSGYEGTFQVFPPVVPVGSHWSIH 60
E++F + PEL + LF + D E V + G + + L SGY +F P +
Sbjct 407 EILFLMSPELILARLFTEKLDDHECVRITGPQMYSLTSGYSRSFSWTGPYM--------- 457
Query 61 GYTPLDSMGRRETTVVAIDAIIVKNEREQYSTPSMKREILKAALGFQGDPFEDAIHATRA 120
T D RR +VAIDA+ KN EQYS ++ RE+ KA +GF G P +
Sbjct 458 DRTKRDVWKRRFRQIVAIDALDFKNPLEQYSRENITRELNKAFVGFCGQP--------KT 509
Query 121 GVATGKWGCVIFGGDNELKTLMQWISAAAAGRELHYKAFGDTRLSHMQVTLHAI 174
+ATG WGC F GD +LK L+Q ++AA R++ Y FG+T L++ +H I
Sbjct 510 AIATGNWGCGAFRGDPKLKALLQLMAAAVVDRDVAYFTFGNTHLANELQKMHDI 563
> dre:559134 poly(ADP-ribose) glycohydrolase 63 kDa-like; K07759
poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=777
Score = 106 bits (264), Expect = 7e-23, Method: Compositional matrix adjust.
Identities = 71/208 (34%), Positives = 105/208 (50%), Gaps = 20/208 (9%)
Query 1 ELIFALRPELHVIMLFMSAMKDDEAVVVKGAESFVLYSGYEGTFQVFPPVVPVGSHWSIH 60
E+ F + PEL V LF + +E +++ G E + YSGY +F+ W +
Sbjct 550 EIRFLINPELIVSRLFTEVLDHNECLIITGTEQYSKYSGYAESFK-----------WEDN 598
Query 61 --GYTPLDSMGRRETTVVAIDAIIVKNEREQYSTPSMKREILKAALGFQGDPFEDAIHAT 118
P D RR T +VA+DA+ +N +Q+ M RE+ KA GF P + ++
Sbjct 599 HKDKIPRDGWQRRCTEIVAMDALHYRNFMDQFQPEKMTRELNKAYCGFMR-PGVNPLNL- 656
Query 119 RAGVATGKWGCVIFGGDNELKTLMQWISAAAAGRELHYKAFGDTRLSHMQVTLHA-IREK 177
+ VATG WGC FGGD LK L+Q ++AA AGR++ Y FGD L LH +++
Sbjct 657 -SAVATGNWGCGAFGGDTRLKALLQLMAAAEAGRDVAYFTFGDEALMRDVQDLHKFLKDN 715
Query 178 FPTTRDLFEAIVQS---VQQREERPNGN 202
T L+ + Q V ++ RPN N
Sbjct 716 CVTVGSLYVYLKQYSSIVSKQAHRPNIN 743
> xla:734659 parg, MGC115697; poly (ADP-ribose) glycohydrolase;
K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=759
Score = 106 bits (264), Expect = 8e-23, Method: Compositional matrix adjust.
Identities = 64/194 (32%), Positives = 99/194 (51%), Gaps = 19/194 (9%)
Query 1 ELIFALRPELHVIMLFMSAMKDDEAVVVKGAESFVLYSGYEGTFQVFPPVVPVGSHWSI- 59
E+ F + PEL V LF + +E +++ GAE + Y+GY T++ W+
Sbjct 553 EIRFLINPELIVSRLFTEVLDSNECLIITGAEQYSEYTGYSETYK-----------WACV 601
Query 60 -HGYTPLDSMGRRETTVVAIDAIIVKNEREQYSTPSMKREILKAALGFQGDPFEDAIHAT 118
+P D RR T +VAIDA + +Q+ +KRE+ KA GF + ++
Sbjct 602 HEDESPRDEWQRRTTEIVAIDAFHFRRPIDQFVPEKIKRELNKAFCGF----YRPEVNPQ 657
Query 119 R-AGVATGKWGCVIFGGDNELKTLMQWISAAAAGRELHYKAFGDTRL-SHMQVTLHAIRE 176
+ VATG WGC FGGD LK L+Q ++AA GR+L Y FGD L + + + E
Sbjct 658 NLSAVATGNWGCGAFGGDPRLKALIQLLAAAEVGRDLVYFTFGDRELMKDIYLMYSFLTE 717
Query 177 KFPTTRDLFEAIVQ 190
K T D++ +++
Sbjct 718 KNKTVGDIYSMLIE 731
> cel:F20C5.1 pme-3; Poly(ADP-ribose) Metabolism Enzyme family
member (pme-3)
Length=764
Score = 91.3 bits (225), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 61/172 (35%), Positives = 90/172 (52%), Gaps = 16/172 (9%)
Query 1 ELIFALRPELHVIMLFMSAMKDDEAVVVKGAESFVLYSGYEGTFQVFPPVVPVGSHWSIH 60
E+ F + PE+ V ML MK EA+ + GA F Y+GY T + + + P S + +
Sbjct 538 EIRFLMCPEMMVGMLLCEKMKQLEAISIVGAYVFSSYTGYGHTLK-WAELQPNHSRQNTN 596
Query 61 GYTPLDSMGRRETTVVAIDAIIVKNER-----EQYSTPSMKREILKAALGF--QGDPFED 113
+ D GR +AIDAI+ K + EQ + ++ RE+ KA++GF QG F
Sbjct 597 EFR--DRFGRLRVETIAIDAILFKGSKLDCQTEQLNKANIIREMKKASIGFMSQGPKF-- 652
Query 114 AIHATRAGVATGKWGCVIFGGDNELKTLMQWISAAAAGRELHYKAFGDTRLS 165
T + TG WGC F GD LK ++Q I+A A R LH+ +FG+ L+
Sbjct 653 ----TNIPIVTGWWGCGAFNGDKPLKFIIQVIAAGVADRPLHFCSFGEPELA 700
> ath:AT2G31865 poly (ADP-ribose) glycohydrolase (PARG) family
protein
Length=522
Score = 91.3 bits (225), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 68/232 (29%), Positives = 106/232 (45%), Gaps = 44/232 (18%)
Query 1 ELIFALRPELHVIMLFMSAMKDDEAVVVKGAESFVLYSGYEGTFQVFPPVVPVGSHWSIH 60
E+ F + PEL M+F+ M +EA+ + G E F Y+GY +FQ G +
Sbjct 285 EIRFVINPELIAGMIFLPRMDANEAIEIVGVERFSGYTGYGPSFQY------AGDY---T 335
Query 61 GYTPLDSMGRRETTVVAIDAIIVKNEREQYSTPSMKREILKAALGF----------QGDP 110
LD RR+T V+AIDA+ QY ++ RE+ KA G+ + DP
Sbjct 336 DNKDLDIFRRRKTRVIAIDAMPDPG-MGQYKLDALIREVNKAFSGYMHQCKYNIDVKHDP 394
Query 111 FEDAIHA-----------------------TRAGVATGKWGCVIFGGDNELKTLMQWISA 147
+ H + GVATG WGC +FGGD ELK ++QW++
Sbjct 395 EASSSHVPLTSDSASQVIESSHRWCIDHEEKKIGVATGNWGCGVFGGDPELKIMLQWLAI 454
Query 148 AAAGRE-LHYKAFGDTRLSHMQVTLHAIREKFPTTRDLFEAIVQSVQQREER 198
+ +GR + Y FG L ++ + + + T DL++ +V+ +R R
Sbjct 455 SQSGRPFMSYYTFGLQALQNLNQVIEMVALQEMTVGDLWKKLVEYSSERLSR 506
> cel:H23L24.5 pme-4; Poly(ADP-ribose) Metabolism Enzyme family
member (pme-4); K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=485
Score = 88.2 bits (217), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 58/172 (33%), Positives = 89/172 (51%), Gaps = 16/172 (9%)
Query 1 ELIFALRPELHVIMLFMSAMKDDEAVVVKGAESFVLYSGYEGTFQVFPPVVPVGSHWSIH 60
E+ F + PE+ V +L +D EA+ + GA F Y+GY T + + + P H + +
Sbjct 272 EIRFMMCPEMMVAILLNDVTQDLEAISIVGAYVFSSYTGYSNTLK-WAKITP--KHSAQN 328
Query 61 GYTPLDSMGRRETTVVAIDAI-----IVKNEREQYSTPSMKREILKAALGF--QGDPFED 113
+ D GR +T VAIDA+ ++ Q +T + RE+ KAA+GF GD F
Sbjct 329 NNSFRDQFGRLQTETVAIDAVRNAGTPLECLLNQLTTEKLTREVRKAAIGFLSAGDGF-- 386
Query 114 AIHATRAGVATGKWGCVIFGGDNELKTLMQWISAAAAGRELHYKAFGDTRLS 165
++ V +G WGC F G+ LK L+Q I+ + R L + FGDT L+
Sbjct 387 ----SKIPVVSGWWGCGAFRGNKPLKFLIQVIACGISDRPLQFCTFGDTELA 434
> mmu:26430 Parg, AI413217; poly (ADP-ribose) glycohydrolase (EC:3.2.1.143);
K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=961
Score = 85.1 bits (209), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 62/178 (34%), Positives = 92/178 (51%), Gaps = 20/178 (11%)
Query 1 ELIFALRPELHVIMLFMSAMKDDEAVVVKGAESFVLYSGYEGTFQVFPPVVPVGSHW--S 58
E+ F + PEL V LF + +E +++ G E + Y+GY T++ W S
Sbjct 749 EIRFLINPELIVSRLFTEVLDHNECLIITGTEQYSEYTGYAETYR-----------WARS 797
Query 59 IHGYTPLDSMGRRETTVVAIDAIIVKNEREQYSTPSMKREILKAALGF--QGDPFEDAIH 116
+ D RR T +VAIDA+ + +Q+ ++RE+ KA GF G P E+
Sbjct 798 HEDGSEKDDWQRRCTEIVAIDALHFRRYLDQFVPEKVRRELNKAYCGFLRPGVPSENL-- 855
Query 117 ATRAGVATGKWGCVIFGGDNELKTLMQWISAAAAGRELHYKAFGDTRLSHMQVTLHAI 174
+ VATG WGC FGGD LK L+Q ++AAAA R++ Y FGD+ L ++H
Sbjct 856 ---SAVATGNWGCGAFGGDARLKALIQILAAAAAERDVVYFTFGDSELMRDIYSMHTF 910
> hsa:8505 PARG, FLJ54459, FLJ60257, FLJ60456, PARG99; poly (ADP-ribose)
glycohydrolase (EC:3.2.1.143); K07759 poly(ADP-ribose)
glycohydrolase [EC:3.2.1.143]
Length=976
Score = 84.3 bits (207), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 62/194 (31%), Positives = 101/194 (52%), Gaps = 19/194 (9%)
Query 1 ELIFALRPELHVIMLFMSAMKDDEAVVVKGAESFVLYSGYEGTFQVFPPVVPVGSHWSI- 59
E+ F + PEL + LF + +E +++ G E + Y+GY T++ WS
Sbjct 756 EIRFLINPELIISRLFTEVLDHNECLIITGTEQYSEYTGYAETYR-----------WSRS 804
Query 60 -HGYTPLDSMGRRETTVVAIDAIIVKNEREQYSTPSMKREILKAALGFQGDPFEDAIHAT 118
+ D RR T +VAIDA+ + +Q+ M+RE+ KA GF + +
Sbjct 805 HEDGSERDDWQRRCTEIVAIDALHFRRYLDQFVPEKMRRELNKAYCGF----LRPGVSSE 860
Query 119 R-AGVATGKWGCVIFGGDNELKTLMQWISAAAAGRELHYKAFGDTRLSHMQVTLHA-IRE 176
+ VATG WGC FGGD LK L+Q ++AAAA R++ Y FGD+ L ++H + E
Sbjct 861 NLSAVATGNWGCGAFGGDARLKALIQILAAAAAERDVVYFTFGDSELMRDIYSMHIFLTE 920
Query 177 KFPTTRDLFEAIVQ 190
+ T D+++ +++
Sbjct 921 RKLTVGDVYKLLLR 934
> ath:AT2G31870 TEJ; TEJ (Sanskrit for 'bright'); poly(ADP-ribose)
glycohydrolase
Length=547
Score = 78.2 bits (191), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 69/258 (26%), Positives = 102/258 (39%), Gaps = 74/258 (28%)
Query 1 ELIFALRPELHVIMLFMSAMKDDEAVVVKGAESFVLYSGYEGTFQVFPPVVPVGSHWSIH 60
E+ F + PEL MLF+ M D+EA+ + GAE F Y+GY +F+ + +
Sbjct 274 EIRFMINPELIAGMLFLPRMDDNEAIEIVGAERFSCYTGYASSFRFAGEYIDKKA----- 328
Query 61 GYTPLDSMGRRETTVVAIDAIIVKNEREQYSTPSMKREILKAALGFQG------------ 108
+D RR T +VAIDA+ R + + REI KA GF
Sbjct 329 ----MDPFKRRRTRIVAIDALCTPKMR-HFKDICLLREINKALCGFLNCSKAWEHQNIFM 383
Query 109 DPFEDAIHATRAGVATG------------------------------------------- 125
D ++ I R G +G
Sbjct 384 DEGDNEIQLVRNGRDSGLLRTETTSHRTPLNDVEMNREKPANNLIRDFYVEGVDNEDHED 443
Query 126 ------KWGCVIFGGDNELKTLMQWISAAAAGRE-LHYKAFGDTRLSHM-QVTLHAIREK 177
WGC +FGGD ELK +QW++A+ R + Y FG L ++ QVT + K
Sbjct 444 DGVATGNWGCGVFGGDPELKATIQWLAASQTRRPFISYYTFGVEALRNLDQVTKWILSHK 503
Query 178 FPTTRDLFEAIVQSVQQR 195
+ T DL+ +++ QR
Sbjct 504 W-TVGDLWNMMLEYSAQR 520
> tgo:TGME49_062760 poly(ADP-ribose) glycohydrolase, putative
Length=952
Score = 49.3 bits (116), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 20/36 (55%), Positives = 26/36 (72%), Gaps = 0/36 (0%)
Query 123 ATGKWGCVIFGGDNELKTLMQWISAAAAGRELHYKA 158
ATG WGC +F GD +LK L+QW++A+ GR L Y A
Sbjct 829 ATGNWGCGVFKGDPQLKFLLQWLAASLVGRRLIYHA 864
Score = 29.6 bits (65), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 17/47 (36%), Positives = 25/47 (53%), Gaps = 0/47 (0%)
Query 1 ELIFALRPELHVIMLFMSAMKDDEAVVVKGAESFVLYSGYEGTFQVF 47
E+ FA PEL ++ LF + +E+ + GA F YSGY +F
Sbjct 502 EIFFATHPELLLLRLFQQRLAINESCAMSGAMQFSRYSGYADSFTCL 548
> hsa:80309 SPHKAP, DKFZp781H143, DKFZp781J171, KIAA1678, MGC132614,
MGC132616, SKIP; SPHK1 interactor, AKAP domain containing
Length=1700
Score = 43.1 bits (100), Expect = 8e-04, Method: Composition-based stats.
Identities = 23/79 (29%), Positives = 41/79 (51%), Gaps = 1/79 (1%)
Query 135 DNELKTLMQWISAAAAG-RELHYKAFGDTRLSHMQVTLHAIREKFPTTRDLFEAIVQSVQ 193
D EL+ +QWI+A+ G +++K + R+ + + K D+F A+VQ +
Sbjct 1621 DAELRATLQWIAASELGIPTIYFKKSQENRIEKFLDVVQLVHRKSWKVGDIFHAVVQYCK 1680
Query 194 QREERPNGNWSLWQSLLNI 212
EE+ +G SL+ LL +
Sbjct 1681 MHEEQKDGRLSLFDWLLEL 1699
> cpv:cgd8_2160 shares a domain with poly(ADP) ribose glycohydrolases,
some protein kinase A anchoring proteins and baculovirus
HzNV Orf103, possible transmembrane domain within N-terminus
Length=441
Score = 40.0 bits (92), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 45/190 (23%), Positives = 79/190 (41%), Gaps = 15/190 (7%)
Query 1 ELIFALRPELHVIMLFMSAMKDDEAVVVKGAESFVLYSGYEGTFQVFPPVVPVGSHWSIH 60
E++ + PE + F+ + D+++V ++G + Y G+ F + + SI
Sbjct 249 EIVMTVVPETLIGRFFLDDLHDEDSVTIRGVMRYSNYKGFGTDKFTFQSIKESEMYMSI- 307
Query 61 GYTPLDSMGRRETTVVAIDAIIVKNEREQYSTPSMKREILKAALGFQGDPFEDAIHATRA 120
+DA+ + +++ REI K D + ++ R
Sbjct 308 -----------PRVYAVVDALSGGSRFREFTLDYALREINKLISALCDDFYGESEERDRN 356
Query 121 GVATGKWGCVIFGGDNELKTLMQWISAAAAGRELHYKAFGDTRLSHMQVTLHAIREKFPT 180
TG WG +FGGDN+ K ++Q I+A R+L Y G RL Q+ I K+ T
Sbjct 357 QFVTGYWGGGVFGGDNQYKFILQLIAACVCNRQLVYSD-GLNRLDVNQI--RQIESKYTT 413
Query 181 TRDLFEAIVQ 190
L +A +
Sbjct 414 CGQLSDAFFK 423
> mmu:77629 Sphkap, 4930544G21Rik, A930009L15Rik, AI852220, mKIAA1678;
SPHK1 interactor, AKAP domain containing
Length=1658
Score = 32.3 bits (72), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 21/79 (26%), Positives = 41/79 (51%), Gaps = 1/79 (1%)
Query 135 DNELKTLMQWISAAAAG-RELHYKAFGDTRLSHMQVTLHAIREKFPTTRDLFEAIVQSVQ 193
D EL+ +QWI+A+ G +++K ++R+ + +++K D+F A+VQ +
Sbjct 1579 DAELRATLQWIAASELGIPTIYFKKSQESRIEKFLDVVKLVQQKSWKVGDIFHAVVQYCK 1638
Query 194 QREERPNGNWSLWQSLLNI 212
E+ SL+ LL +
Sbjct 1639 LHAEQKERTPSLFDWLLEL 1657
> mmu:16451 Jak1, AA960307, C130039L05Rik, MGC37919; Janus kinase
1 (EC:2.7.10.2); K11217 Janus kinase 1 [EC:2.7.10.2]
Length=1153
Score = 31.2 bits (69), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 18/76 (23%), Positives = 34/76 (44%), Gaps = 3/76 (3%)
Query 117 ATRAGVATGKWGCVIFGGDNELKTLMQWISAAAAGRELHYKAFGDTRLSHMQVTLHAIRE 176
+ G+ +W C F DN L T+ + + G + +K F + + +LH +
Sbjct 457 GSEEGMYVLRWSCTDF--DNILMTVTCFEKSEVLGGQKQFKNF-QIEVQKGRYSLHGSMD 513
Query 177 KFPTTRDLFEAIVQSV 192
FP+ RDL + + +
Sbjct 514 HFPSLRDLMNHLKKQI 529
> xla:380169 cyb5r3, MGC52778, dia1; cytochrome b5 reductase 3
(EC:1.6.2.2); K00326 cytochrome-b5 reductase [EC:1.6.2.2]
Length=301
Score = 30.4 bits (67), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 28/49 (57%), Gaps = 5/49 (10%)
Query 8 PELHVIMLFMSAMKDDEAVVVKGAESFVLYSGYEGTFQVFP----PVVP 52
PE + ++ +++ DE + +G ++YSG +GTFQ+ P P VP
Sbjct 122 PEGGKMSQYLDSLRKDETIDFRGPSGLLVYSG-KGTFQIRPDKKSPPVP 169
> dre:100034387 sphkap, si:dkey-223n17.6, si:dkey-99o15.1; SPHK1
interactor, AKAP domain containing
Length=1596
Score = 29.6 bits (65), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 19/60 (31%), Positives = 29/60 (48%), Gaps = 1/60 (1%)
Query 135 DNELKTLMQWISAAAAG-RELHYKAFGDTRLSHMQVTLHAIREKFPTTRDLFEAIVQSVQ 193
D EL+ +QWISA+ G L+++ L+ + L +K DLF A+ Q Q
Sbjct 1513 DGELRAALQWISASELGVPALYFRKTHQHNLTKLHRVLQLAGQKAWRVGDLFSAVAQFCQ 1572
> bbo:BBOV_I004650 19.m02247; tRNA nucleotidyltransferase/poly(A)
polymerase family domain containing protein
Length=461
Score = 29.6 bits (65), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 33/128 (25%), Positives = 53/128 (41%), Gaps = 19/128 (14%)
Query 49 PVVPVGSHWSIHGYTPLDSMGRRETTVVAIDAIIVKNEREQYS---TPSMKREILKAALG 105
P++ +G TP + RR+ T+ A+ I KN E Y+ +K E+++
Sbjct 155 PIMKIG--------TPFEDAMRRDFTINALFYNISKNAIEDYTGKGIDDLKAEVIRTCSP 206
Query 106 FQGDPFEDAIHATRAGVATGKWG------CVIFGGDNE-LKTLMQWISAAAAGRELHYKA 158
G +D + RA + G + G D E L L Q ++ A +EL
Sbjct 207 AFGTFLDDPLRVIRAARFAARLGYALDSDIIKSGSDPEVLAQLSQKVAKARIAQELDDML 266
Query 159 F-GDTRLS 165
GDT L+
Sbjct 267 IKGDTTLA 274
Lambda K H
0.320 0.135 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7073640200
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40