bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0463_orf1
Length=107
Score E
Sequences producing significant alignments: (Bits) Value
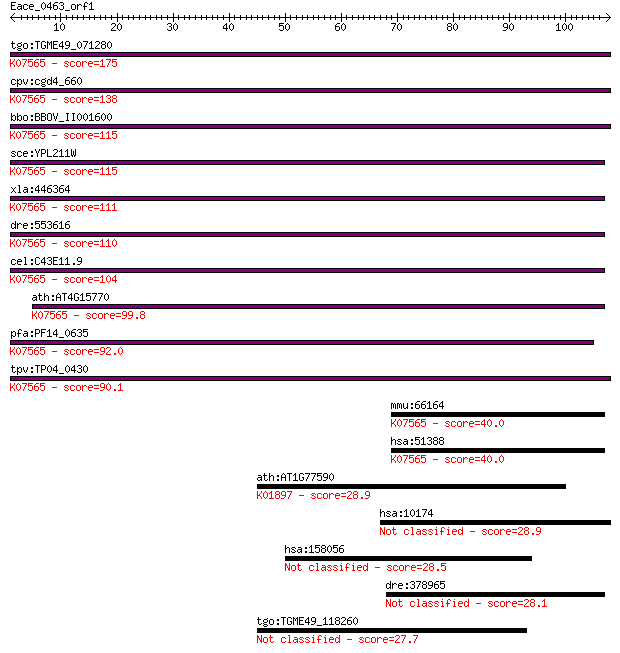
tgo:TGME49_071280 60S ribosome subunit biogenesis protein NIP7... 175 3e-44
cpv:cgd4_660 Nip7 like PUA domain containing protein involved ... 138 4e-33
bbo:BBOV_II001600 18.m06124; 60S ribosome subunit biogenesis p... 115 3e-26
sce:YPL211W NIP7; Nip7p; K07565 60S ribosome subunit biogenesi... 115 4e-26
xla:446364 nip7, MGC83122; nuclear import 7 homolog; K07565 60... 111 7e-25
dre:553616 nip7, MGC110384, zgc:110384; nuclear import 7 homol... 110 1e-24
cel:C43E11.9 hypothetical protein; K07565 60S ribosome subunit... 104 6e-23
ath:AT4G15770 RNA binding / protein binding; K07565 60S riboso... 99.8 2e-21
pfa:PF14_0635 RNA binding protein, putative; K07565 60S riboso... 92.0 4e-19
tpv:TP04_0430 hypothetical protein; K07565 60S ribosome subuni... 90.1 1e-18
mmu:66164 Nip7, 1110017C15Rik, 6330509M23Rik, AA408773, AA4100... 40.0 0.002
hsa:51388 NIP7, FLJ10296, HSPC031, KD93; nuclear import 7 homo... 40.0 0.002
ath:AT1G77590 LACS9; LACS9 (LONG CHAIN ACYL-COA SYNTHETASE 9);... 28.9 4.4
hsa:10174 SORBS3, SCAM-1, SCAM1, SH3D4, VINEXIN; sorbin and SH... 28.9 4.6
hsa:158056 MAMDC4, AEGP, DKFZp434M1411; MAM domain containing 4 28.5
dre:378965 fem1c; fem-1 homolog c (C.elegans) 28.1 7.7
tgo:TGME49_118260 hypothetical protein 27.7 8.5
> tgo:TGME49_071280 60S ribosome subunit biogenesis protein NIP7,
putative ; K07565 60S ribosome subunit biogenesis protein
NIP7
Length=181
Score = 175 bits (443), Expect = 3e-44, Method: Compositional matrix adjust.
Identities = 76/107 (71%), Positives = 92/107 (85%), Gaps = 0/107 (0%)
Query 1 KGRKFRLGITALHLIARYAPHKVWLNPGGEQAFVYGNHIVKRHVGRLTADMPSGAGVCVL 60
K RKFRL +TAL LIARYA +++WL PGGEQ++VYGNH++KRH+GR+TADMP AGVC+
Sbjct 75 KSRKFRLQVTALDLIARYAKYRIWLKPGGEQSYVYGNHVIKRHIGRITADMPQNAGVCIC 134
Query 61 SLNGDLPLGFGTSAKSTAEIHTAPTEAISFYHHADVGEYLREEADLV 107
SLN DLPLGFG +AK+T +IH A TEAI+ YH AD+GEYLREEADLV
Sbjct 135 SLNNDLPLGFGVAAKATQDIHAADTEAIAVYHVADIGEYLREEADLV 181
> cpv:cgd4_660 Nip7 like PUA domain containing protein involved
in ribosomal biogenesis ; K07565 60S ribosome subunit biogenesis
protein NIP7
Length=183
Score = 138 bits (348), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 61/107 (57%), Positives = 81/107 (75%), Gaps = 1/107 (0%)
Query 1 KGRKFRLGITALHLIARYAPHKVWLNPGGEQAFVYGNHIVKRHVGRLTADMPSGAGVCVL 60
K KFRLGIT+L IAR P+KVW+ PGGEQ+++YGNH++KRH+ R+T + + GV V
Sbjct 78 KNNKFRLGITSLQYIARLTPNKVWVKPGGEQSYIYGNHVIKRHISRMTDGLSNNVGVVVY 137
Query 61 SLNGDLPLGFGTSAKSTAEIHTAPTEAISFYHHADVGEYLREEADLV 107
S++ +LPLGFG AKS+A++ A E I+ YH DVGEYLREEADL+
Sbjct 138 SMD-ELPLGFGVLAKSSADMKNADPETIAVYHQTDVGEYLREEADLL 183
> bbo:BBOV_II001600 18.m06124; 60S ribosome subunit biogenesis
protein NIP7; K07565 60S ribosome subunit biogenesis protein
NIP7
Length=180
Score = 115 bits (288), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 52/107 (48%), Positives = 78/107 (72%), Gaps = 3/107 (2%)
Query 1 KGRKFRLGITALHLIARYAPHKVWLNPGGEQAFVYGNHIVKRHVGRLTADMPSGAGVCVL 60
K ++FRL ITALH +A Y+ KVWL G EQ+FVYGN++ K+H+G+++ D+P GV V+
Sbjct 77 KSKQFRLHITALHYLAHYSKSKVWLKTG-EQSFVYGNNVAKKHIGQMSEDIPQNGGVVVM 135
Query 61 SLNGDLPLGFGTSAKSTAEIHTAPTEAISFYHHADVGEYLREEADLV 107
N LPLGFG ++KST + T+ + ++ +H AD+GEYLR EA+++
Sbjct 136 YNN--LPLGFGVTSKSTEALKTSLPDDVAVFHQADIGEYLRHEAEII 180
> sce:YPL211W NIP7; Nip7p; K07565 60S ribosome subunit biogenesis
protein NIP7
Length=181
Score = 115 bits (287), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 53/106 (50%), Positives = 75/106 (70%), Gaps = 1/106 (0%)
Query 1 KGRKFRLGITALHLIARYAPHKVWLNPGGEQAFVYGNHIVKRHVGRLTADMPSGAGVCVL 60
K KFRL IT+L ++A++A +K+W+ P GE F+YGNH++K HVG+++ D+P AGV V
Sbjct 75 KTGKFRLHITSLTVLAKHAKYKIWIKPNGEMPFLYGNHVLKAHVGKMSDDIPEHAGVIVF 134
Query 61 SLNGDLPLGFGTSAKSTAEIHTAPTEAISFYHHADVGEYLREEADL 106
++N D+PLGFG SAKST+E I + AD+GEYLR+E L
Sbjct 135 AMN-DVPLGFGVSAKSTSESRNMQPTGIVAFRQADIGEYLRDEDTL 179
> xla:446364 nip7, MGC83122; nuclear import 7 homolog; K07565
60S ribosome subunit biogenesis protein NIP7
Length=180
Score = 111 bits (277), Expect = 7e-25, Method: Compositional matrix adjust.
Identities = 54/106 (50%), Positives = 71/106 (66%), Gaps = 1/106 (0%)
Query 1 KGRKFRLGITALHLIARYAPHKVWLNPGGEQAFVYGNHIVKRHVGRLTADMPSGAGVCVL 60
K +KFRL +TAL +A YA +KVW+ PG EQ+F+YGNH++K +GR+T + GV V
Sbjct 75 KTQKFRLHVTALDYLAPYAKYKVWVKPGAEQSFLYGNHVLKSGLGRITENTSQYQGVVVY 134
Query 61 SLNGDLPLGFGTSAKSTAEIHTAPTEAISFYHHADVGEYLREEADL 106
S+ D+PLGFG +AKST E AI +H ADVGEY+R E L
Sbjct 135 SM-ADVPLGFGVAAKSTQECRKLDPMAIVVFHQADVGEYIRHEDTL 179
> dre:553616 nip7, MGC110384, zgc:110384; nuclear import 7 homolog
(S. cerevisiae); K07565 60S ribosome subunit biogenesis
protein NIP7
Length=180
Score = 110 bits (275), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 53/106 (50%), Positives = 70/106 (66%), Gaps = 1/106 (0%)
Query 1 KGRKFRLGITALHLIARYAPHKVWLNPGGEQAFVYGNHIVKRHVGRLTADMPSGAGVCVL 60
K +KFRL ITAL +A YA KVW+ PG EQ+F+YGNHI+K +GR+T + GV V
Sbjct 75 KTQKFRLHITALDFLAPYAKFKVWVKPGSEQSFLYGNHIMKSGLGRITENTDKYQGVLVY 134
Query 61 SLNGDLPLGFGTSAKSTAEIHTAPTEAISFYHHADVGEYLREEADL 106
S+ D+PLGFG +A++T E AI +H AD+GEY+R E L
Sbjct 135 SM-ADVPLGFGVAARTTQECRKVDPMAIVVFHQADIGEYIRSEDTL 179
> cel:C43E11.9 hypothetical protein; K07565 60S ribosome subunit
biogenesis protein NIP7
Length=180
Score = 104 bits (260), Expect = 6e-23, Method: Compositional matrix adjust.
Identities = 52/106 (49%), Positives = 68/106 (64%), Gaps = 1/106 (0%)
Query 1 KGRKFRLGITALHLIARYAPHKVWLNPGGEQAFVYGNHIVKRHVGRLTADMPSGAGVCVL 60
K +KF L ITAL +A YA KVWL P EQ F+YGN+I+K + R+T P+ AG+ V
Sbjct 75 KSKKFHLQITALDYLAPYAKFKVWLKPNAEQQFLYGNNILKSGIARMTDGTPTHAGIVVY 134
Query 61 SLNGDLPLGFGTSAKSTAEIHTAPTEAISFYHHADVGEYLREEADL 106
S+ D+PLGFG SAK T++ A A+ H D+GEYLR E+ L
Sbjct 135 SMT-DVPLGFGVSAKGTSDSKRADPTALVVLHQCDLGEYLRNESHL 179
> ath:AT4G15770 RNA binding / protein binding; K07565 60S ribosome
subunit biogenesis protein NIP7
Length=187
Score = 99.8 bits (247), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 49/102 (48%), Positives = 66/102 (64%), Gaps = 1/102 (0%)
Query 5 FRLGITALHLIARYAPHKVWLNPGGEQAFVYGNHIVKRHVGRLTADMPSGAGVCVLSLNG 64
F L I +L+++A A HKVWL P E +F+YGNH++K +GR+T + G GV V S++
Sbjct 87 FHLTIMSLNILAANAKHKVWLKPTSEMSFLYGNHVLKGGLGRITDSIVPGDGVVVFSMS- 145
Query 65 DLPLGFGTSAKSTAEIHTAPTEAISFYHHADVGEYLREEADL 106
D+PLGFG +AKST + I H AD+GEYLR E DL
Sbjct 146 DVPLGFGIAAKSTQDCRKLDPNGIVVLHQADIGEYLRGEDDL 187
> pfa:PF14_0635 RNA binding protein, putative; K07565 60S ribosome
subunit biogenesis protein NIP7
Length=179
Score = 92.0 bits (227), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 38/104 (36%), Positives = 64/104 (61%), Gaps = 1/104 (0%)
Query 1 KGRKFRLGITALHLIARYAPHKVWLNPGGEQAFVYGNHIVKRHVGRLTADMPSGAGVCVL 60
K F + ITA+ + ++ HK+WL GE+ F++GN+++K H+ +++ ++ G G+ VL
Sbjct 75 KTNNFFIKITAISFLNQFCIHKIWLKESGEKNFLFGNNVLKSHLLKISDNIKKGDGIMVL 134
Query 61 SLNGDLPLGFGTSAKSTAEIHTAPTEAISFYHHADVGEYLREEA 104
S+N D P+GFG S ++T +I H +D+GEYLR E
Sbjct 135 SMN-DNPIGFGISVRNTQDIKILNVTDTVLIHQSDIGEYLRCET 177
> tpv:TP04_0430 hypothetical protein; K07565 60S ribosome subunit
biogenesis protein NIP7
Length=178
Score = 90.1 bits (222), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 42/107 (39%), Positives = 71/107 (66%), Gaps = 4/107 (3%)
Query 1 KGRKFRLGITALHLIARYAPHKVWLNPGGEQAFVYGNHIVKRHVGRLTADMPSGAGVCVL 60
K ++FRL ITAL ++ Y+ +KVWL +Q F+YG +++K+HV +++ + AGV V+
Sbjct 76 KSKQFRLHITALEYLSYYSKNKVWLKM--DQPFLYGGNVLKKHVDQISEGIEQNAGVVVM 133
Query 61 SLNGDLPLGFGTSAKSTAEIHTAPTEAISFYHHADVGEYLREEADLV 107
S N +PLGFG ++KST + + + I+ + AD+G YLR E +++
Sbjct 134 SKN--IPLGFGITSKSTENLRNSLPDDIAVINQADIGHYLRHEEEII 178
> mmu:66164 Nip7, 1110017C15Rik, 6330509M23Rik, AA408773, AA410017,
PEachy; nuclear import 7 homolog (S. cerevisiae); K07565
60S ribosome subunit biogenesis protein NIP7
Length=133
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 18/38 (47%), Positives = 23/38 (60%), Gaps = 0/38 (0%)
Query 69 GFGTSAKSTAEIHTAPTEAISFYHHADVGEYLREEADL 106
GFG +AKST + AI +H AD+GEY+R E L
Sbjct 95 GFGVAAKSTQDCRKVDPMAIVVFHQADIGEYVRHEETL 132
> hsa:51388 NIP7, FLJ10296, HSPC031, KD93; nuclear import 7 homolog
(S. cerevisiae); K07565 60S ribosome subunit biogenesis
protein NIP7
Length=133
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 18/38 (47%), Positives = 23/38 (60%), Gaps = 0/38 (0%)
Query 69 GFGTSAKSTAEIHTAPTEAISFYHHADVGEYLREEADL 106
GFG +AKST + AI +H AD+GEY+R E L
Sbjct 95 GFGVAAKSTQDCRKVDPMAIVVFHQADIGEYVRHEETL 132
> ath:AT1G77590 LACS9; LACS9 (LONG CHAIN ACYL-COA SYNTHETASE 9);
long-chain-fatty-acid-CoA ligase (EC:6.2.1.3); K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=691
Score = 28.9 bits (63), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 13/55 (23%), Positives = 29/55 (52%), Gaps = 1/55 (1%)
Query 45 GRLTADMPSGAGVCVLSLNGDLPLGFGTSAKSTAEIHTAPTEAISFYHHADVGEY 99
G LT+D P G V+ ++ LG+ + + T E++ + + +++ D+G +
Sbjct 487 GYLTSDKPMPRGEIVIG-GSNITLGYFKNEEKTKEVYKVDEKGMRWFYTGDIGRF 540
> hsa:10174 SORBS3, SCAM-1, SCAM1, SH3D4, VINEXIN; sorbin and
SH3 domain containing 3
Length=329
Score = 28.9 bits (63), Expect = 4.6, Method: Composition-based stats.
Identities = 15/41 (36%), Positives = 21/41 (51%), Gaps = 0/41 (0%)
Query 67 PLGFGTSAKSTAEIHTAPTEAISFYHHADVGEYLREEADLV 107
PL GTS+ +T++IH P A+ Y + E E D V
Sbjct 257 PLDLGTSSPNTSQIHWTPYRAMYQYRPQNEDELELREGDRV 297
> hsa:158056 MAMDC4, AEGP, DKFZp434M1411; MAM domain containing
4
Length=1137
Score = 28.5 bits (62), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 27/46 (58%), Gaps = 3/46 (6%)
Query 50 DMPSGAGVCVLSLNGDLPLGFGTSAK--STAEIHTAPTEAISFYHH 93
D +G G VL L+ PL +G SA S ++ APTE +SF++H
Sbjct 607 DHTTGQGHFVL-LDPTDPLAWGHSAHLLSRPQVPAAPTECLSFWYH 651
> dre:378965 fem1c; fem-1 homolog c (C.elegans)
Length=618
Score = 28.1 bits (61), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 21/40 (52%), Gaps = 1/40 (2%)
Query 68 LGFGTSAKSTAEIHTAPTEAISFYHHADVGEYLRE-EADL 106
LG G S +T ++ P A F H D+ YL E +ADL
Sbjct 103 LGHGASVNNTTLTNSTPLRAACFDGHLDIVRYLVEHQADL 142
> tgo:TGME49_118260 hypothetical protein
Length=959
Score = 27.7 bits (60), Expect = 8.5, Method: Composition-based stats.
Identities = 19/55 (34%), Positives = 26/55 (47%), Gaps = 7/55 (12%)
Query 45 GRLTADMPSGAGVCVLSLNGDLPLGFGTSA------KSTAEIHTAPTEA-ISFYH 92
GRL A S GVCV + PL G+S + + +H P+ A +SF H
Sbjct 818 GRLAAVGDSAGGVCVFDIPSGRPLAIGSSPSMQKREERFSPLHFPPSIASLSFCH 872
Lambda K H
0.321 0.138 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2007980300
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40