bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0469_orf2
Length=185
Score E
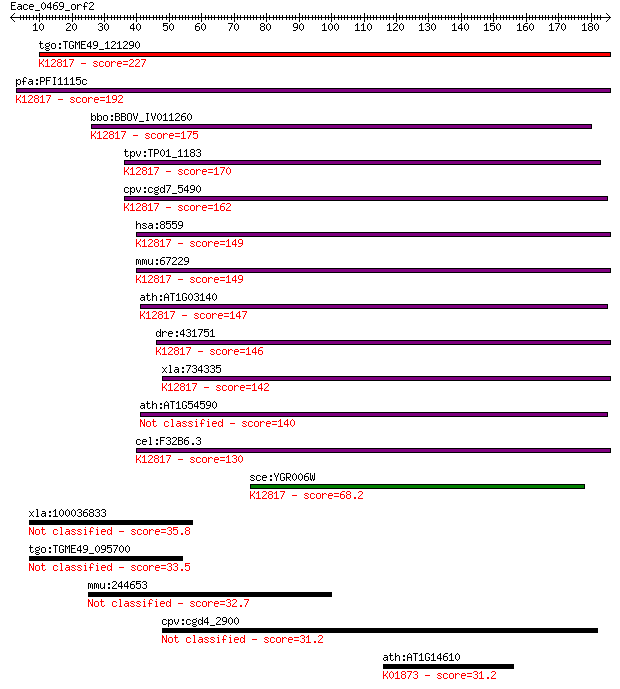
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_121290 pre-mRNA splicing factor 18, putative ; K128... 227 2e-59
pfa:PFI1115c pre-mRNA splicing factor, putative; K12817 pre-mR... 192 5e-49
bbo:BBOV_IV011260 23.m06381; pre-mRNA splicing factor 18; K128... 175 8e-44
tpv:TP01_1183 pre-mRNA splicing factor protein; K12817 pre-mRN... 170 2e-42
cpv:cgd7_5490 PRP18 (SFM+PRP18 domains) ; K12817 pre-mRNA-spli... 162 9e-40
hsa:8559 PRPF18, FLJ10210, PRP18, hPrp18; PRP18 pre-mRNA proce... 149 5e-36
mmu:67229 Prpf18, 2810441A10Rik; PRP18 pre-mRNA processing fac... 149 5e-36
ath:AT1G03140 splicing factor Prp18 family protein; K12817 pre... 147 2e-35
dre:431751 prpf18, zgc:91830; PRP18 pre-mRNA processing factor... 146 5e-35
xla:734335 prpf18, MGC85069; PRP18 pre-mRNA processing factor ... 142 6e-34
ath:AT1G54590 splicing factor Prp18 family protein 140 2e-33
cel:F32B6.3 hypothetical protein; K12817 pre-mRNA-splicing fac... 130 2e-30
sce:YGR006W PRP18; Prp18p; K12817 pre-mRNA-splicing factor 18 68.2
xla:100036833 mks1; Meckel syndrome, type 1 35.8
tgo:TGME49_095700 UBA/TS-N domain-containing protein 33.5 0.42
mmu:244653 Hydin, 1700034M11Rik, 4930545D19Rik, 4932703P14, A8... 32.7 0.74
cpv:cgd4_2900 polyketide synthase 31.2 1.9
ath:AT1G14610 TWN2; TWN2 (TWIN 2); ATP binding / aminoacyl-tRN... 31.2 2.0
> tgo:TGME49_121290 pre-mRNA splicing factor 18, putative ; K12817
pre-mRNA-splicing factor 18
Length=373
Score = 227 bits (578), Expect = 2e-59, Method: Compositional matrix adjust.
Identities = 107/176 (60%), Positives = 131/176 (74%), Gaps = 0/176 (0%)
Query 10 ANKGKKETNSESTDAKQSQNEETTEKSKEAVVIAWARKMLSLWEDELKHRSEDEKATAEG 69
A G E + ++ + SKE VV W R ML WE ELK R E++K +AEG
Sbjct 186 AGSGSDEEDGTDGSGVGRGGQQERQPSKETVVTRWIRTMLQEWEAELKARPEEKKNSAEG 245
Query 70 RQQTALHRQTKKDLKPLFKKLKQRDLEADILEKLFSIVQLCDERRYRDAHGAFMLLAIGN 129
+ T+L RQT+KDLKPL +KL+ ++LE DIL+KL +VQ C+ER+YR AH +MLLAIGN
Sbjct 246 KLMTSLQRQTRKDLKPLLRKLRHKELENDILQKLHIMVQCCEERKYRSAHDTYMLLAIGN 305
Query 130 AAWPMGVTMVGIHERAGRSKLNTSQVAHILNDETTRKYIQMFKRLMSFAQRKFPAN 185
AAWP+GVTMVGIHER GRSKL +SQVAHILNDETTRKYIQMFKRLMSF QR++PA+
Sbjct 306 AAWPVGVTMVGIHERVGRSKLFSSQVAHILNDETTRKYIQMFKRLMSFCQRRYPAD 361
> pfa:PFI1115c pre-mRNA splicing factor, putative; K12817 pre-mRNA-splicing
factor 18
Length=343
Score = 192 bits (488), Expect = 5e-49, Method: Compositional matrix adjust.
Identities = 94/183 (51%), Positives = 133/183 (72%), Gaps = 6/183 (3%)
Query 3 EEEFDEDANKGKKETNSESTDAKQSQNEETTEKSKEAVVIAWARKMLSLWEDELKHRSED 62
E+E ++ ++K K + NS + +++ NE+ + KE ++ W ++ + W +E+++ +
Sbjct 155 EDEIEKGSDK-KDKNNSNVSISEKENNEKGKKIDKEKIIHEWIKRTMKEWNEEIENIVDS 213
Query 63 EKATAEGRQQTALHRQTKKDLKPLFKKLKQRDLEADILEKLFSIVQLCDERRYRDAHGAF 122
+K + A + QT KDLKPL KKLKQ+ LE+DIL+K+++IV C E+ ++ AH A+
Sbjct 214 KKKIKQ-----ATYLQTHKDLKPLEKKLKQKTLESDILDKIYNIVSCCQEKNFKAAHDAY 268
Query 123 MLLAIGNAAWPMGVTMVGIHERAGRSKLNTSQVAHILNDETTRKYIQMFKRLMSFAQRKF 182
MLLAIGNAAWPMGVTMVGIHERAGRSK+ S+VAHILNDETTRKYIQM KRL+SF QRK+
Sbjct 269 MLLAIGNAAWPMGVTMVGIHERAGRSKIYASEVAHILNDETTRKYIQMIKRLLSFCQRKY 328
Query 183 PAN 185
N
Sbjct 329 CTN 331
> bbo:BBOV_IV011260 23.m06381; pre-mRNA splicing factor 18; K12817
pre-mRNA-splicing factor 18
Length=285
Score = 175 bits (444), Expect = 8e-44, Method: Compositional matrix adjust.
Identities = 87/165 (52%), Positives = 117/165 (70%), Gaps = 11/165 (6%)
Query 26 QSQNEETTEKSKEAVVI--------AWARKMLSLWEDELKHRSED---EKATAEGRQQTA 74
Q+Q +E K+++ + + +W L+ WE + + E+ E AE R++ A
Sbjct 119 QAQEKENIAKNQQNIFVEAIIGRIASWVSTTLNKWEQHILSQREEFLQEGNVAEARRRDA 178
Query 75 LHRQTKKDLKPLFKKLKQRDLEADILEKLFSIVQLCDERRYRDAHGAFMLLAIGNAAWPM 134
+ QTKKDL+PL LK + LE +ILEK++++V C+++ Y+ AH A+MLLAIGNAAWPM
Sbjct 179 MLAQTKKDLQPLINMLKSQTLEEEILEKMYNVVICCEKKDYQAAHEAYMLLAIGNAAWPM 238
Query 135 GVTMVGIHERAGRSKLNTSQVAHILNDETTRKYIQMFKRLMSFAQ 179
GVTMVGIHERAGRSK+ TS+VAHILNDE TRKYIQM KRL+SFAQ
Sbjct 239 GVTMVGIHERAGRSKIFTSEVAHILNDEKTRKYIQMIKRLLSFAQ 283
> tpv:TP01_1183 pre-mRNA splicing factor protein; K12817 pre-mRNA-splicing
factor 18
Length=327
Score = 170 bits (431), Expect = 2e-42, Method: Compositional matrix adjust.
Identities = 84/150 (56%), Positives = 105/150 (70%), Gaps = 3/150 (2%)
Query 36 SKEAVVIAWARKMLSLWEDELKHRSED---EKATAEGRQQTALHRQTKKDLKPLFKKLKQ 92
SK V W +ML WE + D E E ++ A+ QTKKD+KPL K +K
Sbjct 163 SKHYKVFEWVSRMLREWESRIVESKGDLIKEGKEFEAKKNEAMLVQTKKDIKPLLKLIKS 222
Query 93 RDLEADILEKLFSIVQLCDERRYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNT 152
L+ +IL+K+ IV C+ +++ AH +MLLAIGNAAWPMGVTMVGIHERAGRSK+ T
Sbjct 223 NKLDQEILDKMEQIVNHCNNGQFKKAHDIYMLLAIGNAAWPMGVTMVGIHERAGRSKIFT 282
Query 153 SQVAHILNDETTRKYIQMFKRLMSFAQRKF 182
S+VAHILNDETTRKYIQMFKRL+SF+Q K+
Sbjct 283 SEVAHILNDETTRKYIQMFKRLISFSQSKY 312
> cpv:cgd7_5490 PRP18 (SFM+PRP18 domains) ; K12817 pre-mRNA-splicing
factor 18
Length=337
Score = 162 bits (409), Expect = 9e-40, Method: Compositional matrix adjust.
Identities = 76/151 (50%), Positives = 104/151 (68%), Gaps = 2/151 (1%)
Query 36 SKEAVVIAWARKMLSLWEDELKHRSEDEKATAEGRQQTALHRQTKKDLKPLFKKLKQRDL 95
S + ++ W L WE LK R + + T +G Q +A + QTK+D++PL L+ D
Sbjct 179 SNQKFILRWIDTQLKEWEKLLKCRKKADSETEKGMQDSAQYYQTKRDIQPLVNSLEASDG 238
Query 96 EAD--ILEKLFSIVQLCDERRYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNTS 153
+ D +L+KLF IV LC++R Y A ++ LAIGNA WPMGVTMVGIHERAGR+K+ +S
Sbjct 239 KVDKEVLDKLFEIVTLCNQRDYNKAQDKYIELAIGNAPWPMGVTMVGIHERAGRTKIFSS 298
Query 154 QVAHILNDETTRKYIQMFKRLMSFAQRKFPA 184
+AH+LNDETTRKYIQMFKRL++ + K P+
Sbjct 299 HIAHVLNDETTRKYIQMFKRLVTHCESKRPS 329
> hsa:8559 PRPF18, FLJ10210, PRP18, hPrp18; PRP18 pre-mRNA processing
factor 18 homolog (S. cerevisiae); K12817 pre-mRNA-splicing
factor 18
Length=342
Score = 149 bits (376), Expect = 5e-36, Method: Compositional matrix adjust.
Identities = 66/146 (45%), Positives = 99/146 (67%), Gaps = 0/146 (0%)
Query 40 VVIAWARKMLSLWEDELKHRSEDEKATAEGRQQTALHRQTKKDLKPLFKKLKQRDLEADI 99
++ + + +L +W EL R + K + +G+ +A +QT+ L+PLF+KL++R+L ADI
Sbjct 187 IITKFLKFLLGVWAKELNAREDYVKRSVQGKLNSATQKQTESYLRPLFRKLRKRNLPADI 246
Query 100 LEKLFSIVQLCDERRYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNTSQVAHIL 159
E + I++ +R Y A+ A++ +AIGNA WP+GVTMVGIH R GR K+ + VAH+L
Sbjct 247 KESITDIIKFMLQREYVKANDAYLQMAIGNAPWPIGVTMVGIHARTGREKIFSKHVAHVL 306
Query 160 NDETTRKYIQMFKRLMSFAQRKFPAN 185
NDET RKYIQ KRLM+ Q+ FP +
Sbjct 307 NDETQRKYIQGLKRLMTICQKHFPTD 332
> mmu:67229 Prpf18, 2810441A10Rik; PRP18 pre-mRNA processing factor
18 homolog (yeast); K12817 pre-mRNA-splicing factor 18
Length=342
Score = 149 bits (376), Expect = 5e-36, Method: Compositional matrix adjust.
Identities = 66/146 (45%), Positives = 99/146 (67%), Gaps = 0/146 (0%)
Query 40 VVIAWARKMLSLWEDELKHRSEDEKATAEGRQQTALHRQTKKDLKPLFKKLKQRDLEADI 99
++ + + +L +W EL R + K + +G+ +A +QT+ L+PLF+KL++R+L ADI
Sbjct 187 IITKFLKFLLGVWAKELNAREDYVKRSVQGKLNSATQKQTESYLRPLFRKLRKRNLPADI 246
Query 100 LEKLFSIVQLCDERRYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNTSQVAHIL 159
E + I++ +R Y A+ A++ +AIGNA WP+GVTMVGIH R GR K+ + VAH+L
Sbjct 247 KESITDIIKFMLQREYVKANDAYLQMAIGNAPWPIGVTMVGIHARTGREKIFSKHVAHVL 306
Query 160 NDETTRKYIQMFKRLMSFAQRKFPAN 185
NDET RKYIQ KRLM+ Q+ FP +
Sbjct 307 NDETQRKYIQGLKRLMTICQKHFPTD 332
> ath:AT1G03140 splicing factor Prp18 family protein; K12817 pre-mRNA-splicing
factor 18
Length=420
Score = 147 bits (371), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 68/144 (47%), Positives = 96/144 (66%), Gaps = 0/144 (0%)
Query 41 VIAWARKMLSLWEDELKHRSEDEKATAEGRQQTALHRQTKKDLKPLFKKLKQRDLEADIL 100
++ + +K+L W+ EL E+ TA+G+Q A +Q + L PLF +++ L ADI
Sbjct 229 ILVFYKKLLIEWKQELDAMENTERRTAKGKQMVATFKQCARYLVPLFNLCRKKGLPADIR 288
Query 101 EKLFSIVQLCDERRYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNTSQVAHILN 160
+ L +V C +R Y A ++ LAIGNA WP+GVTMVGIHER+ R K+ T+ VAHI+N
Sbjct 289 QALMVMVNHCIKRDYLAAMDHYIKLAIGNAPWPIGVTMVGIHERSAREKIYTNSVAHIMN 348
Query 161 DETTRKYIQMFKRLMSFAQRKFPA 184
DETTRKY+Q KRLM+F QR++P
Sbjct 349 DETTRKYLQSVKRLMTFCQRRYPT 372
> dre:431751 prpf18, zgc:91830; PRP18 pre-mRNA processing factor
18 homolog (yeast); K12817 pre-mRNA-splicing factor 18
Length=342
Score = 146 bits (368), Expect = 5e-35, Method: Compositional matrix adjust.
Identities = 66/140 (47%), Positives = 96/140 (68%), Gaps = 0/140 (0%)
Query 46 RKMLSLWEDELKHRSEDEKATAEGRQQTALHRQTKKDLKPLFKKLKQRDLEADILEKLFS 105
R +L +W +L R + K + +G+ +A +QT+ L+PLF+KL++++L ADI E +
Sbjct 193 RFLLGVWAKDLNSREDHIKRSVQGKLASATQKQTESYLEPLFRKLRKKNLPADIKESITD 252
Query 106 IVQLCDERRYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNTSQVAHILNDETTR 165
I++ ER Y A+ A++ +AIGNA WP+GVTMVGIH R GR K+ + VAH+LNDET R
Sbjct 253 IIKFMLEREYVKANDAYLQMAIGNAPWPIGVTMVGIHARTGREKIFSKHVAHVLNDETQR 312
Query 166 KYIQMFKRLMSFAQRKFPAN 185
KYIQ KRLM+ Q+ FP +
Sbjct 313 KYIQGLKRLMTICQKHFPTD 332
> xla:734335 prpf18, MGC85069; PRP18 pre-mRNA processing factor
18 homolog; K12817 pre-mRNA-splicing factor 18
Length=342
Score = 142 bits (358), Expect = 6e-34, Method: Compositional matrix adjust.
Identities = 65/138 (47%), Positives = 93/138 (67%), Gaps = 0/138 (0%)
Query 48 MLSLWEDELKHRSEDEKATAEGRQQTALHRQTKKDLKPLFKKLKQRDLEADILEKLFSIV 107
+L +W EL R + K + G+ +A +QT+ LKPLF+KL++++L ADI E + I+
Sbjct 195 LLGVWAKELNAREDYVKRSVHGKLASATQKQTESYLKPLFRKLRKKNLPADIKESITDII 254
Query 108 QLCDERRYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNTSQVAHILNDETTRKY 167
+ +R Y A+ A++ +AIGNA WP+GVTMVGIH R GR K+ + VAH+LNDET RKY
Sbjct 255 KFMLQREYVKANDAYLQMAIGNAPWPIGVTMVGIHARTGREKIFSKHVAHVLNDETQRKY 314
Query 168 IQMFKRLMSFAQRKFPAN 185
IQ KRLM+ Q+ F +
Sbjct 315 IQGLKRLMTICQKYFSTD 332
> ath:AT1G54590 splicing factor Prp18 family protein
Length=256
Score = 140 bits (354), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 69/145 (47%), Positives = 95/145 (65%), Gaps = 1/145 (0%)
Query 41 VIAWARKMLSLWEDELKHRSEDEKATAEGRQQTALHRQTKKDLKPLFKKLKQRDLEADIL 100
++ + +K+L W+ EL+ E+ TA G+Q A Q + L PLF + + L ADI
Sbjct 64 ILVFCKKLLLEWKQELEAMENTERRTAIGKQMLATFNQCARYLTPLFHLCRNKCLPADIR 123
Query 101 EKLFSIVQLCDERRYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNT-SQVAHIL 159
+ L +V +R Y DA F+ LAIGNA WP+GVTMVGIHER+ R K++T S VAHI+
Sbjct 124 QGLMVMVNCWIKRDYLDATAQFIKLAIGNAPWPIGVTMVGIHERSAREKISTSSSVAHIM 183
Query 160 NDETTRKYIQMFKRLMSFAQRKFPA 184
N+ETTRKY+Q KRLM+F QR++ A
Sbjct 184 NNETTRKYLQSVKRLMTFCQRRYSA 208
> cel:F32B6.3 hypothetical protein; K12817 pre-mRNA-splicing factor
18
Length=352
Score = 130 bits (327), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 64/147 (43%), Positives = 91/147 (61%), Gaps = 1/147 (0%)
Query 40 VVIAWARKMLSLWEDELKHRSEDEKATAEGRQQTALHRQTKKDLKPLFKKLKQRDLEADI 99
++++ R +L+ W +L R D K TA+G + A H+QT LK L +++ + DI
Sbjct 194 IILSICRYILARWAKDLNDRPLDVKKTAQGMHEAAHHKQTMMHLKSLMTSMERYNCNNDI 253
Query 100 LEKLFSIVQL-CDERRYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNTSQVAHI 158
L I +L +R Y +A+ A+M +AIGNA WP+GVT GIH+R G +K S +AH+
Sbjct 254 RHHLAKICRLLVIDRNYLEANNAYMEMAIGNAPWPVGVTRSGIHQRPGSAKSYVSNIAHV 313
Query 159 LNDETTRKYIQMFKRLMSFAQRKFPAN 185
LNDET RKYIQ FKRLM+ Q FP +
Sbjct 314 LNDETQRKYIQAFKRLMTKMQEYFPTD 340
> sce:YGR006W PRP18; Prp18p; K12817 pre-mRNA-splicing factor 18
Length=251
Score = 68.2 bits (165), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 43/105 (40%), Positives = 63/105 (60%), Gaps = 2/105 (1%)
Query 75 LHRQTKKDLKPLFKKLKQRDLEADILEKLFSIV-QLCDERRYRDAHGAFMLLAIGNAAWP 133
L TKK L PL +L++ L D+L L +++ L + A ++M L+IGN AWP
Sbjct 135 LFLDTKKALFPLLLQLRRNQLAPDLLISLATVLYHLQQPKEINLAVQSYMKLSIGNVAWP 194
Query 134 MGVTMVGIHERAGRSKLNTSQ-VAHILNDETTRKYIQMFKRLMSF 177
+GVT VGIH R+ SK+ + A+I+ DE TR +I KRL++F
Sbjct 195 IGVTSVGIHARSAHSKIQGGRNAANIMIDERTRLWITSIKRLITF 239
> xla:100036833 mks1; Meckel syndrome, type 1
Length=561
Score = 35.8 bits (81), Expect = 0.086, Method: Composition-based stats.
Identities = 19/50 (38%), Positives = 32/50 (64%), Gaps = 1/50 (2%)
Query 7 DEDANKGKKETNSESTDAKQSQNEETTEKSKEAVVIAWARKMLSLWEDEL 56
+EDAN+G E + ++ ++S+ + E+ KE VVI+W K+ S +E EL
Sbjct 44 EEDANRGGLELQALHSNPEESRGHQPNEE-KEEVVISWQEKLFSQFEFEL 92
> tgo:TGME49_095700 UBA/TS-N domain-containing protein
Length=5096
Score = 33.5 bits (75), Expect = 0.42, Method: Composition-based stats.
Identities = 19/47 (40%), Positives = 28/47 (59%), Gaps = 3/47 (6%)
Query 7 DEDANKGKKETNSESTDAKQSQNEETTEKSKEAVVIAWARKMLSLWE 53
DE+ KG+K N+E AK S EE EK +EA + W R+++ +E
Sbjct 1866 DEEDTKGEKRGNAEEGQAKDS--EEDAEKRREA-FLCWMRELMKQFE 1909
> mmu:244653 Hydin, 1700034M11Rik, 4930545D19Rik, 4932703P14,
A830061H17, AC069308.21gm4, hy-3, hy3, hyrh; hydrocephalus inducing
Length=5154
Score = 32.7 bits (73), Expect = 0.74, Method: Composition-based stats.
Identities = 19/75 (25%), Positives = 43/75 (57%), Gaps = 1/75 (1%)
Query 25 KQSQNEETTEKSKEAVVIAWARKMLSLWEDELKHRSEDEKATAEGRQQTALHRQTKKDLK 84
++++ E+ K KEA+ R + +L EDE + +EK + + AL + K++L+
Sbjct 2315 EKAKKEQEENKRKEALAKEKER-LQTLDEDEYDALTAEEKVAFDRDVRQALRERKKRELE 2373
Query 85 PLFKKLKQRDLEADI 99
L K+++++ L+ ++
Sbjct 2374 RLAKEMQEKKLQQEL 2388
> cpv:cgd4_2900 polyketide synthase
Length=13413
Score = 31.2 bits (69), Expect = 1.9, Method: Composition-based stats.
Identities = 30/140 (21%), Positives = 61/140 (43%), Gaps = 6/140 (4%)
Query 48 MLSLWEDELKHRSEDEKATAE---GRQQTALHRQTKKDLKPLFKKLKQRDLEADILEKLF 104
+L + E+ + H + + K G ++A + K L+ L +DL D L K+F
Sbjct 10116 ILFITENTIPHNNNNHKFIGSDIWGLVRSARYELDKYSLRLLDLDCFTKDLSEDNLSKIF 10175
Query 105 SIVQLCDERRYRDAHGAFMLLAIGNAAWPM-GVTMVGIHERAGRSKLNTSQVAHI--LND 161
S + +E + +G + + + P+ G + + ER + L T +I +N
Sbjct 10176 SCILSSNESEFIFRNGELLFPRLSKISIPINGPLNIRLAERGALTNLLTEMQTNIPYINS 10235
Query 162 ETTRKYIQMFKRLMSFAQRK 181
+ I F +L ++++ K
Sbjct 10236 DKKLDLIDCFDKLENYSEMK 10255
> ath:AT1G14610 TWN2; TWN2 (TWIN 2); ATP binding / aminoacyl-tRNA
ligase/ nucleotide binding / valine-tRNA ligase (EC:6.1.1.9);
K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=1108
Score = 31.2 bits (69), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 25/44 (56%), Gaps = 4/44 (9%)
Query 116 RDAHGAFMLLAIGNAAWPM----GVTMVGIHERAGRSKLNTSQV 155
RDAHG M ++GN P+ GVT+ G+H+R L+ +V
Sbjct 689 RDAHGRKMSKSLGNVIDPLEVINGVTLEGLHKRLEEGNLDPKEV 732
Lambda K H
0.313 0.126 0.352
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5106150148
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40