bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0471_orf1
Length=198
Score E
Sequences producing significant alignments: (Bits) Value
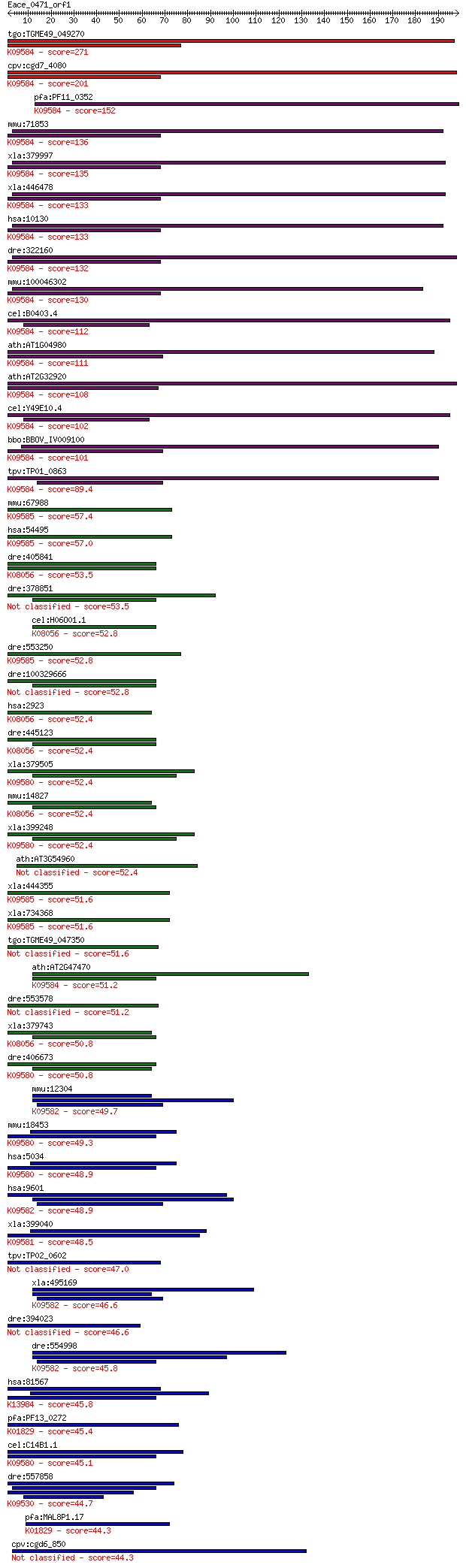
tgo:TGME49_049270 thioredoxin, putative (EC:5.3.4.1); K09584 p... 271 9e-73
cpv:cgd7_4080 protein disulfide isomerase, signal peptide, ER ... 201 9e-52
pfa:PF11_0352 PDI-11; protein disulfide isomerase; K09584 prot... 152 7e-37
mmu:71853 Pdia6, 1700015E05Rik, AL023058, C77895, CaBP5, P5, T... 136 6e-32
xla:379997 pdia6-b, MGC52744, erp5, pdip5, txndc7; protein dis... 135 1e-31
xla:446478 pdia6-a, MGC79068, erp5, pdia6, pdip5, txndc7; prot... 133 3e-31
hsa:10130 PDIA6, ERP5, P5, TXNDC7; protein disulfide isomerase... 133 4e-31
dre:322160 pdip5, pdi-p5, wu:fb51h08, wu:fc09e09; protein disu... 132 9e-31
mmu:100046302 protein disulfide-isomerase A6-like; K09584 prot... 130 2e-30
cel:B0403.4 tag-320; Temporarily Assigned Gene name family mem... 112 7e-25
ath:AT1G04980 ATPDIL2-2 (PDI-LIKE 2-2); protein disulfide isom... 111 1e-24
ath:AT2G32920 ATPDIL2-3 (PDI-LIKE 2-3); protein disulfide isom... 108 9e-24
cel:Y49E10.4 hypothetical protein; K09584 protein disulfide-is... 102 8e-22
bbo:BBOV_IV009100 23.m05770; protein disulfide isomerase relat... 101 2e-21
tpv:TP01_0863 protein disulfide isomerase; K09584 protein disu... 89.4 8e-18
mmu:67988 Tmx3, 6430411B10Rik, A730024F05Rik, AV259382, Txndc1... 57.4 3e-08
hsa:54495 TMX3, FLJ20793, KIAA1830, PDIA13, TXNDC10; thioredox... 57.0 4e-08
dre:405841 MGC77086; zgc:77086; K08056 protein disulfide isome... 53.5 5e-07
dre:378851 pdia3, Grp58, sb:cb825; protein disulfide isomerase... 53.5 5e-07
cel:H06O01.1 pdi-3; Protein Disulfide Isomerase family member ... 52.8 7e-07
dre:553250 MGC152808; zgc:152808 (EC:5.3.4.1); K09585 thioredo... 52.8 7e-07
dre:100329666 protein disulfide-isomerase A3-like 52.8 9e-07
hsa:2923 PDIA3, ER60, ERp57, ERp60, ERp61, GRP57, GRP58, HsT17... 52.4 9e-07
dre:445123 zgc:100906 (EC:5.3.4.1); K08056 protein disulfide i... 52.4 9e-07
xla:379505 hypothetical protein MGC64309; K09580 protein disul... 52.4 1e-06
mmu:14827 Pdia3, 58kDa, ERp57, ERp60, ERp61, Erp, Grp58, PDI, ... 52.4 1e-06
xla:399248 p4hb; prolyl 4-hydroxylase, beta polypeptide (EC:5.... 52.4 1e-06
ath:AT3G54960 ATPDIL1-3 (PDI-LIKE 1-3); protein disulfide isom... 52.4 1e-06
xla:444355 tmx3-a, pdia13, tmx3, txndc10, txndc10a; thioredoxi... 51.6 2e-06
xla:734368 tmx3-b, pdia13, txndc10, txndc10b; thioredoxin-rela... 51.6 2e-06
tgo:TGME49_047350 thioredoxin, putative (EC:5.3.4.1) 51.6 2e-06
ath:AT2G47470 UNE5; UNE5 (UNFERTILIZED EMBRYO SAC 5); protein ... 51.2 2e-06
dre:553578 tmx3, MGC110025, txndc10, zgc:110025; thioredoxin-r... 51.2 2e-06
xla:379743 pdia3, MGC53247, erp57, grp58; protein disulfide is... 50.8 3e-06
dre:406673 p4hb, psmb3, zgc:92596; procollagen-proline, 2-oxog... 50.8 3e-06
mmu:12304 Pdia4, AI987846, Cai, ERp-72, Erp72; protein disulfi... 49.7 7e-06
mmu:18453 P4hb, ERp59, PDI, Pdia1, Thbp; prolyl 4-hydroxylase,... 49.3 9e-06
hsa:5034 P4HB, DSI, ERBA2L, GIT, P4Hbeta, PDI, PDIA1, PHDB, PO... 48.9 1e-05
hsa:9601 PDIA4, ERP70, ERP72, ERp-72; protein disulfide isomer... 48.9 1e-05
xla:399040 pdia2, XPDIp, pdi; protein disulfide isomerase fami... 48.5 1e-05
tpv:TP02_0602 protein disulfide isomerase 47.0 5e-05
xla:495169 pdia4; protein disulfide isomerase family A, member... 46.6 5e-05
dre:394023 pdia2, MGC55398, P4hb, zgc:55398; protein disulfide... 46.6 5e-05
dre:554998 pdia4, MGC113965, MGC136625, cai, wu:fd20c07, zgc:1... 45.8 8e-05
hsa:81567 TXNDC5, ENDOPDI, ERP46, HCC-2, MGC3178, PDIA15, STRF... 45.8 9e-05
pfa:PF13_0272 thioredoxin-related protein, putative (EC:5.3.4.... 45.4 1e-04
cel:C14B1.1 pdi-1; Protein Disulfide Isomerase family member (... 45.1 2e-04
dre:557858 dnajc10, MGC162218, zgc:162218; DnaJ (Hsp40) homolo... 44.7 2e-04
pfa:MAL8P1.17 PfPDI-8; protein disulfide isomerase (EC:5.3.4.1... 44.3 3e-04
cpv:cgd6_850 thioredoxin; protein disulfide isomerase A6, sign... 44.3 3e-04
> tgo:TGME49_049270 thioredoxin, putative (EC:5.3.4.1); K09584
protein disulfide-isomerase A6 [EC:5.3.4.1]
Length=428
Score = 271 bits (694), Expect = 9e-73, Method: Compositional matrix adjust.
Identities = 121/196 (61%), Positives = 158/196 (80%), Gaps = 0/196 (0%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTAD 60
W+EV+ LKGKVKV KVD+T E++LAS +GI+ FPTLKLFPAG KS +VKDYEG RT +
Sbjct 210 WEEVATALKGKVKVGKVDATVEKVLASTYGIRGFPTLKLFPAGEKSVGLVKDYEGARTTE 269
Query 61 EIVKFAMEFYGAKAEADQLLNEEQFRSECGETLCILALLPDVLDSGEEGRKNYLQTLNQV 120
++K+AMEF+ +QLLNE QFR CG+ LC+LA LP +LDS E R YL TLN+V
Sbjct 270 ALLKYAMEFFSVNVTTEQLLNESQFRKACGDQLCVLAFLPHILDSKTEKRNEYLATLNRV 329
Query 121 VKASVTVPVKFLWLQGGDNFDLEEQLHLAFGWPAVVAVHLARGKFSVHRGNFTRESINSF 180
V+AS +P+ F W QGGD ++ EEQL+LAFG+PAVVAVHL++GK+++HRG+F++ESIN+F
Sbjct 330 VRASFHMPIAFFWSQGGDQYEFEEQLNLAFGYPAVVAVHLSKGKYAIHRGDFSQESINTF 389
Query 181 LQQLLAGRAPVSDLPK 196
L QLLAG+AP+S+LPK
Sbjct 390 LTQLLAGKAPISELPK 405
Score = 33.9 bits (76), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 22/76 (28%), Positives = 37/76 (48%), Gaps = 3/76 (3%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTAD 60
+++ + L+G V + V S+Q ++G+Q FPT+K F DY R A
Sbjct 71 FEKAAKALRGIVTLVAV---SDQSAMGEYGVQGFPTVKAFVGRGGKPPKTFDYNQGRDAA 127
Query 61 EIVKFAMEFYGAKAEA 76
+++FA+ G A A
Sbjct 128 SLIEFAVMHAGKLARA 143
> cpv:cgd7_4080 protein disulfide isomerase, signal peptide, ER
retention motif ; K09584 protein disulfide-isomerase A6 [EC:5.3.4.1]
Length=451
Score = 201 bits (512), Expect = 9e-52, Method: Compositional matrix adjust.
Identities = 90/197 (45%), Positives = 134/197 (68%), Gaps = 0/197 (0%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTAD 60
W+E+ G+VK+AK+D+T M+A ++ IQ FPTL +FPAG K +Y G RTA+
Sbjct 221 WEELGSMADGRVKIAKLDATQHTMMAHRYKIQGFPTLLMFPAGEKREITPVNYNGPRTAN 280
Query 61 EIVKFAMEFYGAKAEADQLLNEEQFRSECGETLCILALLPDVLDSGEEGRKNYLQTLNQV 120
++ +FA++F + A Q++++E F + C + LC++A LP + DS + R+ YL+ V
Sbjct 281 DLFEFAIKFQSSSASIKQMISQEVFENTCTKGLCVIAFLPHIADSSDSEREKYLKIYKDV 340
Query 121 VKASVTVPVKFLWLQGGDNFDLEEQLHLAFGWPAVVAVHLARGKFSVHRGNFTRESINSF 180
V AS + ++FLW +GG FD EE+L+LAFG+PAVVA++ + +FS HRG+FT ES+NSF
Sbjct 341 VSASAAMTIRFLWSEGGSQFDFEEKLNLAFGYPAVVAINNEKQRFSTHRGSFTVESLNSF 400
Query 181 LQQLLAGRAPVSDLPKL 197
+ L GRAPV LPKL
Sbjct 401 IIALTTGRAPVDPLPKL 417
Score = 38.1 bits (87), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 39/67 (58%), Gaps = 6/67 (8%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTAD 60
+++ + LKG V V +D S+ +++GIQ FPT+K+F + + KD+ G R A+
Sbjct 86 YEKAAKALKGIVPVVAIDDQSDM---AEYGIQGFPTVKVF---TEHSVKPKDFTGPRRAE 139
Query 61 EIVKFAM 67
++ A+
Sbjct 140 SVLNAAL 146
> pfa:PF11_0352 PDI-11; protein disulfide isomerase; K09584 protein
disulfide-isomerase A6 [EC:5.3.4.1]
Length=423
Score = 152 bits (384), Expect = 7e-37, Method: Compositional matrix adjust.
Identities = 68/186 (36%), Positives = 115/186 (61%), Gaps = 0/186 (0%)
Query 13 KVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIVKFAMEFYGA 72
K+AK+D+T EQ A + I +P+ +LFP+G K DY RT D++ F +++Y
Sbjct 219 KIAKIDATVEQRTAQTYQINHYPSFRLFPSGNKKPHTAIDYNDSRTVDDLYHFFLKYYVE 278
Query 73 KAEADQLLNEEQFRSECGETLCILALLPDVLDSGEEGRKNYLQTLNQVVKASVTVPVKFL 132
K E QL +++ F C + +C+LALLP+ D+ K+Y+ L+ V+K +PV L
Sbjct 279 KKELIQLTSQQVFDEFCEKDVCLLALLPNKEDTEPSYFKSYVHILSNVIKDVNHLPVTLL 338
Query 133 WLQGGDNFDLEEQLHLAFGWPAVVAVHLARGKFSVHRGNFTRESINSFLQQLLAGRAPVS 192
W Q GD D+ ++L+L FG+P V+A+ L++ FS+ +GN++ +SI +F+ Q++ G++ V
Sbjct 339 WTQAGDQLDIVQKLNLTFGFPTVIALSLSKNVFSILKGNYSEQSIKNFITQMMMGKSSVD 398
Query 193 DLPKLK 198
+L K
Sbjct 399 NLVPFK 404
> mmu:71853 Pdia6, 1700015E05Rik, AL023058, C77895, CaBP5, P5,
Txndc7; protein disulfide isomerase associated 6 (EC:5.3.4.1);
K09584 protein disulfide-isomerase A6 [EC:5.3.4.1]
Length=445
Score = 136 bits (342), Expect = 6e-32, Method: Compositional matrix adjust.
Identities = 77/197 (39%), Positives = 120/197 (60%), Gaps = 12/197 (6%)
Query 3 EVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEI 62
EV Q KGKVK+A VD+T Q+LAS++GI+ FPT+K+F G DY+G RT +I
Sbjct 211 EVKEQTKGKVKLAAVDATMNQVLASRYGIKGFPTIKIFQKGESPV----DYDGGRTRSDI 266
Query 63 VKFAMEFYGAKAEADQLL---NEEQFRSECGE-TLCILALLPDVLDSGEEGRKNYLQTLN 118
V A++ + A +LL NE+ + C E LC++A+LP +LD+G GR +YL+ L
Sbjct 267 VSRALDLFSDNAPPPELLEIINEDIAKKTCEEHQLCVVAVLPHILDTGAAGRNSYLEVLL 326
Query 119 QVVKASVTVPVKFLWLQGGDNFDLEEQLHL-AFGWPAVVAVHLARGKFSVHRGNFTRESI 177
++ +LW + G ++LE L + FG+PA+ A++ + KF++ +G+F+ + I
Sbjct 327 KLADKYKKKMWGWLWTEAGAQYELENALGIGGFGYPAMAAINARKMKFALLKGSFSEQGI 386
Query 178 NSFLQQLLAGR---APV 191
N FL++L GR APV
Sbjct 387 NEFLRELSFGRGSTAPV 403
Score = 50.8 bits (120), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 25/67 (37%), Positives = 38/67 (56%), Gaps = 3/67 (4%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTAD 60
W + + LK VKV V++ Q L ++G+Q FPT+K+F A +DY+G RT +
Sbjct 70 WKKAATALKDVVKVGAVNADKHQSLGGQYGVQGFPTIKIFGANKNKP---EDYQGGRTGE 126
Query 61 EIVKFAM 67
IV A+
Sbjct 127 AIVDAAL 133
> xla:379997 pdia6-b, MGC52744, erp5, pdip5, txndc7; protein disulfide
isomerase family A, member 6 (EC:5.3.4.1); K09584 protein
disulfide-isomerase A6 [EC:5.3.4.1]
Length=442
Score = 135 bits (339), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 73/195 (37%), Positives = 118/195 (60%), Gaps = 9/195 (4%)
Query 3 EVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEI 62
EV + GKVK+A VD+T Q+LAS++GI+ FPT+K+F G + DY+G RT +I
Sbjct 208 EVKEKTNGKVKLAAVDATVSQVLASRYGIRGFPTIKIFQKGEEPV----DYDGGRTKPDI 263
Query 63 VKFAMEFYGAKA---EADQLLNEEQFRSECGE-TLCILALLPDVLDSGEEGRKNYLQTLN 118
V A++ + A E +++LN + + C E LCI+A+LP +LD+G GR +YL+ +
Sbjct 264 VARAVDLFSENAPPPEINEILNGDIVKKTCEEHQLCIVAVLPHILDTGAAGRNSYLEVML 323
Query 119 QVVKASVTVPVKFLWLQGGDNFDLEEQLHL-AFGWPAVVAVHLARGKFSVHRGNFTRESI 177
++ +LW + G DLE L + FG+PA+ A++ + KF++ +G+F+ + I
Sbjct 324 KMADKYKKKMWGWLWAEAGTQMDLETSLGIGGFGYPAMAAINARKMKFALLKGSFSEQGI 383
Query 178 NSFLQQLLAGRAPVS 192
N FL++L GR S
Sbjct 384 NEFLRELSYGRGSTS 398
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 26/67 (38%), Positives = 39/67 (58%), Gaps = 3/67 (4%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTAD 60
W + + LKG VKV V++ Q L ++G++ FPT+K+F A DY+G RTAD
Sbjct 65 WKKAATALKGVVKVGAVNADQHQSLGGQYGVRGFPTIKIFGANKNKP---DDYQGGRTAD 121
Query 61 EIVKFAM 67
I+ A+
Sbjct 122 AIIDAAL 128
> xla:446478 pdia6-a, MGC79068, erp5, pdia6, pdip5, txndc7; protein
disulfide isomerase family A, member 6 (EC:5.3.4.1); K09584
protein disulfide-isomerase A6 [EC:5.3.4.1]
Length=442
Score = 133 bits (335), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 72/195 (36%), Positives = 118/195 (60%), Gaps = 9/195 (4%)
Query 3 EVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEI 62
EV + GKVK+A VD+T Q+LAS++GI+ FPT+K+F G + DY+G R +I
Sbjct 208 EVKEKTNGKVKLAAVDATVSQVLASRYGIRGFPTIKIFQKGEEPV----DYDGGRNRADI 263
Query 63 VKFAMEFYGAKA---EADQLLNEEQFRSECGE-TLCILALLPDVLDSGEEGRKNYLQTLN 118
V A++ + A E +++LN + + C E LCI+A+LP +LD+G GR +YL+ +
Sbjct 264 VARALDLFSENAPPPEINEILNGDIVKKTCDEHQLCIVAVLPHILDTGAAGRNSYLEVML 323
Query 119 QVVKASVTVPVKFLWLQGGDNFDLEEQLHL-AFGWPAVVAVHLARGKFSVHRGNFTRESI 177
++ + +LW + G DLE L + FG+PA+ A++ + KF++ +G+F+ + I
Sbjct 324 KMAEKYKKKMWGWLWTEAGAQMDLETSLGIGGFGYPAMAAINARKIKFALLKGSFSEQGI 383
Query 178 NSFLQQLLAGRAPVS 192
N FL++L GR S
Sbjct 384 NEFLRELSYGRGSTS 398
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 27/67 (40%), Positives = 39/67 (58%), Gaps = 3/67 (4%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTAD 60
W + + LKG VKV V++ Q L ++G++ FPT+K+F A DY+G RTAD
Sbjct 65 WKKAATALKGVVKVGAVNADQHQSLGGQYGVRGFPTIKVFGANKNKP---DDYQGGRTAD 121
Query 61 EIVKFAM 67
IV A+
Sbjct 122 AIVDAAL 128
> hsa:10130 PDIA6, ERP5, P5, TXNDC7; protein disulfide isomerase
family A, member 6 (EC:5.3.4.1); K09584 protein disulfide-isomerase
A6 [EC:5.3.4.1]
Length=440
Score = 133 bits (334), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 77/197 (39%), Positives = 119/197 (60%), Gaps = 12/197 (6%)
Query 3 EVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEI 62
EV Q KGKVK+A VD+T Q+LAS++GI+ FPT+K+F G DY+G RT +I
Sbjct 206 EVKEQTKGKVKLAAVDATVNQVLASRYGIRGFPTIKIFQKGESPV----DYDGGRTRSDI 261
Query 63 VKFAMEFYGAKAEADQLL---NEEQFRSECGE-TLCILALLPDVLDSGEEGRKNYLQTLN 118
V A++ + A +LL NE+ + C E LC++A+LP +LD+G GR +YL+ L
Sbjct 262 VSRALDLFSDNAPPPELLEIINEDIAKRTCEEHQLCVVAVLPHILDTGAAGRNSYLEVLL 321
Query 119 QVVKASVTVPVKFLWLQGGDNFDLEEQLHL-AFGWPAVVAVHLARGKFSVHRGNFTRESI 177
++ +LW + G +LE L + FG+PA+ A++ + KF++ +G+F+ + I
Sbjct 322 KLADKYKKKMWGWLWTEAGAQSELETALGIGGFGYPAMAAINARKMKFALLKGSFSEQGI 381
Query 178 NSFLQQLLAGR---APV 191
N FL++L GR APV
Sbjct 382 NEFLRELSFGRGSTAPV 398
Score = 50.1 bits (118), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 24/67 (35%), Positives = 37/67 (55%), Gaps = 3/67 (4%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTAD 60
W + + LK VKV VD+ L ++G+Q FPT+K+F + +DY+G RT +
Sbjct 65 WKKAATALKDVVKVGAVDADKHHSLGGQYGVQGFPTIKIFGSNKNRP---EDYQGGRTGE 121
Query 61 EIVKFAM 67
IV A+
Sbjct 122 AIVDAAL 128
> dre:322160 pdip5, pdi-p5, wu:fb51h08, wu:fc09e09; protein disulfide
isomerase-related protein (provisional) (EC:5.3.4.1);
K09584 protein disulfide-isomerase A6 [EC:5.3.4.1]
Length=440
Score = 132 bits (332), Expect = 9e-31, Method: Compositional matrix adjust.
Identities = 76/205 (37%), Positives = 119/205 (58%), Gaps = 14/205 (6%)
Query 3 EVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEI 62
EV Q KGKV++A D+T Q LAS+FGI+ FPT+K+F G + +DY+G RT +I
Sbjct 206 EVKEQTKGKVRLAAEDATVHQGLASRFGIRGFPTIKVFRKGEEP----EDYQGGRTRSDI 261
Query 63 VKFAMEFYGAKAEADQL---LNEEQFRSECGE-TLCILALLPDVLDSGEEGRKNYLQTLN 118
V A+E Y A +L LNE + C + LCI+A+LP +LD+G GR +YL+ +
Sbjct 262 VARALELYSDNIPAPELQEVLNEGILKKTCEDYQLCIIAVLPHILDTGASGRNSYLEVMK 321
Query 119 QVVKASVTVPVKFLWLQGGDNFDLEEQLHL-AFGWPAVVAVHLARGKFSVHRGNFTRESI 177
+ + +LW + G +LE L + FG+PA+ A++ + KF++ +G+F+ I
Sbjct 322 TMAEKHKKKMWGWLWTEAGAQMELEASLGIGGFGYPAMAAINSRKMKFALLKGSFSETGI 381
Query 178 NSFLQQLLAGRAPVSD-----LPKL 197
+ FL++L GR + LPK+
Sbjct 382 HEFLRELSVGRGSTATVGGGALPKI 406
Score = 50.1 bits (118), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 26/67 (38%), Positives = 38/67 (56%), Gaps = 3/67 (4%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTAD 60
W + + LKG VKV VD+ L ++G++ FPT+K+F G K +DY+G RT
Sbjct 65 WKKAATALKGIVKVGAVDADQHNSLGGQYGVRGFPTIKIF-GGNKHKP--EDYQGGRTNQ 121
Query 61 EIVKFAM 67
IV A+
Sbjct 122 AIVDAAL 128
> mmu:100046302 protein disulfide-isomerase A6-like; K09584 protein
disulfide-isomerase A6 [EC:5.3.4.1]
Length=391
Score = 130 bits (328), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 71/185 (38%), Positives = 113/185 (61%), Gaps = 9/185 (4%)
Query 3 EVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEI 62
EV Q KGKVK+A VD+T Q+LAS++GI+ FPT+K+F G DY+G RT +I
Sbjct 211 EVKEQTKGKVKLAAVDATVNQVLASRYGIKGFPTIKIFQKGESPV----DYDGGRTRSDI 266
Query 63 VKFAMEFYGAKAEADQLL---NEEQFRSECGE-TLCILALLPDVLDSGEEGRKNYLQTLN 118
V A++ + A +LL NE+ + C E LC++A+LP +LD+G GR +YL+ L
Sbjct 267 VSRALDLFSDNAPPPELLEIINEDIAKKTCEEHQLCVVAVLPHILDTGAAGRNSYLEVLL 326
Query 119 QVVKASVTVPVKFLWLQGGDNFDLEEQLHL-AFGWPAVVAVHLARGKFSVHRGNFTRESI 177
++ +LW + G ++LE L + FG+PA+ A++ + KF++ +G+F+ + I
Sbjct 327 KLADKYKKKMWGWLWTEAGAQYELENALGIGGFGYPAMAAINARKMKFALLKGSFSEQGI 386
Query 178 NSFLQ 182
N FL+
Sbjct 387 NEFLR 391
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 25/67 (37%), Positives = 38/67 (56%), Gaps = 3/67 (4%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTAD 60
W + + LK VKV V++ Q L ++G+Q FPT+K+F A +DY+G RT +
Sbjct 70 WKKAATALKDVVKVGAVNADKHQSLGGQYGVQGFPTIKIFGANKNKP---EDYQGGRTGE 126
Query 61 EIVKFAM 67
IV A+
Sbjct 127 AIVDAAL 133
> cel:B0403.4 tag-320; Temporarily Assigned Gene name family member
(tag-320); K09584 protein disulfide-isomerase A6 [EC:5.3.4.1]
Length=440
Score = 112 bits (281), Expect = 7e-25, Method: Compositional matrix adjust.
Identities = 64/199 (32%), Positives = 108/199 (54%), Gaps = 6/199 (3%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTAD 60
W + +LKGKV++ +D+T ++A+KF I+ FPT+K F G + +DY+G R +
Sbjct 204 WKAAASELKGKVRLGALDATVHTVVANKFAIRGFPTIKYFAPG-SDVSDAQDYDGGRQSS 262
Query 61 EIVKFAMEFYGAKAEADQL---LNEEQFRSECGET-LCILALLPDVLDSGEEGRKNYLQT 116
+IV +A A ++ +N++ C E LCI A LP +LD E R NYL
Sbjct 263 DIVAWASARAQENMPAPEVFEGINQQVVEDACKEKQLCIFAFLPHILDCQSECRNNYLAM 322
Query 117 LNQVVKASVTVPVKFLWLQGGDNFDLEEQLHL-AFGWPAVVAVHLARGKFSVHRGNFTRE 175
L + + ++W++G LEE + FG+PA+ A++ + K++V +G+F ++
Sbjct 323 LKEQSEKFKKNLWGWIWVEGAAQPALEESFEVGGFGYPAMTALNFRKNKYAVLKGSFGKD 382
Query 176 SINSFLQQLLAGRAPVSDL 194
I+ FL+ L G+ S L
Sbjct 383 GIHEFLRDLSYGKGRTSSL 401
Score = 48.5 bits (114), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 26/55 (47%), Positives = 30/55 (54%), Gaps = 3/55 (5%)
Query 8 LKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEI 62
LKG KV VD T Q + + +Q FPTLK+F A K DY GQRTA I
Sbjct 71 LKGVAKVGAVDMTQHQSVGGPYNVQGFPTLKIFGADKKKPT---DYNGQRTAQAI 122
> ath:AT1G04980 ATPDIL2-2 (PDI-LIKE 2-2); protein disulfide isomerase;
K09584 protein disulfide-isomerase A6 [EC:5.3.4.1]
Length=447
Score = 111 bits (278), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 63/192 (32%), Positives = 105/192 (54%), Gaps = 8/192 (4%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTAD 60
W + + LKGKVK+ V+ +EQ + S+F +Q FPT+ +F G ++ V YEG R+A
Sbjct 207 WKKAANNLKGKVKLGHVNCDAEQSIKSRFKVQGFPTILVF--GSDKSSPVP-YEGARSAS 263
Query 61 EIVKFA---MEFYGAKAEADQLLNEEQFRSECGE-TLCILALLPDVLDSGEEGRKNYLQT 116
I FA +E AE +L + +CG +C ++ LPD+LDS EGR YL+
Sbjct 264 AIESFALEQLESNAGPAEVTELTGPDVMEDKCGSAAICFVSFLPDILDSKAEGRNKYLEM 323
Query 117 LNQVVKASVTVPVKFLWLQGGDNFDLEEQLHL-AFGWPAVVAVHLARGKFSVHRGNFTRE 175
L V P F+W+ G DLE+++ + +G+PA+VA++ +G ++ + F +
Sbjct 324 LLSVADKFKKDPYGFVWVAAGKQPDLEKRVGVGGYGYPAMVALNAKKGAYAPLKSGFEVK 383
Query 176 SINSFLQQLLAG 187
+ F+++ G
Sbjct 384 HLKDFVKEAAKG 395
Score = 49.7 bits (117), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 41/68 (60%), Gaps = 4/68 (5%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTAD 60
W++V+ LKG VA +D+ + + ++ +G++ FPT+K+F G DY+G R A
Sbjct 72 WEKVASTLKGIATVAAIDADAHKSVSQDYGVRGFPTIKVFVPGKPPI----DYQGARDAK 127
Query 61 EIVKFAME 68
I +FA++
Sbjct 128 SISQFAIK 135
> ath:AT2G32920 ATPDIL2-3 (PDI-LIKE 2-3); protein disulfide isomerase;
K09584 protein disulfide-isomerase A6 [EC:5.3.4.1]
Length=440
Score = 108 bits (271), Expect = 9e-24, Method: Compositional matrix adjust.
Identities = 63/205 (30%), Positives = 110/205 (53%), Gaps = 11/205 (5%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTAD 60
W + L+GKVK+ V+ EQ + S+F +Q FPT+ +F GP ++ YEG R+A
Sbjct 202 WKRAAKNLQGKVKLGHVNCDVEQSIMSRFKVQGFPTILVF--GPDKSSPYP-YEGARSAS 258
Query 61 EIVKFAMEFYGAKA---EADQLLNEEQFRSECGET-LCILALLPDVLDSGEEGRKNYLQT 116
I FA E + A E +L + +CG +C ++ LPD+LDS EGR YL+
Sbjct 259 AIESFASELVESSAGPVEVTELTGPDVMEKKCGSAAICFISFLPDILDSKAEGRNKYLEM 318
Query 117 LNQVVKASVTVPVKFLWLQGGDNFDLEEQLHL-AFGWPAVVAVHLARGKFSVHRGNFTRE 175
L V + P F+W+ DLE+++++ +G+PA+VA+++ +G ++ + F +
Sbjct 319 LLSVAEKFKKQPYSFMWVAAVTQMDLEKRVNVGGYGYPAMVAMNVKKGVYAPLKSAFELQ 378
Query 176 SINSFLQQLLA---GRAPVSDLPKL 197
+ F++ G P++ P++
Sbjct 379 HLLEFVKDAGTGGKGNVPMNGTPEI 403
Score = 50.4 bits (119), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 26/66 (39%), Positives = 37/66 (56%), Gaps = 4/66 (6%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTAD 60
W++V+ LKG VA +D+ + Q A +GI+ FPT+K+F G DY+G R A
Sbjct 70 WEKVANILKGVATVAAIDADAHQSAAQDYGIKGFPTIKVFVPGKAPI----DYQGARDAK 125
Query 61 EIVKFA 66
I FA
Sbjct 126 SIANFA 131
> cel:Y49E10.4 hypothetical protein; K09584 protein disulfide-isomerase
A6 [EC:5.3.4.1]
Length=436
Score = 102 bits (254), Expect = 8e-22, Method: Compositional matrix adjust.
Identities = 53/199 (26%), Positives = 103/199 (51%), Gaps = 5/199 (2%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTAD 60
W + + ++ G+VK +D+T+ + +A KFGI+ FPT+K F G S + +DY+G RT+
Sbjct 194 WKKAAEEMGGRVKFGALDATAHESIAQKFGIRGFPTIKFFAPGTSSASDAEDYQGGRTST 253
Query 61 EIVKFAMEFY---GAKAEADQLLNEEQFRSECGET-LCILALLPDVLDSGEEGRKNYLQT 116
+++ +A Y GA E + + + C + LCI LP + D + RK +
Sbjct 254 DLISYAESKYDDFGAAPEVVEGTGKAVVETVCKDKQLCIFTFLPSIFDCQSKCRKQKIDM 313
Query 117 LNQVVKASVTVPVKFLWLQGGDNFDLEEQLHLA-FGWPAVVAVHLARGKFSVHRGNFTRE 175
LN++ ++W++GG +++ + +G+P ++A+ + +S G F+ +
Sbjct 314 LNELATIFKKRSFGWVWMEGGAQENVQRAFEIGDYGFPVLIAMSPKKMMYSTQIGQFSVD 373
Query 176 SINSFLQQLLAGRAPVSDL 194
I FL + G+ V ++
Sbjct 374 GIKEFLNAVNYGKGRVLEI 392
Score = 43.9 bits (102), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 22/55 (40%), Positives = 33/55 (60%), Gaps = 2/55 (3%)
Query 8 LKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEI 62
LKG ++ +D+T Q + K+ I+ +PT+K+F A KS + DY G RTA I
Sbjct 71 LKGIAEIGAIDATVHQKIPLKYSIKGYPTIKIFGATEKSKPI--DYNGPRTAKGI 123
> bbo:BBOV_IV009100 23.m05770; protein disulfide isomerase related
protein (EC:5.3.4.1); K09584 protein disulfide-isomerase
A6 [EC:5.3.4.1]
Length=395
Score = 101 bits (251), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 54/184 (29%), Positives = 101/184 (54%), Gaps = 8/184 (4%)
Query 7 QLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIVKFA 66
Q GKVKV +D+T LA+++G++ FPT+ LFP G KS Y+G R A++I++FA
Sbjct 200 QSSGKVKVGSIDATVYTALAARYGVKGFPTIFLFPQGVKSPTTAIRYKGPRKAEDILQFA 259
Query 67 MEFYGAKAEADQLLNEEQFRSECGETLCILALLPDVLDSGEEGRKNYLQTLNQVVKASVT 126
+Y ++ + + C LC+L +P E +L T++ V++ +
Sbjct 260 KSYYRNMGPPVKVDSVSDLKQRCSRPLCLLFFIP------ETSMDEHLSTISLVMEKHSS 313
Query 127 VPVKFLWLQGGDNFDLEEQLHLAFGWPAVVAVHLARGKFSVHRG-NFTRESINSFLQQLL 185
+P +F + G + E L + + PAV A++L++ +SV R T +++ +F+ ++L
Sbjct 314 LPFEFCYTTAGRHLQWERVLGV-YSTPAVFALNLSKNVYSVMRKEKLTFDNVKTFVSEIL 372
Query 186 AGRA 189
+G++
Sbjct 373 SGQS 376
Score = 39.7 bits (91), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 38/69 (55%), Gaps = 4/69 (5%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPA-GPKSTAMVKDYEGQRTA 59
++ V+ +K V V V +S + ++FGI +FP+ K+F GP + V DY G+
Sbjct 65 YETVATCMKDVVNVYAVKDSS---VMARFGISSFPSFKVFLGRGPSAKPDVVDYNGKLAV 121
Query 60 DEIVKFAME 68
++V F M+
Sbjct 122 PDLVTFTMK 130
> tpv:TP01_0863 protein disulfide isomerase; K09584 protein disulfide-isomerase
A6 [EC:5.3.4.1]
Length=387
Score = 89.4 bits (220), Expect = 8e-18, Method: Compositional matrix adjust.
Identities = 59/193 (30%), Positives = 98/193 (50%), Gaps = 17/193 (8%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTAD 60
W + + KG VKV +VD TS Q L ++F ++ +PT+ LF G K+ +YEGQRTA
Sbjct 192 WMSLPKKSKG-VKVGRVDCTSHQSLCAQFNVKGYPTILLFNKGEKNPKTAMNYEGQRTAA 250
Query 61 EIVKFAMEFYGAKAEADQLLNEEQFRSECGETLCILALL-PDVLDSGEEGRKNYLQTLNQ 119
+I+ FA + A + + + +C LC+L P + + KN+
Sbjct 251 DILAFAKKNDKALSPPTHATLVAELKEKCSGPLCLLFFFKPSTKEENLKTLKNF------ 304
Query 120 VVKASVTVPVKFLWLQGGDNFDLEEQLHLAFG---WPAVVAVHLARGKFSVHRGNFTRES 176
+ T P + G+N EQ FG +PAVV ++LA+G + F++E+
Sbjct 305 --ASKHTAPFALAYSLVGEN----EQWERVFGLKEFPAVVGLNLAKGVYLPLNSEFSKEN 358
Query 177 INSFLQQLLAGRA 189
+N F++ +L+G+A
Sbjct 359 LNKFVKSILSGKA 371
Score = 31.6 bits (70), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 16/56 (28%), Positives = 34/56 (60%), Gaps = 1/56 (1%)
Query 14 VAKVDSTSEQMLASKFGIQAFPTLKLFPA-GPKSTAMVKDYEGQRTADEIVKFAME 68
+ +V + ++ ++ K+ +++FP+LKLF G +S V D + R D++V F ++
Sbjct 76 LVQVVAVKDENVSKKYKVKSFPSLKLFLGNGKESEPDVVDVDEGRDLDDLVSFTLK 131
> mmu:67988 Tmx3, 6430411B10Rik, A730024F05Rik, AV259382, Txndc10,
mKIAA1830; thioredoxin-related transmembrane protein 3
(EC:5.3.4.1); K09585 thioredoxin domain-containing protein 10
[EC:5.3.4.1]
Length=456
Score = 57.4 bits (137), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 29/75 (38%), Positives = 46/75 (61%), Gaps = 8/75 (10%)
Query 1 WDEVSVQLKG---KVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQR 57
W+EV +++K VKV K+D+TS +AS+FG++ +PT+KL + +Y G R
Sbjct 66 WNEVGLEMKSIGSPVKVGKMDATSYSSIASEFGVRGYPTIKLLKGD-----LAYNYRGPR 120
Query 58 TADEIVKFAMEFYGA 72
T D+I++FA GA
Sbjct 121 TKDDIIEFAHRVSGA 135
> hsa:54495 TMX3, FLJ20793, KIAA1830, PDIA13, TXNDC10; thioredoxin-related
transmembrane protein 3 (EC:5.3.4.1); K09585 thioredoxin
domain-containing protein 10 [EC:5.3.4.1]
Length=454
Score = 57.0 bits (136), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 29/75 (38%), Positives = 46/75 (61%), Gaps = 8/75 (10%)
Query 1 WDEVSVQLKG---KVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQR 57
W+EV +++K VKV K+D+TS +AS+FG++ +PT+KL + +Y G R
Sbjct 63 WNEVGLEMKSIGSPVKVGKMDATSYSSIASEFGVRGYPTIKLLKGD-----LAYNYRGPR 117
Query 58 TADEIVKFAMEFYGA 72
T D+I++FA GA
Sbjct 118 TKDDIIEFAHRVSGA 132
> dre:405841 MGC77086; zgc:77086; K08056 protein disulfide isomerase
family A, member 3 [EC:5.3.4.1]
Length=488
Score = 53.5 bits (127), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 26/65 (40%), Positives = 39/65 (60%), Gaps = 4/65 (6%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTAD 60
++ + +LKG + +AKVD T +FG+ +PTLK+F G +S A Y+G RTAD
Sbjct 59 YEAAATKLKGTLALAKVDCTVNSETCERFGVNGYPTLKIFRNGEESGA----YDGPRTAD 114
Query 61 EIVKF 65
IV +
Sbjct 115 GIVSY 119
Score = 34.7 bits (78), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 18/67 (26%), Positives = 36/67 (53%), Gaps = 5/67 (7%)
Query 1 WDEVSVQLKGK--VKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRT 58
+ E+ +L G + +AK+D+T+ + + +Q FPT+ P+G K + YEG R
Sbjct 407 YKELGEKLSGNPNIVIAKMDATAND-VPPNYDVQGFPTIYFVPSGQKDQP--RRYEGGRE 463
Query 59 ADEIVKF 65
++ + +
Sbjct 464 VNDFITY 470
> dre:378851 pdia3, Grp58, sb:cb825; protein disulfide isomerase
family A, member 3 (EC:5.3.4.1)
Length=494
Score = 53.5 bits (127), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 32/91 (35%), Positives = 48/91 (52%), Gaps = 6/91 (6%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTAD 60
++ + +LKG V +AKVD T+ + K+G+ +PTLK+F G S Y+G RTAD
Sbjct 57 YEAAATRLKGIVPLAKVDCTANSKVCGKYGVSGYPTLKIFRDGEDSGG----YDGPRTAD 112
Query 61 EIVKFAMEFYGAKAEADQLLNEEQFRSECGE 91
IV + G + +L NE F G+
Sbjct 113 GIVSHLKKQAGPASV--ELKNEADFEKYIGD 141
Score = 33.5 bits (75), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 17/54 (31%), Positives = 29/54 (53%), Gaps = 3/54 (5%)
Query 12 VKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIVKF 65
+ +AK+D+T+ + S + + FPT+ PAG K K YEG R + + +
Sbjct 419 IVIAKMDATAND-VPSPYEVSGFPTIYFSPAGRKQNP--KKYEGGREVSDFISY 469
> cel:H06O01.1 pdi-3; Protein Disulfide Isomerase family member
(pdi-3); K08056 protein disulfide isomerase family A, member
3 [EC:5.3.4.1]
Length=488
Score = 52.8 bits (125), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 26/54 (48%), Positives = 35/54 (64%), Gaps = 4/54 (7%)
Query 12 VKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIVKF 65
V + KVD T+E+ + KFG++ FPTLK+F G +DY+G R AD IVKF
Sbjct 73 VALVKVDCTTEKTVCDKFGVKGFPTLKIFRNG----VPAQDYDGPRDADGIVKF 122
> dre:553250 MGC152808; zgc:152808 (EC:5.3.4.1); K09585 thioredoxin
domain-containing protein 10 [EC:5.3.4.1]
Length=484
Score = 52.8 bits (125), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 27/79 (34%), Positives = 45/79 (56%), Gaps = 8/79 (10%)
Query 1 WDEVSVQLK---GKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQR 57
W+EV +L V+V K+D+T+ +AS+FG++ +PT+KL + +Y+G R
Sbjct 56 WEEVGAELSRSGSPVRVGKMDATAYSGMASEFGVRGYPTIKLLKGD-----LAYNYKGPR 110
Query 58 TADEIVKFAMEFYGAKAEA 76
T D+I++FA G A
Sbjct 111 TKDDIIEFANRVAGPAVRA 129
> dre:100329666 protein disulfide-isomerase A3-like
Length=494
Score = 52.8 bits (125), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 24/65 (36%), Positives = 41/65 (63%), Gaps = 4/65 (6%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTAD 60
++ + +LKG V +AKVD T+ + +G+ +PTLK+F G +S++ Y+G R+AD
Sbjct 65 FESAASRLKGTVTLAKVDCTANTEICKHYGVNGYPTLKIFRNGHESSS----YDGPRSAD 120
Query 61 EIVKF 65
IV +
Sbjct 121 GIVDY 125
Score = 33.9 bits (76), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 20/54 (37%), Positives = 29/54 (53%), Gaps = 3/54 (5%)
Query 12 VKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIVKF 65
+ +AK+D+T + A + +Q FPT+ AG KS K YEG R + V F
Sbjct 426 IVIAKMDATVNDVPAG-YDVQGFPTIYFAAAGRKSEP--KRYEGAREVKDFVNF 476
> hsa:2923 PDIA3, ER60, ERp57, ERp60, ERp61, GRP57, GRP58, HsT17083,
P58, PI-PLC; protein disulfide isomerase family A, member
3 (EC:5.3.4.1); K08056 protein disulfide isomerase family
A, member 3 [EC:5.3.4.1]
Length=505
Score = 52.4 bits (124), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 26/63 (41%), Positives = 40/63 (63%), Gaps = 4/63 (6%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTAD 60
++ + +LKG V +AKVD T+ +K+G+ +PTLK+F G ++ A Y+G RTAD
Sbjct 67 YEAAATRLKGIVPLAKVDCTANTNTCNKYGVSGYPTLKIFRDGEEAGA----YDGPRTAD 122
Query 61 EIV 63
IV
Sbjct 123 GIV 125
> dre:445123 zgc:100906 (EC:5.3.4.1); K08056 protein disulfide
isomerase family A, member 3 [EC:5.3.4.1]
Length=493
Score = 52.4 bits (124), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 24/65 (36%), Positives = 41/65 (63%), Gaps = 4/65 (6%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTAD 60
++ + +LKG V +AKVD T+ + +G+ +PTLK+F G +S++ Y+G R+AD
Sbjct 64 FESAASRLKGTVTLAKVDCTANTEICKHYGVNGYPTLKIFRNGQESSS----YDGPRSAD 119
Query 61 EIVKF 65
IV +
Sbjct 120 GIVDY 124
Score = 33.9 bits (76), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 20/54 (37%), Positives = 29/54 (53%), Gaps = 3/54 (5%)
Query 12 VKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIVKF 65
+ +AK+D+T + A + +Q FPT+ AG KS K YEG R + V F
Sbjct 425 IVIAKMDATVNDVPAG-YDVQGFPTIYFAAAGRKSEP--KRYEGAREVKDFVNF 475
> xla:379505 hypothetical protein MGC64309; K09580 protein disulfide-isomerase
A1 [EC:5.3.4.1]
Length=505
Score = 52.4 bits (124), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 34/84 (40%), Positives = 48/84 (57%), Gaps = 8/84 (9%)
Query 1 WDEVSVQLKG--KVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRT 58
WD++ + K + +AK+DST ++ A K I +FPTLK FPAGP V DY G+RT
Sbjct 407 WDQLGEKYKNHDSIIIAKMDSTVNEIEAVK--IHSFPTLKFFPAGP---GKVADYNGERT 461
Query 59 ADEIVKFAMEFYGAKAEADQLLNE 82
+ KF +E G AD+ L +
Sbjct 462 LEGFSKF-LESGGQDGAADEDLED 484
Score = 46.2 bits (108), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 24/63 (38%), Positives = 38/63 (60%), Gaps = 2/63 (3%)
Query 12 VKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIVKFAMEFYG 71
+++ KVD+T E LA +FG++ +PT+K F G KS+ K+Y R A +IV + + G
Sbjct 77 IRLGKVDATEESDLAQEFGVRGYPTIKFFKNGDKSSP--KEYSAGREAADIVNWLKKRTG 134
Query 72 AKA 74
A
Sbjct 135 PAA 137
> mmu:14827 Pdia3, 58kDa, ERp57, ERp60, ERp61, Erp, Grp58, PDI,
PDI-Q2, PI-PLC, PLC[a], Plca; protein disulfide isomerase
associated 3 (EC:5.3.4.1); K08056 protein disulfide isomerase
family A, member 3 [EC:5.3.4.1]
Length=505
Score = 52.4 bits (124), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 26/63 (41%), Positives = 40/63 (63%), Gaps = 4/63 (6%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTAD 60
++ + +LKG V +AKVD T+ +K+G+ +PTLK+F G ++ A Y+G RTAD
Sbjct 67 YEAAATRLKGIVPLAKVDCTANTNTCNKYGVSGYPTLKIFRDGEEAGA----YDGPRTAD 122
Query 61 EIV 63
IV
Sbjct 123 GIV 125
Score = 32.7 bits (73), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 17/54 (31%), Positives = 31/54 (57%), Gaps = 3/54 (5%)
Query 12 VKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIVKF 65
+ +AK+D+T+ + S + ++ FPT+ PA K T K YEG R ++ + +
Sbjct 429 IVIAKMDATAND-VPSPYEVKGFPTIYFSPANKKLTP--KKYEGGRELNDFISY 479
> xla:399248 p4hb; prolyl 4-hydroxylase, beta polypeptide (EC:5.3.4.1);
K09580 protein disulfide-isomerase A1 [EC:5.3.4.1]
Length=506
Score = 52.4 bits (124), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 34/84 (40%), Positives = 49/84 (58%), Gaps = 7/84 (8%)
Query 1 WDEVSVQLKG--KVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRT 58
WD++ + K + +AK+DST+ ++ A K I +FPTLK FPAGP V DY G+RT
Sbjct 407 WDQLGEKYKDHESIIIAKMDSTANEIEAVK--IHSFPTLKFFPAGPGKK--VVDYNGERT 462
Query 59 ADEIVKFAMEFYGAKAEADQLLNE 82
+ KF +E G AD+ L +
Sbjct 463 QEGFSKF-LESGGQDGAADEDLED 485
Score = 44.3 bits (103), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 23/63 (36%), Positives = 37/63 (58%), Gaps = 2/63 (3%)
Query 12 VKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIVKFAMEFYG 71
+++ KVD+T E LA +FG++ +PT+K F G KS+ K+Y R A + V + + G
Sbjct 77 IRLGKVDATEESDLAQEFGVRGYPTIKFFKNGDKSSP--KEYSAGREAADFVNWLKKRTG 134
Query 72 AKA 74
A
Sbjct 135 PAA 137
> ath:AT3G54960 ATPDIL1-3 (PDI-LIKE 1-3); protein disulfide isomerase
Length=518
Score = 52.4 bits (124), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 34/86 (39%), Positives = 49/86 (56%), Gaps = 11/86 (12%)
Query 5 SVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIVK 64
+ +LKG +AK+D+T E LA K+ IQ FPT+ LF G M K YEG+RT D IV
Sbjct 142 ATELKGLAALAKIDATEEGDLAQKYEIQGFPTVFLFVDG----EMRKTYEGERTKDGIVT 197
Query 65 F-------AMEFYGAKAEADQLLNEE 83
+ ++ K EA+++L+ E
Sbjct 198 WLKKKASPSIHNITTKEEAERVLSAE 223
> xla:444355 tmx3-a, pdia13, tmx3, txndc10, txndc10a; thioredoxin-related
transmembrane protein 3 (EC:5.3.4.1); K09585 thioredoxin
domain-containing protein 10 [EC:5.3.4.1]
Length=452
Score = 51.6 bits (122), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 25/74 (33%), Positives = 43/74 (58%), Gaps = 8/74 (10%)
Query 1 WDEVSVQLK---GKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQR 57
W+EV ++++ ++V K+D+T +AS+FG++ FPT+K M +Y G R
Sbjct 63 WNEVGIEIRTSGSPIRVGKIDATVYSSIASEFGVRGFPTIKALKGD-----MAYNYRGPR 117
Query 58 TADEIVKFAMEFYG 71
T ++IV+FA G
Sbjct 118 TKEDIVEFANRVAG 131
> xla:734368 tmx3-b, pdia13, txndc10, txndc10b; thioredoxin-related
transmembrane protein 3 (EC:5.3.4.1); K09585 thioredoxin
domain-containing protein 10 [EC:5.3.4.1]
Length=452
Score = 51.6 bits (122), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 24/74 (32%), Positives = 44/74 (59%), Gaps = 8/74 (10%)
Query 1 WDEVSVQLKGK---VKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQR 57
W++V ++++ ++V K+D+T +AS+FG++ FPT+K+ M +Y G R
Sbjct 63 WNDVGIEIRSSGSPIRVGKMDATVHSSIASEFGVRGFPTIKVLKGD-----MAYNYRGPR 117
Query 58 TADEIVKFAMEFYG 71
T ++IV+FA G
Sbjct 118 TKEDIVEFANRVAG 131
> tgo:TGME49_047350 thioredoxin, putative (EC:5.3.4.1)
Length=312
Score = 51.6 bits (122), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 24/66 (36%), Positives = 40/66 (60%), Gaps = 5/66 (7%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTAD 60
W++++ L GK+ VAK+D+TS + A +F IQ FPTL G + +Y G+R+ +
Sbjct 167 WEDLAKALSGKINVAKLDATSNSITAKRFKIQGFPTLYYLANGK-----MYEYRGERSVE 221
Query 61 EIVKFA 66
++ FA
Sbjct 222 KLKAFA 227
> ath:AT2G47470 UNE5; UNE5 (UNFERTILIZED EMBRYO SAC 5); protein
disulfide isomerase; K09584 protein disulfide-isomerase A6
[EC:5.3.4.1]
Length=323
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 36/124 (29%), Positives = 60/124 (48%), Gaps = 11/124 (8%)
Query 12 VKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIVKFAMEFYG 71
V +A +D+ + + L K+G+ FPTLK F PK DY+G R D+ V F E G
Sbjct 194 VVIANLDADAHKALGEKYGVSGFPTLKFF---PKDNKAGHDYDGGRDLDDFVSFINEKSG 250
Query 72 AKAEADQLLNEEQFRSECGETLCILALLPDVLDSGEEGRKNYLQTLNQ---VVKASVTVP 128
++ + Q S+ G + AL+ +++ + E+ +K L + + +K S T
Sbjct 251 TSRDS-----KGQLTSKAGIVESLDALVKELVAASEDEKKAVLSRIEEEASTLKGSTTRY 305
Query 129 VKFL 132
V L
Sbjct 306 VTLL 309
Score = 40.4 bits (93), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 17/54 (31%), Positives = 33/54 (61%), Gaps = 3/54 (5%)
Query 12 VKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIVKF 65
V +AKVD ++ + +K+G+ +PT++ FP G + + YEG R A+ + ++
Sbjct 75 VLIAKVDCDEQKSVCTKYGVSGYPTIQWFPKG---SLEPQKYEGPRNAEALAEY 125
> dre:553578 tmx3, MGC110025, txndc10, zgc:110025; thioredoxin-related
transmembrane protein 3 (EC:5.3.4.1)
Length=434
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 26/69 (37%), Positives = 40/69 (57%), Gaps = 8/69 (11%)
Query 1 WDEVSVQLKG---KVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQR 57
W EV +LK V V K+D+T+ +A++F I+ +PT+KLF + DY+G R
Sbjct 55 WTEVGAELKSLGSPVNVGKIDTTAHTSIATEFNIRGYPTIKLFKGD-----LSFDYKGPR 109
Query 58 TADEIVKFA 66
T D I++F
Sbjct 110 TKDGIIEFT 118
> xla:379743 pdia3, MGC53247, erp57, grp58; protein disulfide
isomerase family A, member 3 (EC:5.3.4.1); K08056 protein disulfide
isomerase family A, member 3 [EC:5.3.4.1]
Length=502
Score = 50.8 bits (120), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 24/63 (38%), Positives = 40/63 (63%), Gaps = 4/63 (6%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTAD 60
++ + +LKG + +AKVD T+ + +K+G+ +PTLK+F G S + Y+G R+AD
Sbjct 63 YEIAATKLKGTLSLAKVDCTANSNICNKYGVSGYPTLKIFRDGEDSGS----YDGPRSAD 118
Query 61 EIV 63
IV
Sbjct 119 GIV 121
Score = 32.7 bits (73), Expect = 0.79, Method: Compositional matrix adjust.
Identities = 17/54 (31%), Positives = 30/54 (55%), Gaps = 3/54 (5%)
Query 12 VKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIVKF 65
+ +AK+D+T+ + ++ ++ FPT+ PAG K K YEG R E + +
Sbjct 426 IVIAKMDATAND-VPPQYEVRGFPTIYFAPAGNKQNP--KRYEGGREVSEFLSY 476
> dre:406673 p4hb, psmb3, zgc:92596; procollagen-proline, 2-oxoglutarate
4-dioxygenase (proline 4-hydroxylase), beta polypeptide
(EC:5.3.4.1); K09580 protein disulfide-isomerase A1 [EC:5.3.4.1]
Length=509
Score = 50.8 bits (120), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 28/67 (41%), Positives = 41/67 (61%), Gaps = 6/67 (8%)
Query 1 WDEVSVQLK--GKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRT 58
WD++ + K + VAK+DST+ ++ A K + +FPTLK FPAG + + DY G+RT
Sbjct 405 WDQLGEKFKDNANIVVAKMDSTANEIEAVK--VHSFPTLKFFPAGDERKVI--DYNGERT 460
Query 59 ADEIVKF 65
D KF
Sbjct 461 LDGFTKF 467
Score = 47.4 bits (111), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 22/52 (42%), Positives = 34/52 (65%), Gaps = 2/52 (3%)
Query 12 VKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIV 63
+++AKVD+T E LA +FG++ +PT+K F G K K+Y R A++IV
Sbjct 75 IRLAKVDATEESELAQEFGVRGYPTIKFFKGGEKGNP--KEYSAGRQAEDIV 124
> mmu:12304 Pdia4, AI987846, Cai, ERp-72, Erp72; protein disulfide
isomerase associated 4 (EC:5.3.4.1); K09582 protein disulfide-isomerase
A4 [EC:5.3.4.1]
Length=641
Score = 49.7 bits (117), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 24/52 (46%), Positives = 33/52 (63%), Gaps = 5/52 (9%)
Query 12 VKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIV 63
+ VAK+D+TS MLASKF + +PT+K+ G DY+G RT +EIV
Sbjct 111 IAVAKIDATSASMLASKFDVSGYPTIKILKKG-----QAVDYDGSRTQEEIV 157
Score = 42.0 bits (97), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/88 (30%), Positives = 44/88 (50%), Gaps = 5/88 (5%)
Query 12 VKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIVKFAMEFYG 71
+ +AKVD+T + LA +F + +PTLK+F G DY G R IV + +E G
Sbjct 226 IPLAKVDATEQTDLAKRFDVSGYPTLKIFRKG-----RPFDYNGPREKYGIVDYMIEQSG 280
Query 72 AKAEADQLLNEEQFRSECGETLCILALL 99
++ L + Q + G+ + I+ L
Sbjct 281 PPSKEILTLKQVQEFLKDGDDVVIIGLF 308
Score = 32.7 bits (73), Expect = 0.80, Method: Compositional matrix adjust.
Identities = 16/55 (29%), Positives = 30/55 (54%), Gaps = 1/55 (1%)
Query 14 VAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIVKFAME 68
+AK+D+T+ + ++ ++ FPT+ P+G K +K G R + + KF E
Sbjct 576 IAKMDATANDITNDQYKVEGFPTIYFAPSGDKKNP-IKFEGGNRDLEHLSKFIDE 629
> mmu:18453 P4hb, ERp59, PDI, Pdia1, Thbp; prolyl 4-hydroxylase,
beta polypeptide (EC:5.3.4.1); K09580 protein disulfide-isomerase
A1 [EC:5.3.4.1]
Length=509
Score = 49.3 bits (116), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 25/64 (39%), Positives = 40/64 (62%), Gaps = 2/64 (3%)
Query 11 KVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIVKFAMEFY 70
++++AKVD+T E LA ++G++ +PT+K F G TA K+Y R AD+IV + +
Sbjct 78 EIRLAKVDATEESDLAQQYGVRGYPTIKFFKNG--DTASPKEYTAGREADDIVNWLKKRT 135
Query 71 GAKA 74
G A
Sbjct 136 GPAA 139
Score = 47.0 bits (110), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 27/67 (40%), Positives = 39/67 (58%), Gaps = 6/67 (8%)
Query 1 WDEVSVQLKG--KVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRT 58
WD++ K + +AK+DST+ ++ A K + +FPTLK FPA T + DY G+RT
Sbjct 409 WDKLGETYKDHENIIIAKMDSTANEVEAVK--VHSFPTLKFFPASADRTVI--DYNGERT 464
Query 59 ADEIVKF 65
D KF
Sbjct 465 LDGFKKF 471
> hsa:5034 P4HB, DSI, ERBA2L, GIT, P4Hbeta, PDI, PDIA1, PHDB,
PO4DB, PO4HB, PROHB; prolyl 4-hydroxylase, beta polypeptide
(EC:5.3.4.1); K09580 protein disulfide-isomerase A1 [EC:5.3.4.1]
Length=508
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 25/64 (39%), Positives = 40/64 (62%), Gaps = 2/64 (3%)
Query 11 KVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIVKFAMEFY 70
++++AKVD+T E LA ++G++ +PT+K F G TA K+Y R AD+IV + +
Sbjct 76 EIRLAKVDATEESDLAQQYGVRGYPTIKFFRNG--DTASPKEYTAGREADDIVNWLKKRT 133
Query 71 GAKA 74
G A
Sbjct 134 GPAA 137
Score = 47.0 bits (110), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 27/67 (40%), Positives = 39/67 (58%), Gaps = 6/67 (8%)
Query 1 WDEVSVQLKG--KVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRT 58
WD++ K + +AK+DST+ ++ A K + +FPTLK FPA T + DY G+RT
Sbjct 407 WDKLGETYKDHENIVIAKMDSTANEVEAVK--VHSFPTLKFFPASADRTVI--DYNGERT 462
Query 59 ADEIVKF 65
D KF
Sbjct 463 LDGFKKF 469
> hsa:9601 PDIA4, ERP70, ERP72, ERp-72; protein disulfide isomerase
family A, member 4 (EC:5.3.4.1); K09582 protein disulfide-isomerase
A4 [EC:5.3.4.1]
Length=645
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 33/103 (32%), Positives = 52/103 (50%), Gaps = 12/103 (11%)
Query 1 WDEVSVQLKGK---VKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQR 57
+++++ LK K + VAK+D+TS +LAS+F + +PT+K+ G DYEG R
Sbjct 101 YEKIANILKDKDPPIPVAKIDATSASVLASRFDVSGYPTIKILKKG-----QAVDYEGSR 155
Query 58 TADEIVKFAMEF----YGAKAEADQLLNEEQFRSECGETLCIL 96
T +EIV E + E +L +E F + IL
Sbjct 156 TQEEIVAKVREVSQPDWTPPPEVTLVLTKENFDEVVNDADIIL 198
Score = 43.1 bits (100), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 27/88 (30%), Positives = 45/88 (51%), Gaps = 5/88 (5%)
Query 12 VKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIVKFAMEFYG 71
+ +AKVD+T+E LA +F + +PTLK+F G DY G R IV + +E G
Sbjct 230 IPLAKVDATAETDLAKRFDVSGYPTLKIFRKG-----RPYDYNGPREKYGIVDYMIEQSG 284
Query 72 AKAEADQLLNEEQFRSECGETLCILALL 99
++ L + Q + G+ + I+ +
Sbjct 285 PPSKEILTLKQVQEFLKDGDDVIIIGVF 312
Score = 33.9 bits (76), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 31/55 (56%), Gaps = 1/55 (1%)
Query 14 VAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIVKFAME 68
+AK+D+T+ + + ++ ++ FPT+ P+G K VK G R + + KF E
Sbjct 580 IAKMDATANDVPSDRYKVEGFPTIYFAPSGDKKNP-VKFEGGDRDLEHLSKFIEE 633
> xla:399040 pdia2, XPDIp, pdi; protein disulfide isomerase family
A, member 2 (EC:5.3.4.1); K09581 protein disulfide isomerase
family A, member 2 [EC:5.3.4.1]
Length=526
Score = 48.5 bits (114), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 28/78 (35%), Positives = 46/78 (58%), Gaps = 3/78 (3%)
Query 11 KVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIVKFAMEFY 70
+V++AKVD T E L+++F + +PTLK F G ++ + DY G+R D +VK+ +
Sbjct 98 EVRLAKVDGTVETDLSTEFNVNGYPTLKFFKGGNRTGHI--DYGGKRDQDGLVKWMLRRM 155
Query 71 GAKAEA-DQLLNEEQFRS 87
G A D + + E+F S
Sbjct 156 GPAAVVLDNVESAEKFTS 173
Score = 37.7 bits (86), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 24/86 (27%), Positives = 47/86 (54%), Gaps = 14/86 (16%)
Query 1 WDEVSVQLKG--KVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRT 58
W+E+ + K V +AK+D+T+ ++ + ++ FP L+ FPAGP+ + +Y +RT
Sbjct 430 WEELGEKYKDHENVIIAKIDATANEIDGLR--VRGFPNLRFFPAGPERKMI--EYTKERT 485
Query 59 ADEIVKFAMEFYGAKAEADQLLNEEQ 84
+E + A ++ +L +EQ
Sbjct 486 --------VELFSAFIDSGGVLPDEQ 503
> tpv:TP02_0602 protein disulfide isomerase
Length=220
Score = 47.0 bits (110), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 25/67 (37%), Positives = 39/67 (58%), Gaps = 4/67 (5%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTAD 60
W+ ++ LKG+V VA VD T L +F I+ +PTL LF G M + G+RT +
Sbjct 74 WESLAKALKGQVNVADVDVTRNLNLGKRFQIRGYPTLLLFHKGK----MYQYEGGERTVE 129
Query 61 EIVKFAM 67
++ +FA+
Sbjct 130 KLSEFAL 136
> xla:495169 pdia4; protein disulfide isomerase family A, member
4 (EC:5.3.4.1); K09582 protein disulfide-isomerase A4 [EC:5.3.4.1]
Length=637
Score = 46.6 bits (109), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 35/99 (35%), Positives = 50/99 (50%), Gaps = 14/99 (14%)
Query 12 VKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIVKFAMEFYG 71
+ +AKVD+T E L SK+G+ FPTLK+F G DY G R IV + E G
Sbjct 223 IPLAKVDATVESSLGSKYGVTGFPTLKIFRKG-----KAFDYNGPREKYGIVDYMTEQAG 277
Query 72 AKAEADQLLNE--EQFRSECGETLCILALLPDVLDSGEE 108
++ Q + + E FR G+ + +L + SGEE
Sbjct 278 PPSKQIQAVKQVHEFFRD--GDDVGVLGVF-----SGEE 309
Score = 36.2 bits (82), Expect = 0.079, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 29/52 (55%), Gaps = 5/52 (9%)
Query 12 VKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIV 63
V VAK+D+T +A ++ I +PT+K+ G DY+G RT + +V
Sbjct 108 VPVAKIDATVATNIAGRYDISGYPTIKILKKG-----QPIDYDGARTQEALV 154
Score = 30.0 bits (66), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 16/55 (29%), Positives = 28/55 (50%), Gaps = 1/55 (1%)
Query 14 VAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIVKFAME 68
+AK+D+T+ + + K+ ++ FPT+ P K +K G R + KF E
Sbjct 573 IAKMDATANDISSDKYKVEGFPTIYFAPQNNKQNP-IKFSGGNRDLEGFSKFIEE 626
> dre:394023 pdia2, MGC55398, P4hb, zgc:55398; protein disulfide
isomerase family A, member 2 (EC:5.3.4.1)
Length=278
Score = 46.6 bits (109), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 26/61 (42%), Positives = 39/61 (63%), Gaps = 5/61 (8%)
Query 1 WDEVSVQLKG---KVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQR 57
+ EV+ QLK +V++AKVD+ E+ LAS+F + +FPTLK F G + A + G+R
Sbjct 95 YAEVAGQLKNASSEVRLAKVDAIEEKELASEFSVDSFPTLKFFKEGNRQNATT--FFGKR 152
Query 58 T 58
T
Sbjct 153 T 153
> dre:554998 pdia4, MGC113965, MGC136625, cai, wu:fd20c07, zgc:113965,
zgc:136625, zgc:56014, zgc:77773; protein disulfide
isomerase associated 4 (EC:5.3.4.1); K09582 protein disulfide-isomerase
A4 [EC:5.3.4.1]
Length=645
Score = 45.8 bits (107), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 32/111 (28%), Positives = 53/111 (47%), Gaps = 5/111 (4%)
Query 12 VKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIVKFAMEFYG 71
+ +AKVD+T+E LA++FG+ +PTLK+F G DY G R IV + + G
Sbjct 230 IPLAKVDATAESDLATRFGVSGYPTLKIFRKG-----KAFDYNGPREKFGIVDYMSDQAG 284
Query 72 AKAEADQLLNEEQFRSECGETLCILALLPDVLDSGEEGRKNYLQTLNQVVK 122
++ Q L + Q G+ I+ + D+ E + +L + K
Sbjct 285 PPSKQVQTLKQVQELLRDGDDAVIVGVFSSDEDAAYEIYQEACNSLREDYK 335
Score = 37.0 bits (84), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 26/89 (29%), Positives = 40/89 (44%), Gaps = 9/89 (10%)
Query 12 VKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIVKFAMEF-- 69
+ VAKVD+T L S+F + +PT+K+ G DY+G R+ IV+ E
Sbjct 115 IPVAKVDATKASGLGSRFEVSGYPTIKILKKGEPL-----DYDGDRSEHAIVERVKEVAQ 169
Query 70 --YGAKAEADQLLNEEQFRSECGETLCIL 96
+ EA +L ++ F IL
Sbjct 170 PDWKPPPEATLVLTKDNFDDVVNNADIIL 198
Score = 30.8 bits (68), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 15/52 (28%), Positives = 28/52 (53%), Gaps = 1/52 (1%)
Query 14 VAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIVKF 65
+AK+D+T+ + + ++ FPT+ P+ K +K G+R +E KF
Sbjct 580 IAKMDATANDVPHDSYKVEGFPTIYFAPSNNKQNP-IKFEGGKRDVEEFSKF 630
> hsa:81567 TXNDC5, ENDOPDI, ERP46, HCC-2, MGC3178, PDIA15, STRF8,
UNQ364; thioredoxin domain containing 5 (endoplasmic reticulum);
K13984 thioredoxin domain-containing protein 5
Length=324
Score = 45.8 bits (107), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 26/71 (36%), Positives = 43/71 (60%), Gaps = 9/71 (12%)
Query 1 WDEVSVQ----LKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQ 56
W+E+S + L G VK+A+VD T+E+ + SK+ ++ +PTL LF G K V ++ G
Sbjct 252 WEELSKKEFPGLAG-VKIAEVDCTAERNICSKYSVRGYPTLLLFRGGKK----VSEHSGG 306
Query 57 RTADEIVKFAM 67
R D + +F +
Sbjct 307 RDLDSLHRFVL 317
Score = 37.7 bits (86), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 37/78 (47%), Gaps = 14/78 (17%)
Query 11 KVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIVKFAMEFY 70
KV VAKVD T+ + S G++ +PTLKLF G ++ Y+G R + + +
Sbjct 5 KVYVAKVDCTAHSDVCSAQGVRGYPTLKLFKPGQEAV----KYQGPRDFQTLENWML--- 57
Query 71 GAKAEADQLLNEEQFRSE 88
Q LNEE E
Sbjct 58 -------QTLNEEPVTPE 68
Score = 33.5 bits (75), Expect = 0.45, Method: Compositional matrix adjust.
Identities = 20/67 (29%), Positives = 36/67 (53%), Gaps = 6/67 (8%)
Query 1 WDEVSVQLKGK--VKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRT 58
W+++++ L+ VK+ KVD T L S ++ +PTL F G K V Y+G+R
Sbjct 119 WEQLALGLEHSETVKIGKVDCTQHYELCSGNQVRGYPTLLWFRDGKK----VDQYKGKRD 174
Query 59 ADEIVKF 65
+ + ++
Sbjct 175 LESLREY 181
> pfa:PF13_0272 thioredoxin-related protein, putative (EC:5.3.4.1);
K01829 protein disulfide-isomerase [EC:5.3.4.1]
Length=208
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 27/77 (35%), Positives = 40/77 (51%), Gaps = 7/77 (9%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQ-RTA 59
W +++ +LKGK+ VAK+D T +F I+ FPTL F G + DY+ R+
Sbjct 67 WAQLATELKGKINVAKIDVTLNSKTRKRFKIEGFPTLLYFKNGK-----MYDYKNHDRSL 121
Query 60 DEIVKFAMEFY-GAKAE 75
+ F +E Y AKA
Sbjct 122 EAFKNFVLETYKNAKAS 138
> cel:C14B1.1 pdi-1; Protein Disulfide Isomerase family member
(pdi-1); K09580 protein disulfide-isomerase A1 [EC:5.3.4.1]
Length=485
Score = 45.1 bits (105), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 25/79 (31%), Positives = 48/79 (60%), Gaps = 7/79 (8%)
Query 1 WDEVSVQLKGK--VKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRT 58
WDE++ + + V +AK+D+T ++ K + +FPTLKL+PAG + DY+G R
Sbjct 403 WDELAEKYESNPNVVIAKLDATLNELADVK--VNSFPTLKLWPAGSSTPV---DYDGDRN 457
Query 59 ADEIVKFAMEFYGAKAEAD 77
++ +F ++ G+ +E++
Sbjct 458 LEKFEEFVNKYAGSASESE 476
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/68 (36%), Positives = 38/68 (55%), Gaps = 8/68 (11%)
Query 1 WDEVSVQLK---GKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQR 57
+DE + LK +K+AKVD+T Q LASKF ++ +PT+ F +G + Y G R
Sbjct 62 YDEAADLLKEEGSDIKLAKVDATENQALASKFEVRGYPTILYFKSGKPTK-----YTGGR 116
Query 58 TADEIVKF 65
+IV +
Sbjct 117 ATAQIVDW 124
> dre:557858 dnajc10, MGC162218, zgc:162218; DnaJ (Hsp40) homolog,
subfamily C, member 10; K09530 DnaJ homolog subfamily C
member 10
Length=791
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 22/73 (30%), Positives = 36/73 (49%), Gaps = 4/73 (5%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTAD 60
W E + ++ G +++ V+ ML GI ++P+L +F AG D RT
Sbjct 167 WREFAKEMDGVIRIGAVNCGDNGMLCRSKGINSYPSLYVFRAGMNPEKYFND----RTKS 222
Query 61 EIVKFAMEFYGAK 73
+ KFAM+F +K
Sbjct 223 SLTKFAMQFVKSK 235
Score = 44.3 bits (103), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 20/63 (31%), Positives = 37/63 (58%), Gaps = 5/63 (7%)
Query 3 EVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEI 62
+ S+QL G++K +D T + L + + I A+PT +F + + + +YEG +AD I
Sbjct 488 KASIQLFGQLKFGTLDCTIHEGLCNTYNIHAYPTTVIF-----NKSSIHEYEGHHSADGI 542
Query 63 VKF 65
++F
Sbjct 543 LEF 545
Score = 34.3 bits (77), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 12/35 (34%), Positives = 20/35 (57%), Gaps = 0/35 (0%)
Query 8 LKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPA 42
+KG V+ KVD + GI+A+PT++ +P
Sbjct 713 MKGTVRAGKVDCQAHYQTCQSAGIKAYPTVRFYPT 747
Score = 34.3 bits (77), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 14/55 (25%), Positives = 22/55 (40%), Gaps = 0/55 (0%)
Query 1 WDEVSVQLKGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEG 55
W ++ L G V V VD ++A+P ++LFP + Y G
Sbjct 594 WRRMARMLSGIVNVGTVDCQKHHSFCQSESVRAYPEIRLFPQNSNRRDQYQTYNG 648
> pfa:MAL8P1.17 PfPDI-8; protein disulfide isomerase (EC:5.3.4.1);
K01829 protein disulfide-isomerase [EC:5.3.4.1]
Length=483
Score = 44.3 bits (103), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 24/63 (38%), Positives = 36/63 (57%), Gaps = 5/63 (7%)
Query 9 KGKVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEGQRTADEIVKFAME 68
K ++K+ +D+TSE LA ++GI +PTL LF K +Y G RTA IV + ++
Sbjct 81 KSEIKLVSIDATSENALAQEYGITGYPTLILFNKKNKI-----NYGGGRTAQSIVDWLLQ 135
Query 69 FYG 71
G
Sbjct 136 MTG 138
> cpv:cgd6_850 thioredoxin; protein disulfide isomerase A6, signal
peptide, possible transmembrane domain in C-terminal region
Length=524
Score = 44.3 bits (103), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 39/135 (28%), Positives = 73/135 (54%), Gaps = 14/135 (10%)
Query 3 EVSVQLKG--KVKVAKVDSTSEQMLASKFGIQAFPTLKLFPAGPKSTAMVKDYEG-QRTA 59
+VS KG KVK+AKVD + E L + + ++PT+++F G ++K Y+ +RT
Sbjct 79 KVSEHYKGNEKVKIAKVDCSVETKLCKEQNVVSYPTMRIFSKG----NLIKQYKRPKRTH 134
Query 60 DEIVKFAMEFYGAKAEADQLLNEEQFRSECGETLCILALLPDVLDSGEEGRKNYLQTLNQ 119
+I+KF + G + + ++ + +Q +E L +L + +S E +N L+ L +
Sbjct 135 TDIIKFIEK--GIQPDIIKIQSYDQI-NELSSDLSAYPILLIMFNSETEINQN-LEFLEE 190
Query 120 VVKAS---VTVPVKF 131
+VK + VT+ V +
Sbjct 191 IVKKNDFEVTIAVTY 205
Lambda K H
0.318 0.134 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5866798756
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40