bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0475_orf1
Length=299
Score E
Sequences producing significant alignments: (Bits) Value
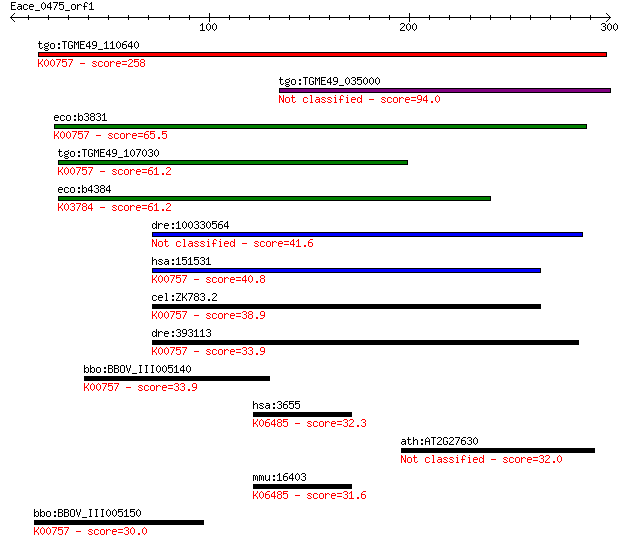
tgo:TGME49_110640 uridine phosphorylase, putative (EC:2.4.2.1)... 258 2e-68
tgo:TGME49_035000 hypothetical protein 94.0 6e-19
eco:b3831 udp, ECK3825, JW3808; uridine phosphorylase (EC:2.4.... 65.5 2e-10
tgo:TGME49_107030 purine nucleoside phosphorylase (EC:2.4.2.3)... 61.2 4e-09
eco:b4384 deoD, ECK4376, JW4347, pup; purine-nucleoside phosph... 61.2 4e-09
dre:100330564 Uridine phosphorylase 1-like 41.6 0.003
hsa:151531 UPP2, UDRPASE2, UP2, UPASE2; uridine phosphorylase ... 40.8 0.006
cel:ZK783.2 upp-1; Uridine PhosPhorylase family member (upp-1)... 38.9 0.024
dre:393113 upp2, MGC55883, uppl, zgc:55883; uridine phosphoryl... 33.9 0.66
bbo:BBOV_III005140 17.m07459; phosphorylase family protein (EC... 33.9 0.84
hsa:3655 ITGA6, CD49f, DKFZp686J01244, FLJ18737, ITGA6B, VLA-6... 32.3 2.4
ath:AT2G27630 ubiquitin carboxyl-terminal hydrolase-related 32.0 2.7
mmu:16403 Itga6, 5033401O05Rik, AI115430, Cd49f; integrin alph... 31.6 3.8
bbo:BBOV_III005150 17.m07460; hypothetical protein; K00757 uri... 30.0 10.0
> tgo:TGME49_110640 uridine phosphorylase, putative (EC:2.4.2.1);
K00757 uridine phosphorylase [EC:2.4.2.3]
Length=303
Score = 258 bits (658), Expect = 2e-68, Method: Compositional matrix adjust.
Identities = 129/289 (44%), Positives = 185/289 (64%), Gaps = 10/289 (3%)
Query 15 VFLTPDGSLYHLGVKAGEINQRILTVGDRERAEIIANAYLTDVKEYEKSRNFRTFSGVY- 73
+FLTPDG YHLGVK G++ I+TVG +RA ++A +LTDV E SR F TF+G Y
Sbjct 9 IFLTPDGRTYHLGVKKGDLASLIVTVGCEQRARVLAAKFLTDVTEVSSSRQFYTFTGKYI 68
Query 74 -KGVPVSVVSIGMGAPNMDFLVRESSYLFKDQPMAFVRVGTCGIFKPNVAPGALCLADKI 132
G +S+V IGMGAP DF +RE+S++ + +P+A VR+G+CGI PG L L+D+
Sbjct 69 KGGQRMSIVCIGMGAPMADFFIREASFVLEGEPLAVVRIGSCGIVDYATRPGTLMLSDES 128
Query 133 YYCQRNYSYFDGNVTEGSSFS----DSPYLISGPVKGDEELIEKIKSNLGKKDDVVVLKG 188
YC RNY +FDG G+ PYL++ PVK D+ L + I++NL + V +G
Sbjct 129 MYCYRNYCHFDGEAALGNKRECGRDPRPYLLTAPVKADKRLTDLIEANLNAQG-AAVKRG 187
Query 189 PAISAETFYSCQGRRFPDFSDENKSLVDTFVDLGVIGCEMESHQLYHLCKQRTDANPKKT 248
SAETF++CQGR P ++D N +L+ DLGV+ EME+HQ++HL QR + P
Sbjct 188 LNCSAETFFACQGRELPLWNDNNANLLKDMTDLGVVSLEMETHQIFHLMHQRQHSIPNGP 247
Query 249 KTRAASVCLGVVNRVDPSTPAHLTRQSEESTKQTVLAAAGAALDALISL 297
K+ AAS+ +G+VNR +P+ AH+T + ++ + LAA AA DAL++L
Sbjct 248 KSYAASLMIGLVNRSNPNLTAHVTAEDQD---KATLAAGEAAFDALVTL 293
> tgo:TGME49_035000 hypothetical protein
Length=161
Score = 94.0 bits (232), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 60/168 (35%), Positives = 90/168 (53%), Gaps = 12/168 (7%)
Query 135 CQRNYSYFDGNVTEGSSFSD-SPYLISGPVKGDEELIEKIKSNLGKK-DDVVVLK-GPAI 191
C +Y++FDG++ +G+ SPY+I+ P D EL +I N+ K DD +L G
Sbjct 3 CYNDYTFFDGSMEDGACLKGVSPYVITRPCAPDHELSNRIVENISKAVDDPSLLHLGLHA 62
Query 192 SAETFYSCQGRRFPDFSDENKSLVDTFVDLGVIGCEMESHQLYHLCKQRTDANPKKTKTR 251
SAE FY+CQG F+D N+ L+D + GV +MESHQL HL AN + +
Sbjct 63 SAENFYACQGHVDNLFNDRNEKLIDQLLSHGVDSMDMESHQLLHL------ANRRFKPVK 116
Query 252 AASVCLGVVNRVDPSTPAHLTRQSEESTKQTVLAAAGAALDALISLPL 299
AA+ +G+ +R + +T E + VL A LDAL+S+ +
Sbjct 117 AAAAHIGISHRTNDQFAHPIT---AEVLNERVLLGGRACLDALVSVQM 161
> eco:b3831 udp, ECK3825, JW3808; uridine phosphorylase (EC:2.4.2.3);
K00757 uridine phosphorylase [EC:2.4.2.3]
Length=253
Score = 65.5 bits (158), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 74/276 (26%), Positives = 116/276 (42%), Gaps = 43/276 (15%)
Query 23 LYHLGVKAGEINQRILTV--GDRERAEIIANAYLTDVKEYEKSRNFRTFSGVYKGVPVSV 80
++HLG+ ++ L + GD +R E IA A + + R F T+ G PV V
Sbjct 6 VFHLGLTKNDLQGATLAIVPGDPDRVEKIA-ALMDKPVKLASHREFTTWRAELDGKPVIV 64
Query 81 VSIGMGAPNMDFLVRESSYLFKDQPMAFVRVGTCGIFKPNVAPGALCLADKIYYCQRNYS 140
S G+G P+ V E + L F+R+GT G +P++ G + +
Sbjct 65 CSTGIGGPSTSIAVEELAQL---GIRTFLRIGTTGAIQPHINVGDVLVTTASVRL----- 116
Query 141 YFDGNVTEGSSFSDSPYLISGPVKGDEE----LIEKIKSNLGKKDDVVVLKGPAISAETF 196
+G+S +P + P D E L+E KS +G V G S++TF
Sbjct 117 -------DGASLHFAP--LEFPAVADFECTTALVEAAKS-IGATTHV----GVTASSDTF 162
Query 197 YSCQGRRFPDFSDEN----KSLVDTFVDLGVIGCEMESHQLYHLCKQRTDANPKKTKTRA 252
Y Q R+ +S K ++ + +GV+ EMES L +C + RA
Sbjct 163 YPGQ-ERYDTYSGRVVRHFKGSMEEWQAMGVMNYEMESATLLTMCASQ--------GLRA 213
Query 253 ASVCLGVVNRVDPSTP-AHLTRQSEESTKQTVLAAA 287
V +VNR P A +Q+E + V+ AA
Sbjct 214 GMVAGVIVNRTQQEIPNAETMKQTESHAVKIVVEAA 249
> tgo:TGME49_107030 purine nucleoside phosphorylase (EC:2.4.2.3);
K00757 uridine phosphorylase [EC:2.4.2.3]
Length=247
Score = 61.2 bits (147), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 47/176 (26%), Positives = 78/176 (44%), Gaps = 10/176 (5%)
Query 25 HLGVKAGEINQRILTVGDRERAEIIANAYLTDVKEYEKSRNFRTFSGVYKGVPVSVVSIG 84
H+ ++ ++ ++ VGD R E +AN +E +R +R+F VY P++V+S G
Sbjct 9 HIRLRKEDVEPVVIIVGDPARTEEVAN-MCEKKQELAYNREYRSFRVVYDSQPITVISHG 67
Query 85 MGAPNMDFLVRESSYLFKDQPMAFVRVGTCGIFKP-NVAPGALCLADKIYYCQRNYSYFD 143
+G P + E +YL +R GTCG KP + G +C + Y N +
Sbjct 68 IGCPGTSIAIEELAYLGAK---VIIRAGTCGSLKPKTLKQGDVC----VTYAAVNETGLI 120
Query 144 GNVT-EGSSFSDSPYLISGPVKGDEELIEKIKSNLGKKDDVVVLKGPAISAETFYS 198
N+ EG +P++ + +EL + S +G D G S YS
Sbjct 121 SNILPEGFPCVATPHVYQALMDAAKELGIEAASGIGVTQDYFYQNGILPSKLEMYS 176
> eco:b4384 deoD, ECK4376, JW4347, pup; purine-nucleoside phosphorylase
(EC:2.4.2.1); K03784 purine-nucleoside phosphorylase
[EC:2.4.2.1]
Length=239
Score = 61.2 bits (147), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 57/215 (26%), Positives = 88/215 (40%), Gaps = 27/215 (12%)
Query 25 HLGVKAGEINQRILTVGDRERAEIIANAYLTDVKEYEKSRNFRTFSGVYKGVPVSVVSIG 84
H+ + G+ +L GD RA+ IA +L D +E R F+G YKG +SV+ G
Sbjct 5 HINAEMGDFADVVLMPGDPLRAKYIAETFLEDAREVNNVRGMLGFTGTYKGRKISVMGHG 64
Query 85 MGAPNMDFLVRESSYLFKDQPMAFVRVGTCGIFKPNVAPGALCLADKIYYCQRNYSYFDG 144
MG P+ +E F + + +RVG+CG P+V + + + C D
Sbjct 65 MGIPSCSIYTKELITDFGVKKI--IRVGSCGAVLPHVKLRDVVIG--MGACT------DS 114
Query 145 NVTEGSSFSDSPYLISGPVKGDEELIEKIKSNLGKKDDVVVLKGPAISAETFYSCQGRRF 204
V F D + ++ K+ LG V G SA+ FYS G F
Sbjct 115 KVNR-IRFKDHDFAAIADFDMVRNAVDAAKA-LGIDARV----GNLFSADLFYSPDGEMF 168
Query 205 PDFSDENKSLVDTFVDLGVIGCEMESHQLYHLCKQ 239
D G++G EME+ +Y + +
Sbjct 169 -----------DVMEKYGILGVEMEAAGIYGVAAE 192
> dre:100330564 Uridine phosphorylase 1-like
Length=309
Score = 41.6 bits (96), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 56/224 (25%), Positives = 97/224 (43%), Gaps = 30/224 (13%)
Query 72 VYKGVPVSVVSIGMGAPNMDFLVRESSYLF---KDQPMAFVRVGTCGIFKPNVAPGALCL 128
+YK PV VS GMG P++ ++ E L + + VR+GT G + PG + +
Sbjct 93 MYKVGPVLSVSHGMGIPSISIMLHELIKLLYHARCTDVTVVRIGTSGGI--GLKPGTVVV 150
Query 129 ADKIYYCQRNYSYFDGNVTEGSSFSDSPYLISGPVKGDEELIEKIKSNLGKKDDVVVLKG 188
Q S F ++ + + P + S + D+EL E++ + + ++ G
Sbjct 151 TK-----QSVDSLFQPHLEQ--TILGKPVVRSTEL--DKELAEELLQCGKELGEFEIIIG 201
Query 189 PAISAETFYSCQGR---RFPDFSDENKS--LVDTFVDLGVIGCEMESHQLYHLCKQRTDA 243
+ FY Q R F +S+E+K L + + GV EMES +CK
Sbjct 202 NTMCTLDFYEGQARLDGAFCSYSEEDKQNYLAEAYA-AGVRNIEMESSVFAAMCKL---- 256
Query 244 NPKKTKTRAASVCLGVVNRV--DPSTPAHLTRQSEESTKQTVLA 285
+ RAA VC+ +++R D T +H + + Q ++
Sbjct 257 ----SNLRAAVVCVTLLDRQKGDQLTSSHEVLNNYQQRPQALVG 296
> hsa:151531 UPP2, UDRPASE2, UP2, UPASE2; uridine phosphorylase
2 (EC:2.4.2.3); K00757 uridine phosphorylase [EC:2.4.2.3]
Length=374
Score = 40.8 bits (94), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 50/204 (24%), Positives = 80/204 (39%), Gaps = 34/204 (16%)
Query 72 VYKGVPVSVVSIGMGAPNMDFLVRESSYLFKDQ---PMAFVRVGTCGIFKPNVAPGALCL 128
+YK PV +S GMG P++ ++ E L + +R+GT G +APG + +
Sbjct 160 MYKTGPVLAISHGMGIPSISIMLHELIKLLHHARCCDVTIIRIGTSGGI--GIAPGTVVI 217
Query 129 ----ADKIYYCQRNYSYFDGNVTEGSSFSDSPYLISGPVKGDEELIEKIKSNLGKKDDVV 184
D + + D VT + D+EL E++ + + +
Sbjct 218 TDIAVDSFFKPRFEQVILDNIVTRSTEL-------------DKELSEELFNCSKEIPNFP 264
Query 185 VLKGPAISAETFYSCQGR---RFPDFSDENK-SLVDTFVDLGVIGCEMESHQLYHLCKQR 240
L G + FY QGR FS E K + GV EMES +C
Sbjct 265 TLVGHTMCTYDFYEGQGRLDGALCSFSREKKLDYLKRAFKAGVRNIEMESTVFAAMCGL- 323
Query 241 TDANPKKTKTRAASVCLGVVNRVD 264
+AA VC+ +++R+D
Sbjct 324 -------CGLKAAVVCVTLLDRLD 340
> cel:ZK783.2 upp-1; Uridine PhosPhorylase family member (upp-1);
K00757 uridine phosphorylase [EC:2.4.2.3]
Length=295
Score = 38.9 bits (89), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 50/202 (24%), Positives = 82/202 (40%), Gaps = 31/202 (15%)
Query 72 VYKGVPVSVVSIGMGAPNMDFLVRESSYLFKDQPM---AFVRVGTCGIFKPNVAPGALCL 128
+YK PV ++ GMG P++ ++ ES L + F+R+GT G
Sbjct 84 IYKTGPVCWINHGMGTPSLSIMLVESFKLMHHAGVKNPTFIRLGTSGGVGVPPG------ 137
Query 129 ADKIYYCQRNYSYFDGNVTEGSSFSD--SPYLISGPVKGDEELIEKIKSNLGKKDDVVVL 186
S N G ++ + I P + D L E + +GK+ + V
Sbjct 138 -------TVVVSTEAMNAELGDTYVQIIAGKRIERPTQLDAALREAL-CEVGKEKSIPVE 189
Query 187 KGPAISAETFYSCQGR---RFPDFSDENK-SLVDTFVDLGVIGCEMESHQLYHLCKQRTD 242
G + A+ FY Q R F D+ +E+K + + LGV EMES
Sbjct 190 TGKTMCADDFYEGQMRLDGYFCDYEEEDKYAFLRKLNALGVRNIEMESTCFASF------ 243
Query 243 ANPKKTKTRAASVCLGVVNRVD 264
+ ++A VC+ ++NR+D
Sbjct 244 --TCRAGFQSAIVCVTLLNRMD 263
> dre:393113 upp2, MGC55883, uppl, zgc:55883; uridine phosphorylase
2 (EC:2.4.2.3); K00757 uridine phosphorylase [EC:2.4.2.3]
Length=313
Score = 33.9 bits (76), Expect = 0.66, Method: Compositional matrix adjust.
Identities = 52/225 (23%), Positives = 90/225 (40%), Gaps = 35/225 (15%)
Query 72 VYKGVPVSVVSIGMGAPNMDFLVRESSYLF---KDQPMAFVRVGTCGIFKPNVAPGALCL 128
+YK PV +S G+G P++ ++ E L + + + R+GT G +A G + +
Sbjct 99 MYKVGPVLSISHGVGVPSISIMLHELIKLLYHARCRGVIIFRIGTSG--GIGLAAGTIVI 156
Query 129 ADKIYYCQRNYSYFD---GNVTEGSSFSDSPYLISGPVKGDEELIEKIKSNLGKKDDVVV 185
DK S+F V G + S L D ++ +++ + D+
Sbjct 157 TDKAV-----DSFFRPQFEQVVLGKVITRSTEL-------DVDIAQELLQYSSELTDMPT 204
Query 186 LKGPAISAETFYSCQGR---RFPDFSDENK-SLVDTFVDLGVIGCEMESHQLYHLCKQRT 241
+ + FY QGR FS E K + GV EMES +C
Sbjct 205 VFANTMCTHDFYEGQGRLDGALCSFSTEEKFEYLRKASKAGVRNIEMESSVFAAMC---- 260
Query 242 DANPKKTKTRAASVCLGVVNRVDP---STPAHLTRQSEESTKQTV 283
+AA VC+ ++NR + STP + + ++ ++ V
Sbjct 261 ----HVCNLKAAVVCVTLLNRFEGDQISTPHDILMEYQQRPQRLV 301
> bbo:BBOV_III005140 17.m07459; phosphorylase family protein (EC:2.4.2.3);
K00757 uridine phosphorylase [EC:2.4.2.3]
Length=253
Score = 33.9 bits (76), Expect = 0.84, Method: Compositional matrix adjust.
Identities = 23/93 (24%), Positives = 45/93 (48%), Gaps = 5/93 (5%)
Query 38 LTVGDRERAEIIANAYLTDVKEYEKSRNFRTFSGVYKGVPVSVVSIGMGAPNMDFLVRES 97
+ GD +R EI + T +++ ++ Y G + + S G+G+ +VRE
Sbjct 24 IICGDHKRFEIFKE-HSTGYRDFGTYLCWKGAELFYDGEYILLASHGVGSHPTYVVVRE- 81
Query 98 SYLFKDQPMAFVRVGTCGIFKPNV-APGALCLA 129
L + +R GTCG ++P++ + G +C+
Sbjct 82 --LIELGVKCIIRSGTCGTYRPDIRSTGDICIC 112
> hsa:3655 ITGA6, CD49f, DKFZp686J01244, FLJ18737, ITGA6B, VLA-6;
integrin, alpha 6; K06485 integrin alpha 6
Length=1073
Score = 32.3 bits (72), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 24/49 (48%), Gaps = 5/49 (10%)
Query 122 APGALCLADKIYYCQRNYSYFDGNVTEGSSFSDSPYLISGPVKGDEELI 170
APG + Q+N ++FD N+ F D PY + G + DE L+
Sbjct 207 APGTYNWKGIVRVEQKNNTFFDMNI-----FEDGPYEVGGETEHDESLV 250
> ath:AT2G27630 ubiquitin carboxyl-terminal hydrolase-related
Length=1122
Score = 32.0 bits (71), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 31/109 (28%), Positives = 50/109 (45%), Gaps = 13/109 (11%)
Query 196 FYSCQGRRFPDFSDENKSL-VDTFVDLGVIGCEMESHQLYHLCKQ--RTDANP------K 246
YS QG F S++ ++L VD LG +GC E +L LC T A+
Sbjct 51 LYSEQGYMFTKLSEKAENLDVDFTFKLGAVGCSSEHEKLLSLCAHGFHTLAHHIGSVMYY 110
Query 247 KTKTRAASVCLGVVNRVD--PSTPAHLTRQSEESTK--QTVLAAAGAAL 291
K + A CL V N+ + S+ A ++ ++ K +T++ A A+
Sbjct 111 KKCLKKAKQCLSVSNQSEDSDSSVAQMSLHKNKTNKDLETLIKNAETAI 159
> mmu:16403 Itga6, 5033401O05Rik, AI115430, Cd49f; integrin alpha
6; K06485 integrin alpha 6
Length=1073
Score = 31.6 bits (70), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 23/49 (46%), Gaps = 5/49 (10%)
Query 122 APGALCLADKIYYCQRNYSYFDGNVTEGSSFSDSPYLISGPVKGDEELI 170
APG + Q+N ++FD N+ F D PY + G DE L+
Sbjct 207 APGTYNWKGIVRVEQKNNTFFDMNI-----FEDGPYEVGGETDHDESLV 250
> bbo:BBOV_III005150 17.m07460; hypothetical protein; K00757 uridine
phosphorylase [EC:2.4.2.3]
Length=260
Score = 30.0 bits (66), Expect = 10.0, Method: Compositional matrix adjust.
Identities = 20/84 (23%), Positives = 40/84 (47%), Gaps = 1/84 (1%)
Query 13 AEVFLTPDGSLYHLGVKAGEINQRILTVGDRERAEIIANAYLTDVKEYEKSRNFRTFSGV 72
A+ + P G + +GV I+ + VGD + E++A + T + + + ++ +
Sbjct 2 AKTAVCPPGIMNKIGVPLDRIHPVAIVVGDPKWLEMLA-SLATRQEFFARYKSLSSMELE 60
Query 73 YKGVPVSVVSIGMGAPNMDFLVRE 96
+ G +S G GA N+ L+ E
Sbjct 61 FNGQTFFALSFGFGATNLHRLLTE 84
Lambda K H
0.318 0.135 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 11902145332
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40