bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
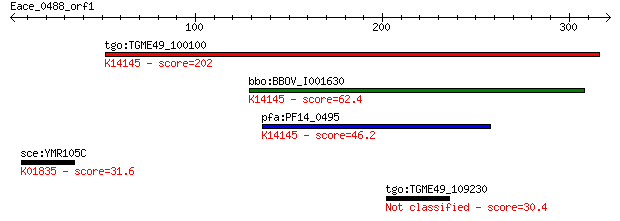
Query= Eace_0488_orf1
Length=321
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_100100 rhoptry neck protein 2 (EC:3.2.1.39); K14145... 202 1e-51
bbo:BBOV_I001630 19.m02019; hypothetical protein; K14145 rhopt... 62.4 2e-09
pfa:PF14_0495 PfRON2; rhoptry neck protein 2; K14145 rhoptry n... 46.2 2e-04
sce:YMR105C PGM2, GAL5; Pgm2p (EC:5.4.2.2); K01835 phosphogluc... 31.6 3.9
tgo:TGME49_109230 hypothetical protein 30.4 8.8
> tgo:TGME49_100100 rhoptry neck protein 2 (EC:3.2.1.39); K14145
rhoptry neck protein 2
Length=1404
Score = 202 bits (515), Expect = 1e-51, Method: Compositional matrix adjust.
Identities = 108/269 (40%), Positives = 152/269 (56%), Gaps = 29/269 (10%)
Query 52 TVDRIFGSRVEILLTCKLAALLGQPHIFLSDAIKPVEALEMVATALGL-----GDVSSTP 106
T R+ G ++ + L CKLAAL G P +FL+ + LE VA ALG+ GD +
Sbjct 315 TSKRLVGMQLGLYLICKLAALFGHPTLFLNPYYTEQQLLEAVAQALGIAPPHRGDFENE- 373
Query 107 LPAEAAAAAAGAARATAGPMFGGLEPFLMASNPLTTGHILTLLIGYIDRDAFFGSSPRKS 166
A A A +A + +E F + NP T GH+LTL+I Y+D ++FFG+SP K
Sbjct 374 --GNEAQATANQHNGSADQLLAAIEIFRLGPNPYTIGHVLTLMIAYLDYESFFGASPSKP 431
Query 167 FYNFTTLVGATGGESGILMLDEMCDRDRGPRHKTSMLARNRRKDRQGRPTARGSSKPWFV 226
F+++ +L + G +G MLDEMCD RGP+ R K W+
Sbjct 432 FHSWVSLAASAGNNTGFAMLDEMCDNHRGPK--------------------RRGQKHWYQ 471
Query 227 RGTRRQHKNKNQPELLRAYCDATEMILNAVMTKQEDIQEEMLKYSLPVEPLVDPATNAAR 286
G R+HKN++ L R CDA E++LN V Q D+ +E+ KY VEPLVDPATN+AR
Sbjct 472 TGGARKHKNRDMLPLHRQLCDALELVLNGVQQIQIDLMDELGKYKTGVEPLVDPATNSAR 531
Query 287 IQTRTYRGEAAVCEFDNSILSPVASPLDP 315
I TRT RG + VC+++ +IL+PV + L+P
Sbjct 532 IHTRTCRGLSPVCDYEATILAPVRA-LEP 559
> bbo:BBOV_I001630 19.m02019; hypothetical protein; K14145 rhoptry
neck protein 2
Length=1365
Score = 62.4 bits (150), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 49/188 (26%), Positives = 86/188 (45%), Gaps = 34/188 (18%)
Query 129 GLEPFLMA-SNPLTTGHILTLLIGYIDRDAFFGSSPRKSFYNFTTLVGATGGESGILMLD 187
G+E F + +NP GH+ T ++ Y + FF + FY + LV + + MLD
Sbjct 357 GMELFCVCGNNPFLLGHLATNMLAYTQYNMFFAGGSGRPFYTWLDLVSS----GNLDMLD 412
Query 188 EMCDRDRGPRHKTSMLARNRRKDRQGRPTARGSSKPWFVRGTRRQHKNKNQPELLR---A 244
MC RGP++K R S V RR+ K+ P+L
Sbjct 413 RMCGTKRGPKYK------------------RSSDGSVTVNKKRRR---KDDPDLDADGVF 451
Query 245 YCDATEMILNAVMTKQEDIQEEMLKYSLPVEPLVDPATNAARIQTR--TYRGEAAV---C 299
C+ E++L ++ +D+ + + K+ +P E + P N R+Q+ + + +AA+ C
Sbjct 452 LCNLLEILLISIDASIDDMGQLLSKHGVPYEHHIGPVANGRRMQSVLCSNKKDAAISVRC 511
Query 300 EFDNSILS 307
+F +S L+
Sbjct 512 DFVSSTLN 519
> pfa:PF14_0495 PfRON2; rhoptry neck protein 2; K14145 rhoptry
neck protein 2
Length=2189
Score = 46.2 bits (108), Expect = 2e-04, Method: Composition-based stats.
Identities = 35/122 (28%), Positives = 52/122 (42%), Gaps = 21/122 (17%)
Query 136 ASNPLTTGHILTLLIGYIDRDAFFGSSPRKSFYNFTTLVGATGGESGILMLDEMCDRDRG 195
ASN H L L + Y+ + +F ++ KSFY+ T++ A S ML+EMC+
Sbjct 1114 ASNIYLLAHFLVLSLAYLSYNEYF-TTGTKSFYSLPTILTANSDNS-FFMLNEMCNIHYN 1171
Query 196 PRHKTSMLARNRRKDRQGRPTARGSSKPWFVRGTRRQHKNKNQPELLRAYCDATEMILNA 255
P +N +KD P + G RR CD E++LNA
Sbjct 1172 PN-------KNFKKDITFIPIESRPKRTTTFYGERR------------LTCDLLELVLNA 1212
Query 256 VM 257
+M
Sbjct 1213 IM 1214
> sce:YMR105C PGM2, GAL5; Pgm2p (EC:5.4.2.2); K01835 phosphoglucomutase
[EC:5.4.2.2]
Length=569
Score = 31.6 bits (70), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 14/28 (50%), Positives = 19/28 (67%), Gaps = 0/28 (0%)
Query 7 PNMTYARSVATRANLERGETGAAAAGGG 34
PN+TYA S+ R + E+ E GAA+ G G
Sbjct 266 PNLTYASSLVKRVDREKIEFGAASDGDG 293
> tgo:TGME49_109230 hypothetical protein
Length=139
Score = 30.4 bits (67), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 21/39 (53%), Positives = 25/39 (64%), Gaps = 5/39 (12%)
Query 202 MLARNRR-KD--RQGRPTARGSSK--PWFVRGTRRQHKN 235
L RNRR KD R+G P +RG+SK PW RGT R+ N
Sbjct 61 FLVRNRRGKDSSRRGAPASRGNSKAAPWQRRGTLRRKFN 99
Lambda K H
0.317 0.131 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 13204534720
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40