bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0497_orf1
Length=120
Score E
Sequences producing significant alignments: (Bits) Value
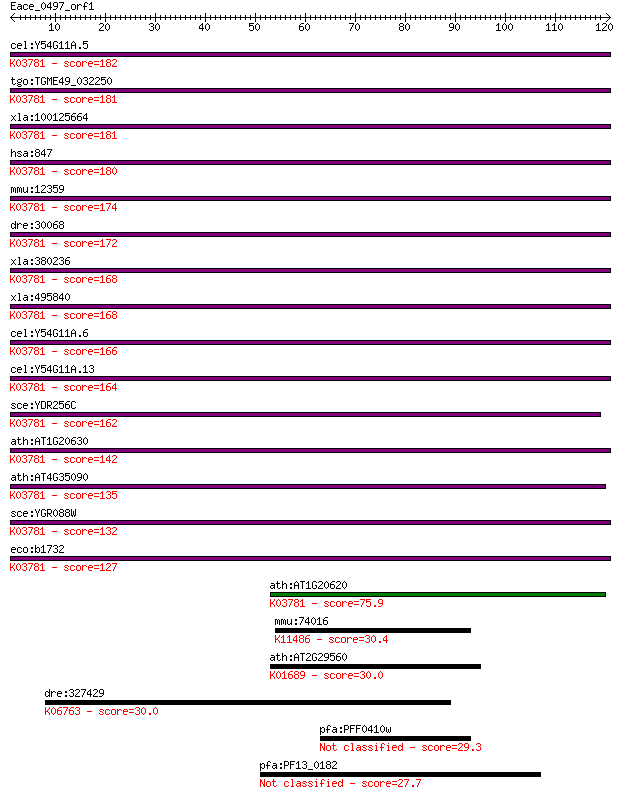
cel:Y54G11A.5 ctl-2; CaTaLase family member (ctl-2); K03781 ca... 182 2e-46
tgo:TGME49_032250 peroxisomal catalase (EC:1.11.1.6); K03781 c... 181 6e-46
xla:100125664 hypothetical protein LOC100125664; K03781 catala... 181 7e-46
hsa:847 CAT, MGC138422, MGC138424; catalase (EC:1.11.1.6); K03... 180 9e-46
mmu:12359 Cat, 2210418N07, Cas-1, Cas1, Cs-1; catalase (EC:1.1... 174 4e-44
dre:30068 cat, fb68a12, wu:fb68a12; catalase (EC:1.11.1.6); K0... 172 3e-43
xla:380236 cat, MGC64369; catalase (EC:1.11.1.6); K03781 catal... 168 4e-42
xla:495840 cat, MGC116544; catalase (EC:1.11.1.6); K03781 cata... 168 4e-42
cel:Y54G11A.6 ctl-1; CaTaLase family member (ctl-1); K03781 ca... 166 2e-41
cel:Y54G11A.13 ctl-3; CaTaLase family member (ctl-3); K03781 c... 164 5e-41
sce:YDR256C CTA1; Catalase A, breaks down hydrogen peroxide in... 162 3e-40
ath:AT1G20630 CAT1; CAT1 (CATALASE 1); catalase (EC:1.11.1.6);... 142 2e-34
ath:AT4G35090 CAT2; CAT2 (CATALASE 2); catalase (EC:1.11.1.6);... 135 3e-32
sce:YGR088W CTT1, SPS101; Ctt1p (EC:1.11.1.6); K03781 catalase... 132 4e-31
eco:b1732 katE, ECK1730, JW1721; hydroperoxidase HPII(III) (ca... 127 6e-30
ath:AT1G20620 CAT3; CAT3 (CATALASE 3); catalase; K03781 catala... 75.9 3e-14
mmu:74016 Phf19, 3110009G19Rik, 3321402G02Rik; PHD finger prot... 30.4 1.3
ath:AT2G29560 enolase, putative (EC:4.2.1.11); K01689 enolase ... 30.0 2.0
dre:327429 cntn5, fi09g01, wu:fi09g01; zgc:55318; K06763 conta... 30.0 2.0
pfa:PFF0410w conserved Plasmodium protein, unknown function 29.3 3.4
pfa:PF13_0182 ubiquitin-activating enzyme, putative 27.7 8.7
> cel:Y54G11A.5 ctl-2; CaTaLase family member (ctl-2); K03781
catalase [EC:1.11.1.6]
Length=500
Score = 182 bits (463), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 82/120 (68%), Positives = 104/120 (86%), Gaps = 0/120 (0%)
Query 1 HAKGAGAHGYFEVTDDITQYCKADIFKEIGLKTEVFVRFSTVGGERGSADTARDPRGFAI 60
HAKGAGAHGYFEVT DI++YCKADIF ++G +T + +RFSTVGGE GSADTARDPRGFAI
Sbjct 71 HAKGAGAHGYFEVTHDISKYCKADIFNKVGKQTPLLIRFSTVGGESGSADTARDPRGFAI 130
Query 61 KFYSREGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQTNLKNNDMMWGFFSLSPESLH 120
KFY+ EG +D+V NNTP+FF+RDP+ F +FIHTQKR+PQT+LK+ +M++ F+ PE+LH
Sbjct 131 KFYTEEGNWDLVGNNTPIFFIRDPIHFPNFIHTQKRNPQTHLKDPNMIFDFWLHRPEALH 190
> tgo:TGME49_032250 peroxisomal catalase (EC:1.11.1.6); K03781
catalase [EC:1.11.1.6]
Length=502
Score = 181 bits (458), Expect = 6e-46, Method: Compositional matrix adjust.
Identities = 84/120 (70%), Positives = 100/120 (83%), Gaps = 0/120 (0%)
Query 1 HAKGAGAHGYFEVTDDITQYCKADIFKEIGLKTEVFVRFSTVGGERGSADTARDPRGFAI 60
HAKG GA GYFEVT DIT++CKA +F++IG +T VF RFSTV GE GSADT RDPRGFA+
Sbjct 64 HAKGGGAFGYFEVTHDITRFCKAKLFEKIGKRTPVFARFSTVAGESGSADTRRDPRGFAL 123
Query 61 KFYSREGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQTNLKNNDMMWGFFSLSPESLH 120
KFY+ EG +DMV NNTP+FFVRD +KF DFIHTQKR PQT+L + +M+W FFSL PES+H
Sbjct 124 KFYTEEGNWDMVGNNTPIFFVRDAIKFPDFIHTQKRHPQTHLHDPNMVWDFFSLVPESVH 183
> xla:100125664 hypothetical protein LOC100125664; K03781 catalase
[EC:1.11.1.6]
Length=503
Score = 181 bits (458), Expect = 7e-46, Method: Compositional matrix adjust.
Identities = 80/120 (66%), Positives = 102/120 (85%), Gaps = 0/120 (0%)
Query 1 HAKGAGAHGYFEVTDDITQYCKADIFKEIGLKTEVFVRFSTVGGERGSADTARDPRGFAI 60
HAKGAGA GYFEVT DIT+YC+A +F+++G +T+V VRFSTV GE GSADT RDPRGFA+
Sbjct 57 HAKGAGAFGYFEVTHDITKYCRAKVFEKVGKRTDVAVRFSTVAGEAGSADTVRDPRGFAV 116
Query 61 KFYSREGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQTNLKNNDMMWGFFSLSPESLH 120
KFY+ EGI+D+V NN P+FF+RDP+ F FIH+QKR+PQT+LK+ D +W F+SL PE+LH
Sbjct 117 KFYTEEGIWDLVGNNIPIFFLRDPMMFPSFIHSQKRNPQTHLKDPDTVWDFWSLRPETLH 176
> hsa:847 CAT, MGC138422, MGC138424; catalase (EC:1.11.1.6); K03781
catalase [EC:1.11.1.6]
Length=527
Score = 180 bits (457), Expect = 9e-46, Method: Compositional matrix adjust.
Identities = 82/120 (68%), Positives = 100/120 (83%), Gaps = 0/120 (0%)
Query 1 HAKGAGAHGYFEVTDDITQYCKADIFKEIGLKTEVFVRFSTVGGERGSADTARDPRGFAI 60
HAKGAGA GYFEVT DIT+Y KA +F+ IG KT + VRFSTV GE GSADT RDPRGFA+
Sbjct 75 HAKGAGAFGYFEVTHDITKYSKAKVFEHIGKKTPIAVRFSTVAGESGSADTVRDPRGFAV 134
Query 61 KFYSREGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQTNLKNNDMMWGFFSLSPESLH 120
KFY+ +G +D+V NNTP+FF+RDP+ F FIH+QKR+PQT+LK+ DM+W F+SL PESLH
Sbjct 135 KFYTEDGNWDLVGNNTPIFFIRDPILFPSFIHSQKRNPQTHLKDPDMVWDFWSLRPESLH 194
> mmu:12359 Cat, 2210418N07, Cas-1, Cas1, Cs-1; catalase (EC:1.11.1.6);
K03781 catalase [EC:1.11.1.6]
Length=527
Score = 174 bits (442), Expect = 4e-44, Method: Compositional matrix adjust.
Identities = 80/120 (66%), Positives = 99/120 (82%), Gaps = 0/120 (0%)
Query 1 HAKGAGAHGYFEVTDDITQYCKADIFKEIGLKTEVFVRFSTVGGERGSADTARDPRGFAI 60
HAKGAGA GYFEVT DIT+Y KA +F+ IG +T + VRFSTV GE GSADT RDPRGFA+
Sbjct 75 HAKGAGAFGYFEVTHDITRYSKAKVFEHIGKRTPIAVRFSTVTGESGSADTVRDPRGFAV 134
Query 61 KFYSREGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQTNLKNNDMMWGFFSLSPESLH 120
KFY+ +G +D+V NNTP+FF+RD + F FIH+QKR+PQT+LK+ DM+W F+SL PESLH
Sbjct 135 KFYTEDGNWDLVGNNTPIFFIRDAILFPSFIHSQKRNPQTHLKDPDMVWDFWSLRPESLH 194
> dre:30068 cat, fb68a12, wu:fb68a12; catalase (EC:1.11.1.6);
K03781 catalase [EC:1.11.1.6]
Length=526
Score = 172 bits (436), Expect = 3e-43, Method: Compositional matrix adjust.
Identities = 79/120 (65%), Positives = 97/120 (80%), Gaps = 0/120 (0%)
Query 1 HAKGAGAHGYFEVTDDITQYCKADIFKEIGLKTEVFVRFSTVGGERGSADTARDPRGFAI 60
HAKGAGA GYFEVT DIT+Y KA +F+ +G T + VRFSTV GE GS+DT RDPRGFA+
Sbjct 75 HAKGAGAFGYFEVTHDITRYSKAKVFEHVGKTTPIAVRFSTVAGEAGSSDTVRDPRGFAV 134
Query 61 KFYSREGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQTNLKNNDMMWGFFSLSPESLH 120
KFY+ EG +D+ NNTP+FF+RD L F FIH+QKR+PQT+LK+ DM+W F+SL PESLH
Sbjct 135 KFYTDEGNWDLTGNNTPIFFIRDTLLFPSFIHSQKRNPQTHLKDPDMVWDFWSLRPESLH 194
> xla:380236 cat, MGC64369; catalase (EC:1.11.1.6); K03781 catalase
[EC:1.11.1.6]
Length=528
Score = 168 bits (425), Expect = 4e-42, Method: Compositional matrix adjust.
Identities = 77/120 (64%), Positives = 96/120 (80%), Gaps = 0/120 (0%)
Query 1 HAKGAGAHGYFEVTDDITQYCKADIFKEIGLKTEVFVRFSTVGGERGSADTARDPRGFAI 60
HAKGAGA GY EVT DIT+Y KA +F+ IG +T + VRFSTV GE GS+DT RDPRGFA+
Sbjct 75 HAKGAGAFGYCEVTHDITKYSKAKVFENIGKRTPIAVRFSTVAGEAGSSDTVRDPRGFAV 134
Query 61 KFYSREGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQTNLKNNDMMWGFFSLSPESLH 120
K Y+ +G +D+ NNTPVFF+RD + F FIH+QKR+PQT+LK+ DM+W F+SL PESLH
Sbjct 135 KMYTEDGNWDLTGNNTPVFFIRDAMLFPSFIHSQKRNPQTHLKDPDMVWDFWSLRPESLH 194
> xla:495840 cat, MGC116544; catalase (EC:1.11.1.6); K03781 catalase
[EC:1.11.1.6]
Length=528
Score = 168 bits (425), Expect = 4e-42, Method: Compositional matrix adjust.
Identities = 73/120 (60%), Positives = 97/120 (80%), Gaps = 0/120 (0%)
Query 1 HAKGAGAHGYFEVTDDITQYCKADIFKEIGLKTEVFVRFSTVGGERGSADTARDPRGFAI 60
HAKGAGA GYFEVT DITQYCKA +F+++G +T V RFSTV GE GS DT RDPRGFA+
Sbjct 75 HAKGAGAFGYFEVTHDITQYCKAKVFEKVGKRTPVAARFSTVAGEAGSPDTIRDPRGFAV 134
Query 61 KFYSREGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQTNLKNNDMMWGFFSLSPESLH 120
K Y+ +G +D+ NNTP+FF+RD + F F+H+Q+R+PQT++K+ +M+W F++L PESLH
Sbjct 135 KMYTEDGNWDLTGNNTPIFFIRDAILFPSFVHSQERNPQTHMKDPNMVWDFWTLRPESLH 194
> cel:Y54G11A.6 ctl-1; CaTaLase family member (ctl-1); K03781
catalase [EC:1.11.1.6]
Length=497
Score = 166 bits (419), Expect = 2e-41, Method: Compositional matrix adjust.
Identities = 73/120 (60%), Positives = 98/120 (81%), Gaps = 0/120 (0%)
Query 1 HAKGAGAHGYFEVTDDITQYCKADIFKEIGLKTEVFVRFSTVGGERGSADTARDPRGFAI 60
HAKGAGAHGYFEVT DIT+YCKAD+F ++G +T + VRFSTV GE GSADT RDPRGF++
Sbjct 71 HAKGAGAHGYFEVTHDITKYCKADMFNKVGKQTPLLVRFSTVAGESGSADTVRDPRGFSL 130
Query 61 KFYSREGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQTNLKNNDMMWGFFSLSPESLH 120
KFY+ EG +D+V NNTP+FF+RD + F +FIH KR+PQT++++ + ++ F+ PES+H
Sbjct 131 KFYTEEGNWDLVGNNTPIFFIRDAIHFPNFIHALKRNPQTHMRDPNALFDFWMNRPESIH 190
> cel:Y54G11A.13 ctl-3; CaTaLase family member (ctl-3); K03781
catalase [EC:1.11.1.6]
Length=512
Score = 164 bits (416), Expect = 5e-41, Method: Compositional matrix adjust.
Identities = 72/120 (60%), Positives = 97/120 (80%), Gaps = 0/120 (0%)
Query 1 HAKGAGAHGYFEVTDDITQYCKADIFKEIGLKTEVFVRFSTVGGERGSADTARDPRGFAI 60
HAKG GAHGYFEVT DIT+YCKAD+F ++G +T + VRFSTV GE GSADT RDPRGF++
Sbjct 86 HAKGGGAHGYFEVTHDITKYCKADMFNKVGKQTPLLVRFSTVAGESGSADTVRDPRGFSL 145
Query 61 KFYSREGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQTNLKNNDMMWGFFSLSPESLH 120
KFY+ EG +D+V NNTP+FF+RD + F +FIH KR+PQT++++ + ++ F+ PES+H
Sbjct 146 KFYTEEGNWDLVGNNTPIFFIRDAIHFPNFIHALKRNPQTHMRDPNALFDFWMNRPESIH 205
> sce:YDR256C CTA1; Catalase A, breaks down hydrogen peroxide
in the peroxisomal matrix formed by acyl-CoA oxidase (Pox1p)
during fatty acid beta-oxidation (EC:1.11.1.6); K03781 catalase
[EC:1.11.1.6]
Length=515
Score = 162 bits (409), Expect = 3e-40, Method: Compositional matrix adjust.
Identities = 76/118 (64%), Positives = 92/118 (77%), Gaps = 1/118 (0%)
Query 1 HAKGAGAHGYFEVTDDITQYCKADIFKEIGLKTEVFVRFSTVGGERGSADTARDPRGFAI 60
HA G+GA GYFEVTDDIT C + +F +IG +T+ RFSTVGG++GSADT RDPRGFA
Sbjct 70 HAHGSGAFGYFEVTDDITDICGSAMFSKIGKRTKCLTRFSTVGGDKGSADTVRDPRGFAT 129
Query 61 KFYSREGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQTNLKNNDMMWGFFSLSPES 118
KFY+ EG D V NNTPVFF+RDP KF FIHTQKR+PQTNL++ DM W F + +PE+
Sbjct 130 KFYTEEGNLDWVYNNTPVFFIRDPSKFPHFIHTQKRNPQTNLRDADMFWDFLT-TPEN 186
> ath:AT1G20630 CAT1; CAT1 (CATALASE 1); catalase (EC:1.11.1.6);
K03781 catalase [EC:1.11.1.6]
Length=492
Score = 142 bits (359), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 69/120 (57%), Positives = 88/120 (73%), Gaps = 0/120 (0%)
Query 1 HAKGAGAHGYFEVTDDITQYCKADIFKEIGLKTEVFVRFSTVGGERGSADTARDPRGFAI 60
HA+GA A G+FEVT DITQ AD + G++T V VRFSTV ERGS +T RDPRGFA+
Sbjct 65 HARGASAKGFFEVTHDITQLTSADFLRGPGVQTPVIVRFSTVIHERGSPETLRDPRGFAV 124
Query 61 KFYSREGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQTNLKNNDMMWGFFSLSPESLH 120
KFY+REG +D+V NN PVFFVRD +KF D +H K +P+++++ N + FFS PESLH
Sbjct 125 KFYTREGNFDLVGNNFPVFFVRDGMKFPDMVHALKPNPKSHIQENWRILDFFSHHPESLH 184
> ath:AT4G35090 CAT2; CAT2 (CATALASE 2); catalase (EC:1.11.1.6);
K03781 catalase [EC:1.11.1.6]
Length=474
Score = 135 bits (341), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 65/119 (54%), Positives = 86/119 (72%), Gaps = 0/119 (0%)
Query 1 HAKGAGAHGYFEVTDDITQYCKADIFKEIGLKTEVFVRFSTVGGERGSADTARDPRGFAI 60
HA+GA A G+FEVT DI+ AD + G++T V VRFSTV ERGS +T RDPRGFA+
Sbjct 65 HARGASAKGFFEVTHDISNLTCADFLRAPGVQTPVIVRFSTVIHERGSPETLRDPRGFAV 124
Query 61 KFYSREGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQTNLKNNDMMWGFFSLSPESL 119
KFY+REG +D+V NN PVFF+RD +KF D +H K +P+++++ N + FFS PESL
Sbjct 125 KFYTREGNFDLVGNNFPVFFIRDGMKFPDMVHALKPNPKSHIQENWRILDFFSHHPESL 183
> sce:YGR088W CTT1, SPS101; Ctt1p (EC:1.11.1.6); K03781 catalase
[EC:1.11.1.6]
Length=562
Score = 132 bits (331), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 63/123 (51%), Positives = 85/123 (69%), Gaps = 3/123 (2%)
Query 1 HAKGAGAHGYFEVTDDITQYCKADIFKEIGLKTEVFVRFSTVGGERGSADTARDPRGFAI 60
HAKG G FE+TD ++ A ++ +G K VRFSTVGGE G+ DTARDPRG +
Sbjct 64 HAKGGGCRLEFELTDSLSDITYAAPYQNVGYKCPGLVRFSTVGGESGTPDTARDPRGVSF 123
Query 61 KFYSREGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQTNL---KNNDMMWGFFSLSPE 117
KFY+ G +D V NNTPVFF+RD +KF FIH+QKRDPQ++L ++ + W + +L+PE
Sbjct 124 KFYTEWGNHDWVFNNTPVFFLRDAIKFPVFIHSQKRDPQSHLNQFQDTTIYWDYLTLNPE 183
Query 118 SLH 120
S+H
Sbjct 184 SIH 186
> eco:b1732 katE, ECK1730, JW1721; hydroperoxidase HPII(III) (catalase)
(EC:1.11.1.6); K03781 catalase [EC:1.11.1.6]
Length=753
Score = 127 bits (320), Expect = 6e-30, Method: Compositional matrix adjust.
Identities = 62/124 (50%), Positives = 80/124 (64%), Gaps = 4/124 (3%)
Query 1 HAKGAGAHGYFEVTDDITQYCKADIFKEIGLKTEVFVRFSTVGGERGSADTARDPRGFAI 60
HA+G+ AHGYF+ ++ KAD + T VFVRFSTV G GSADT RD RGFA
Sbjct 128 HARGSAAHGYFQPYKSLSDITKADFLSDPNKITPVFVRFSTVQGGAGSADTVRDIRGFAT 187
Query 61 KFYSREGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQTNLKN----NDMMWGFFSLSP 116
KFY+ EGI+D+V NNTP+FF++D KF DF+H K +P + +D W + SL P
Sbjct 188 KFYTEEGIFDLVGNNTPIFFIQDAHKFPDFVHAVKPEPHWAIPQGQSAHDTFWDYVSLQP 247
Query 117 ESLH 120
E+LH
Sbjct 248 ETLH 251
> ath:AT1G20620 CAT3; CAT3 (CATALASE 3); catalase; K03781 catalase
[EC:1.11.1.6]
Length=377
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 35/67 (52%), Positives = 49/67 (73%), Gaps = 0/67 (0%)
Query 53 RDPRGFAIKFYSREGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQTNLKNNDMMWGFF 112
RD RGFA+KFY+REG +D+V NNTPVFF+RD ++F D +H K +P+TN++ + +
Sbjct 2 RDIRGFAVKFYTREGNFDLVGNNTPVFFIRDGIQFPDVVHALKPNPKTNIQEYWRILDYM 61
Query 113 SLSPESL 119
S PESL
Sbjct 62 SHLPESL 68
> mmu:74016 Phf19, 3110009G19Rik, 3321402G02Rik; PHD finger protein
19; K11486 polycomb-like protein 3
Length=578
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 21/39 (53%), Gaps = 3/39 (7%)
Query 54 DPRGFAIKFYSREGIYDMVCNNTPVFFVRDPLKFVDFIH 92
+P F +FY + VCN P + R PL++VD +H
Sbjct 228 EPMVFGDRFYL---FFCSVCNQGPEYIERLPLRWVDIVH 263
> ath:AT2G29560 enolase, putative (EC:4.2.1.11); K01689 enolase
[EC:4.2.1.11]
Length=475
Score = 30.0 bits (66), Expect = 2.0, Method: Composition-based stats.
Identities = 14/45 (31%), Positives = 22/45 (48%), Gaps = 3/45 (6%)
Query 53 RDPRGFAIKFYSREGIYDM---VCNNTPVFFVRDPLKFVDFIHTQ 94
+ P F S E + DM +CN+ P+ + DP D+ HT+
Sbjct 304 KSPNKSGQNFKSAEDMIDMYKEICNDYPIVSIEDPFDKEDWEHTK 348
> dre:327429 cntn5, fi09g01, wu:fi09g01; zgc:55318; K06763 contactin
5
Length=1056
Score = 30.0 bits (66), Expect = 2.0, Method: Composition-based stats.
Identities = 24/88 (27%), Positives = 39/88 (44%), Gaps = 7/88 (7%)
Query 8 HGYFEVTDDITQYCKADIFKEIGLKTEVFVRFSTVG---GERGSADTARDPRGFAI---- 60
H EV D C+A+ I L + ++F+ +G G+ A + R+ +G +
Sbjct 118 HNASEVIDYGRYQCRAENSIGIVLSRDALLQFAYLGPFSGKTRGAVSVREGQGVVLMCAP 177
Query 61 KFYSREGIYDMVCNNTPVFFVRDPLKFV 88
+S E IY V N P F D +F+
Sbjct 178 PSHSPEIIYSWVFNELPSFVAEDSRRFI 205
> pfa:PFF0410w conserved Plasmodium protein, unknown function
Length=1948
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 11/30 (36%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 63 YSREGIYDMVCNNTPVFFVRDPLKFVDFIH 92
Y E IYD++ + + V++ LK+V+FI+
Sbjct 782 YRNENIYDLIIEYSKQYIVQNDLKYVEFIN 811
> pfa:PF13_0182 ubiquitin-activating enzyme, putative
Length=1838
Score = 27.7 bits (60), Expect = 8.7, Method: Composition-based stats.
Identities = 18/58 (31%), Positives = 24/58 (41%), Gaps = 2/58 (3%)
Query 51 TARDPRGFAIKF--YSREGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQTNLKNND 106
T F IK Y RE +C NT F R + + DF +T D T N++
Sbjct 278 TVNQKTNFNIKLNNYCRENKKKFICVNTCGLFGRVFIDYGDFYYTNNIDTNTTYDNHN 335
Lambda K H
0.323 0.140 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2018002440
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40