bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0500_orf2
Length=140
Score E
Sequences producing significant alignments: (Bits) Value
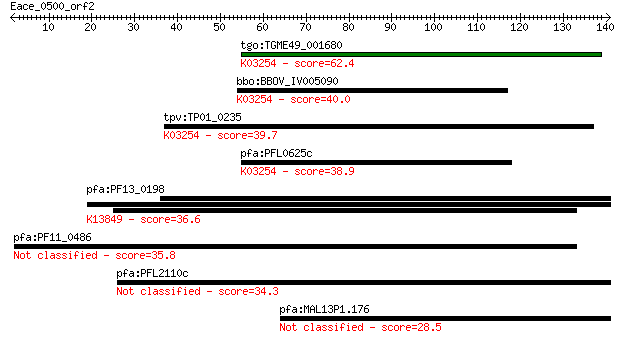
tgo:TGME49_001680 eukaryotic translation initiation factor 3 s... 62.4 4e-10
bbo:BBOV_IV005090 23.m05782; translation initiation factor 3 s... 40.0 0.003
tpv:TP01_0235 eukaryotic translation initiation factor 3 subun... 39.7 0.003
pfa:PFL0625c eukaryotic translation initiation factor 3 subuni... 38.9 0.005
pfa:PF13_0198 PfRh2a; reticulocyte binding protein 2 homolog A... 36.6 0.024
pfa:PF11_0486 MAEBL, putative 35.8 0.047
pfa:PFL2110c hypothetical protein 34.3 0.14
pfa:MAL13P1.176 PfRh2b; reticulocyte binding protein 2, homolog B 28.5 7.2
> tgo:TGME49_001680 eukaryotic translation initiation factor 3
subunit 10, putative (EC:3.1.2.15); K03254 translation initiation
factor 3 subunit A
Length=1033
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 39/84 (46%), Positives = 59/84 (70%), Gaps = 0/84 (0%)
Query 55 EEKAAFEGIQEDKELWVSRQLQQRQEQFAVAAAAQRERLIQMLQLQKIQRARERRDEEIK 114
EEK FE ++EDKE WV+ ++ RQ+ F A QRERL+ +L+ KIQRAR R++E +K
Sbjct 784 EEKQIFEVVKEDKEKWVAEKIAVRQKDFEQQAQEQRERLLHVLRQAKIQRARARKEEHLK 843
Query 115 RQKRLEEERRIEAEQKRQMEEMER 138
+ K E +R+E EQK+ +E++E+
Sbjct 844 KLKIEVERKRMEEEQKKYLEQLEK 867
> bbo:BBOV_IV005090 23.m05782; translation initiation factor 3
subunit 10; K03254 translation initiation factor 3 subunit
A
Length=991
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 29/64 (45%), Positives = 40/64 (62%), Gaps = 2/64 (3%)
Query 54 KEEK-AAFEGIQEDKELWVSRQLQQRQEQFAVAAAAQRERLIQMLQLQKIQRARERRDEE 112
KE+K A E I E KE W++ + +RQ + QR+R ++L KIQRARERRD E
Sbjct 792 KEQKNALIEAIPE-KEKWINDTMARRQAIYDAEIEEQRKRFTEILIKDKIQRARERRDYE 850
Query 113 IKRQ 116
++RQ
Sbjct 851 LRRQ 854
> tpv:TP01_0235 eukaryotic translation initiation factor 3 subunit
10; K03254 translation initiation factor 3 subunit A
Length=1107
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 36/103 (34%), Positives = 63/103 (61%), Gaps = 7/103 (6%)
Query 37 AEKSRRETRERDAKKHQKEEKAAFEGIQEDKELWVSRQLQQRQEQFAVAAAAQRERLIQM 96
+KSR + +++A+ ++EK F+ + +K+ WV +QLQ R+++ AQ R +M
Sbjct 779 VQKSREDKYKQNAEA-MRQEKILFKSFEAEKQQWVEKQLQSRKKEHQSKVDAQNARFKKM 837
Query 97 LQLQKIQRARERRDEEIKRQKRLEEERR---IEAEQKRQMEEM 136
L +KI RA+ R ++E Q RLE++R+ + EQKR+ EE+
Sbjct 838 LVEEKIARAKMRYNQE---QLRLEQKRKEEQLRLEQKRKEEEL 877
> pfa:PFL0625c eukaryotic translation initiation factor 3 subunit
10, putative; K03254 translation initiation factor 3 subunit
A
Length=1377
Score = 38.9 bits (89), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 28/73 (38%), Positives = 44/73 (60%), Gaps = 10/73 (13%)
Query 55 EEKAAFEGIQEDKELWVS----------RQLQQRQEQFAVAAAAQRERLIQMLQLQKIQR 104
E+KA FE ++KE +V +++ R E+F AQ+ERL +L+ +KIQR
Sbjct 889 EQKADFEAALKEKEEYVKFSIDIEEFTKNEMKLRVEEFEKNLKAQKERLYNILKEEKIQR 948
Query 105 ARERRDEEIKRQK 117
A+ERRD+ I++ K
Sbjct 949 AKERRDQHIRKLK 961
> pfa:PF13_0198 PfRh2a; reticulocyte binding protein 2 homolog
A; K13849 reticulocyte-binding protein
Length=3130
Score = 36.6 bits (83), Expect = 0.024, Method: Composition-based stats.
Identities = 27/105 (25%), Positives = 59/105 (56%), Gaps = 1/105 (0%)
Query 36 RAEKSRRETRERDAKKHQKEEKAAFEGIQEDKELWVSRQLQQRQEQFAVAAAAQRERLIQ 95
+ ++ R +E + K+ ++E + Q KE + RQ Q+R ++ ++ERL +
Sbjct 2740 KRQEQERLQKEEELKRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQK 2799
Query 96 MLQLQKIQRARERRDEEIKRQKRLEEERRIEAEQKRQMEEMERQK 140
+L++ ++ R R+++ + QK EE +R E E+ ++ E ++RQ+
Sbjct 2800 EEELKRQEQERLEREKQEQLQKE-EELKRQEQERLQKEEALKRQE 2843
Score = 36.2 bits (82), Expect = 0.035, Method: Composition-based stats.
Identities = 40/129 (31%), Positives = 70/129 (54%), Gaps = 8/129 (6%)
Query 19 APRRAAKVRRRTKTQRSRAEKSRRETRERDAKKHQKEEKAAFEGIQEDKELWVSRQLQQR 78
A +R + R + + + R E+ R E RE+ + ++EE E + KE + RQ Q+R
Sbjct 2738 ALKRQEQERLQKEEELKRQEQERLE-REKQEQLQKEEELKRQEQERLQKEEALKRQEQER 2796
Query 79 QEQFAVAAAAQRERLI--QMLQLQKIQRARERRDEEIKRQ---KRLEEER-RIEAEQKRQ 132
++ ++ERL + QLQK + + + E ++++ KR E+ER + E E KRQ
Sbjct 2797 LQKEEELKRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQKEEELKRQ 2856
Query 133 MEE-MERQK 140
+E +ER+K
Sbjct 2857 EQERLERKK 2865
Score = 35.0 bits (79), Expect = 0.065, Method: Composition-based stats.
Identities = 30/114 (26%), Positives = 63/114 (55%), Gaps = 6/114 (5%)
Query 25 KVRRRTKTQRSRAEKSRRETRERDAKKH--QKEEKAAFEGIQEDKELWVSRQLQQRQEQF 82
++ R + Q + E+ +R+ +ER K+ +++E+ + +E K R +++QEQ
Sbjct 2760 RLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQKEEELKRQEQERLEREKQEQL 2819
Query 83 AVAAAAQR---ERLIQMLQLQKIQRARERRDEEIKRQKRLEEER-RIEAEQKRQ 132
+R ERL + L++ ++ R +++EE+KRQ++ ER +IE ++ Q
Sbjct 2820 QKEEELKRQEQERLQKEEALKRQEQERLQKEEELKRQEQERLERKKIELAEREQ 2873
> pfa:PF11_0486 MAEBL, putative
Length=2055
Score = 35.8 bits (81), Expect = 0.047, Method: Composition-based stats.
Identities = 37/134 (27%), Positives = 69/134 (51%), Gaps = 7/134 (5%)
Query 2 SSKNERATVADESAAFGAPRRAAKVRRRTKTQRSRAEKSRRETRERDAKKHQKEEKAAFE 61
+ K E + +ES A +RA RR + +R AE +RR R A+ ++ E A
Sbjct 1087 ARKTETGRIEEESKKKEAMKRAEDARRIEEARR--AEDARRIEEARRAEDARRVEIA--R 1142
Query 62 GIQEDKELWVSRQLQ--QRQEQFAVAAAAQRERLIQMLQLQKIQRARERRDEE-IKRQKR 118
+++ + + +SR+ + +R E A +R L + ++I+ AR +E I+ +R
Sbjct 1143 RVEDARRIEISRRAEDAKRIEAARRAIEVRRAELRKAEDARRIEAARRYENERRIEEARR 1202
Query 119 LEEERRIEAEQKRQ 132
E+E+RIEA ++ +
Sbjct 1203 YEDEKRIEAVKRAE 1216
> pfa:PFL2110c hypothetical protein
Length=1846
Score = 34.3 bits (77), Expect = 0.14, Method: Composition-based stats.
Identities = 24/120 (20%), Positives = 69/120 (57%), Gaps = 5/120 (4%)
Query 26 VRRRTKTQRSRAEKSRRETRERDAKKHQKEEKAAFE-GIQEDKELWVSRQLQQRQEQFAV 84
+RR + + S E + E + R+ +K +E+K E + E+++L +++++ Q+
Sbjct 1486 IRRGEEEKMSADENMKEEQKMREEQKVGEEQKVGEEQKVGEEQKLREEQKMREEQKMREE 1545
Query 85 AAAAQRERLIQMLQLQKIQRARE----RRDEEIKRQKRLEEERRIEAEQKRQMEEMERQK 140
+ +++ + ++++ Q+ RE R +++++ ++++ EE+++ EQK + E+ R++
Sbjct 1546 QKMREEQKMREEQKVREEQKMREEQKMREEQKMREEQKVREEQKLREEQKMREEQKMREE 1605
> pfa:MAL13P1.176 PfRh2b; reticulocyte binding protein 2, homolog
B
Length=3179
Score = 28.5 bits (62), Expect = 7.2, Method: Composition-based stats.
Identities = 25/92 (27%), Positives = 48/92 (52%), Gaps = 15/92 (16%)
Query 64 QEDKELWVSRQLQQRQEQFAVAAAAQRERLIQMLQLQKIQRARERRDEEIKRQKR----- 118
QE + L +Q Q ++E ++E+ + L++ ++ R +++EE+KRQ++
Sbjct 2628 QEKERLEREKQEQLKKEALKKQEQERQEQQQKEEALKRQEQERLQKEEELKRQEQERLER 2687
Query 119 -------LEEERRI---EAEQKRQMEEMERQK 140
EEE R E +Q+R ++E+E QK
Sbjct 2688 EKQEQLQKEEELRKKEQEKQQQRNIQELEEQK 2719
Lambda K H
0.312 0.121 0.306
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2552834388
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40