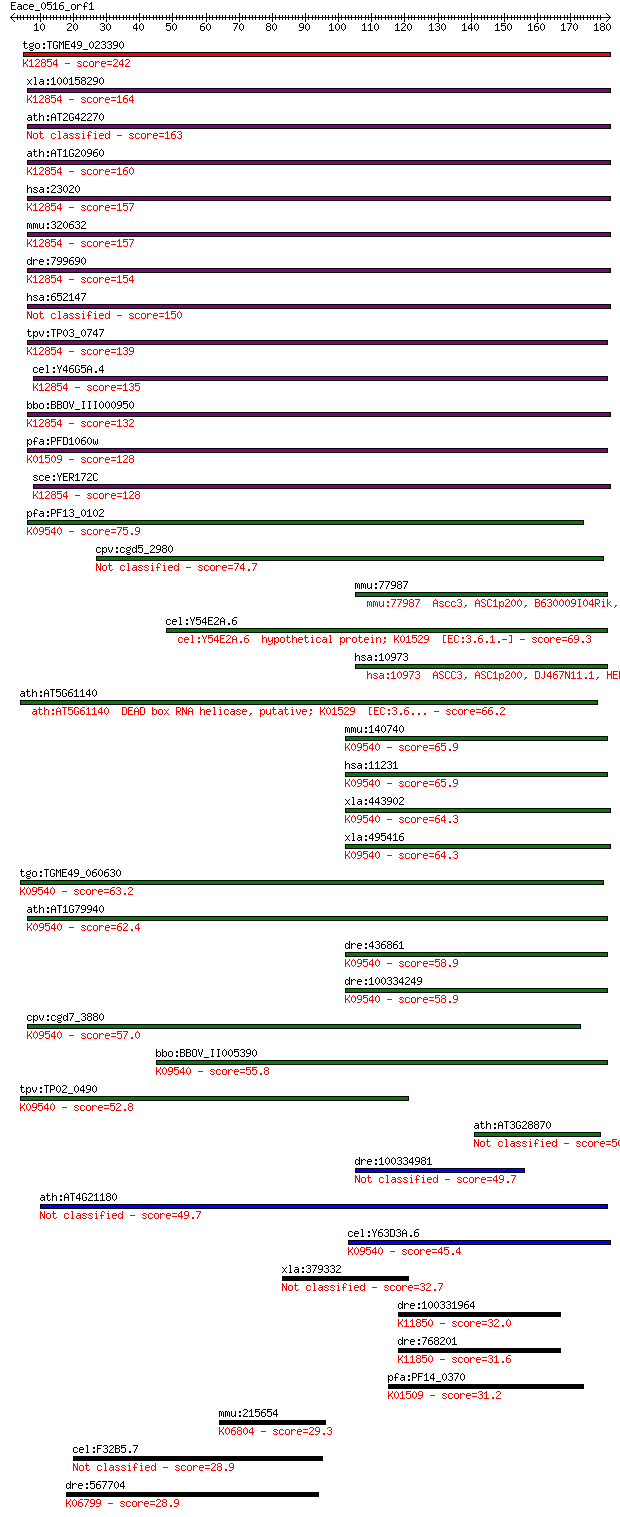
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0516_orf1
Length=181
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_023390 sec63 domain-containing DEAD/DEAH box helica... 242 5e-64
xla:100158290 snrnp200, ascc3l1; small nuclear ribonucleoprote... 164 1e-40
ath:AT2G42270 U5 small nuclear ribonucleoprotein helicase, put... 163 4e-40
ath:AT1G20960 emb1507 (embryo defective 1507); ATP binding / A... 160 3e-39
hsa:23020 SNRNP200, ASCC3L1, BRR2, FLJ11521, HELIC2, RP33, U5-... 157 2e-38
mmu:320632 Snrnp200, A330064G03Rik, Ascc3l1, BC011390, HELIC2,... 157 3e-38
dre:799690 fb63a09; wu:fb63a09 (EC:3.6.4.13); K12854 pre-mRNA-... 154 2e-37
hsa:652147 u5 small nuclear ribonucleoprotein 200 kDa helicase... 150 2e-36
tpv:TP03_0747 ATP-dependent RNA helicase; K12854 pre-mRNA-spli... 139 7e-33
cel:Y46G5A.4 hypothetical protein; K12854 pre-mRNA-splicing he... 135 8e-32
bbo:BBOV_III000950 17.m07111; sec63 domain containing protein ... 132 5e-31
pfa:PFD1060w U5 small nuclear ribonucleoprotein-specific prote... 128 9e-30
sce:YER172C BRR2, PRP44, RSS1, SLT22, SNU246; Brr2p (EC:3.6.1.... 128 1e-29
pfa:PF13_0102 DnaJ/SEC63 protein, putative; K09540 translocati... 75.9 7e-14
cpv:cgd5_2980 hypothetical protein 74.7 2e-13
mmu:77987 Ascc3, ASC1p200, B630009I04Rik, BC023451, D430001L07... 71.6 1e-12
cel:Y54E2A.6 hypothetical protein; K01529 [EC:3.6.1.-] 69.3 6e-12
hsa:10973 ASCC3, ASC1p200, DJ467N11.1, HELIC1, MGC26074, RNAH,... 69.3 7e-12
ath:AT5G61140 DEAD box RNA helicase, putative; K01529 [EC:3.6... 66.2 6e-11
mmu:140740 Sec63, 5730478J10Rik, AI649014, AW319215; SEC63-lik... 65.9 7e-11
hsa:11231 SEC63, ERdj2, PRO2507, SEC63L; SEC63 homolog (S. cer... 65.9 8e-11
xla:443902 MGC80164 protein; K09540 translocation protein SEC63 64.3 2e-10
xla:495416 sec63, MGC132056; SEC63 homolog; K09540 translocati... 64.3 2e-10
tgo:TGME49_060630 DnaJ domain-containing protein ; K09540 tran... 63.2 4e-10
ath:AT1G79940 ATERDJ2A; heat shock protein binding / unfolded ... 62.4 8e-10
dre:436861 sec63, zgc:92718; SEC63-like (S. cerevisiae); K0954... 58.9 8e-09
dre:100334249 SEC63-like protein-like; K09540 translocation pr... 58.9 8e-09
cpv:cgd7_3880 DNAJ domain protein sec63 ortholog, 4 transmembr... 57.0 4e-08
bbo:BBOV_II005390 18.m06448; DnaJ domain containing protein; K... 55.8 9e-08
tpv:TP02_0490 hypothetical protein; K09540 translocation prote... 52.8 6e-07
ath:AT3G28870 hypothetical protein 50.1 4e-06
dre:100334981 hypothetical protein LOC100334981 49.7 5e-06
ath:AT4G21180 ATERDJ2B; heat shock protein binding / unfolded ... 49.7 6e-06
cel:Y63D3A.6 dnj-29; DNaJ domain (prokaryotic heat shock prote... 45.4 1e-04
xla:379332 MGC52970; similar to RU2S 32.7 0.78
dre:100331964 ubiquitin specific peptidase 29-like; K11850 ubi... 32.0 1.2
dre:768201 usp37, MGC153999, wu:fi15b04, wu:fi38d03, zgc:15288... 31.6 1.4
pfa:PF14_0370 DEAD/DEAH box helicase, putative; K01509 adenosi... 31.2 2.0
mmu:215654 Cdh12, Cdhb, MGC92959; cadherin 12; K06804 cadherin... 29.3 7.3
cel:F32B5.7 hypothetical protein 28.9 9.5
dre:567704 cdh7; cadherin 7, type 2; K06799 cadherin 7, type 2 28.9
> tgo:TGME49_023390 sec63 domain-containing DEAD/DEAH box helicase,
putative ; K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2198
Score = 242 bits (618), Expect = 5e-64, Method: Compositional matrix adjust.
Identities = 108/177 (61%), Positives = 147/177 (83%), Gaps = 1/177 (0%)
Query 5 ASALRQLPHFSTDLVKAANAMEVKDVFDFMNMEDGQREKLLASLTQAQLREVAKASNRYP 64
SAL+QLPHF+ +LV+ A M V D+FD MNM++ +REKLL LT +QL++VAKASNRYP
Sbjct 2022 CSALKQLPHFTDELVEQAKEMGVDDIFDLMNMDEKEREKLLKPLTPSQLKDVAKASNRYP 2081
Query 65 VISLEFELSKTENISPGENIQCTVQLERDLADGDTVGPVYAPLFPKEKEEQWWLVIGQSA 124
V+++EF++SK +++ P EN+QCTV LERD A+ +T G VYAP FP+EKEEQWWLV+G+++
Sbjct 2082 VVNVEFQVSKKDDVLPNENLQCTVTLERDCAE-ETSGAVYAPYFPREKEEQWWLVVGRAS 2140
Query 125 SNGLNAIKRISVTKQSSTIKLAFEAPETPGKHQFVLFLMCDSYIGADQEHKFEIRVR 181
SN L AIKR+S+ K ++T+ L+FEAPET GKH +VL+LM DSY+G DQE+KF++RVR
Sbjct 2141 SNSLAAIKRLSLNKPTTTVTLSFEAPETDGKHSYVLYLMGDSYVGGDQEYKFDVRVR 2197
> xla:100158290 snrnp200, ascc3l1; small nuclear ribonucleoprotein
200kDa (U5); K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=457
Score = 164 bits (416), Expect = 1e-40, Method: Compositional matrix adjust.
Identities = 82/176 (46%), Positives = 120/176 (68%), Gaps = 6/176 (3%)
Query 6 SALRQLPHFSTDLVKAANAMEVKDVFDFMNMEDGQREKLLASLTQAQLREVAKASNRYPV 65
S LRQLPHFS++ +K EV+ VFD M MED +R +LL L+ +Q+ +VA+ NRYP
Sbjct 279 SYLRQLPHFSSEHIKRCTDKEVESVFDIMEMEDEERSELL-QLSDSQMADVARFCNRYPN 337
Query 66 ISLEFELSKTENISPGENIQCTVQLERDLADGDTVGPVYAPLFPKEKEEQWWLVIGQSAS 125
I L +E+++ ++I G + VQLER+ + GPV APLFP+++EE WW+VIG S S
Sbjct 338 IELSYEVAERDSIRSGGAVVVLVQLERE---EEVTGPVIAPLFPQKREEGWWVVIGDSKS 394
Query 126 NGLNAIKRISVTKQSSTIKLAFEAPETPGKHQFVLFLMCDSYIGADQEHKFEIRVR 181
N L +IKR+++ +Q + +KL F AP T G H + L+ M D+Y+G DQE+KF + V+
Sbjct 395 NSLISIKRLTL-QQKAKVKLDFVAPAT-GNHNYTLYFMSDAYMGCDQEYKFSVDVK 448
> ath:AT2G42270 U5 small nuclear ribonucleoprotein helicase, putative
Length=2172
Score = 163 bits (412), Expect = 4e-40, Method: Compositional matrix adjust.
Identities = 78/179 (43%), Positives = 117/179 (65%), Gaps = 5/179 (2%)
Query 6 SALRQLPHFSTDLVKAAN---AMEVKDVFDFMNMEDGQREKLLASLTQAQLREVAKASNR 62
S L QLPHF+ DL K + ++ +FD + MED +R++LL ++ AQL ++A+ NR
Sbjct 1986 SMLLQLPHFTKDLAKRCHENPGNNIETIFDLVEMEDDKRQELL-QMSDAQLLDIARFCNR 2044
Query 63 YPVISLEFELSKTENISPGENIQCTVQLERDLADGDTVGPVYAPLFPKEKEEQWWLVIGQ 122
+P I L +E+ + +SPG++I V LERD+ VGPV AP +PK KEE WWLV+G+
Sbjct 2045 FPNIDLTYEIVGSNEVSPGKDITLQVLLERDMEGRTEVGPVDAPRYPKTKEEGWWLVVGE 2104
Query 123 SASNGLNAIKRISVTKQSSTIKLAFEAPETPGKHQFVLFLMCDSYIGADQEHKFEIRVR 181
+ +N L AIKRIS+ +++ +KL F P G+ + L+ MCDSY+G DQE+ F + V+
Sbjct 2105 AKTNQLMAIKRISLQRKAQ-VKLEFAVPTETGEKSYTLYFMCDSYLGCDQEYSFTVDVK 2162
> ath:AT1G20960 emb1507 (embryo defective 1507); ATP binding /
ATP-dependent helicase/ helicase/ nucleic acid binding / nucleoside-triphosphatase/
nucleotide binding; K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2171
Score = 160 bits (404), Expect = 3e-39, Method: Compositional matrix adjust.
Identities = 77/179 (43%), Positives = 115/179 (64%), Gaps = 5/179 (2%)
Query 6 SALRQLPHFSTDLVKAAN---AMEVKDVFDFMNMEDGQREKLLASLTQAQLREVAKASNR 62
S L QLPHF+ DL K ++ VFD + MED +R++LL ++ AQL ++A+ NR
Sbjct 1985 SMLLQLPHFTKDLAKRCQENPGKNIETVFDLVEMEDEERQELL-KMSDAQLLDIARFCNR 2043
Query 63 YPVISLEFELSKTENISPGENIQCTVQLERDLADGDTVGPVYAPLFPKEKEEQWWLVIGQ 122
+P I L +E+ +E ++PG+ + V LERD+ VGPV + +PK KEE WWLV+G
Sbjct 2044 FPNIDLTYEIVGSEEVNPGKEVTLQVMLERDMEGRTEVGPVDSLRYPKTKEEGWWLVVGD 2103
Query 123 SASNGLNAIKRISVTKQSSTIKLAFEAPETPGKHQFVLFLMCDSYIGADQEHKFEIRVR 181
+ +N L AIKR+S+ ++ +KL F AP PG+ + L+ MCDSY+G DQE+ F + V+
Sbjct 2104 TKTNQLLAIKRVSLQRKVK-VKLDFTAPSEPGEKSYTLYFMCDSYLGCDQEYSFSVDVK 2161
> hsa:23020 SNRNP200, ASCC3L1, BRR2, FLJ11521, HELIC2, RP33, U5-200KD;
small nuclear ribonucleoprotein 200kDa (U5) (EC:3.6.4.13);
K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2136
Score = 157 bits (396), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 79/176 (44%), Positives = 116/176 (65%), Gaps = 6/176 (3%)
Query 6 SALRQLPHFSTDLVKAANAMEVKDVFDFMNMEDGQREKLLASLTQAQLREVAKASNRYPV 65
S L+QLPHF+++ +K V+ VFD M MED +R LL LT +Q+ +VA+ NRYP
Sbjct 1958 SYLKQLPHFTSEHIKRCTDKGVESVFDIMEMEDEERNALL-QLTDSQIADVARFCNRYPN 2016
Query 66 ISLEFELSKTENISPGENIQCTVQLERDLADGDTVGPVYAPLFPKEKEEQWWLVIGQSAS 125
I L +E+ ++I G + VQLER+ + GPV APLFP+++EE WW+VIG + S
Sbjct 2017 IELSYEVVDKDSIRSGGPVVVLVQLERE---EEVTGPVIAPLFPQKREEGWWVVIGDAKS 2073
Query 126 NGLNAIKRISVTKQSSTIKLAFEAPETPGKHQFVLFLMCDSYIGADQEHKFEIRVR 181
N L +IKR+++ +Q + +KL F AP T G H + L+ M D+Y+G DQE+KF + V+
Sbjct 2074 NSLISIKRLTL-QQKAKVKLDFVAPAT-GAHNYTLYFMSDAYMGCDQEYKFSVDVK 2127
> mmu:320632 Snrnp200, A330064G03Rik, Ascc3l1, BC011390, HELIC2,
KIAA0788, U5-200-KD, U5-200KD; small nuclear ribonucleoprotein
200 (U5) (EC:3.6.4.13); K12854 pre-mRNA-splicing helicase
BRR2 [EC:3.6.4.13]
Length=2136
Score = 157 bits (396), Expect = 3e-38, Method: Compositional matrix adjust.
Identities = 79/176 (44%), Positives = 116/176 (65%), Gaps = 6/176 (3%)
Query 6 SALRQLPHFSTDLVKAANAMEVKDVFDFMNMEDGQREKLLASLTQAQLREVAKASNRYPV 65
S L+QLPHF+++ +K V+ VFD M MED +R LL LT +Q+ +VA+ NRYP
Sbjct 1958 SYLKQLPHFTSEHIKRCTDKGVESVFDIMEMEDEERNALL-QLTDSQIADVARFCNRYPN 2016
Query 66 ISLEFELSKTENISPGENIQCTVQLERDLADGDTVGPVYAPLFPKEKEEQWWLVIGQSAS 125
I L +E+ ++I G + VQLER+ + GPV APLFP+++EE WW+VIG + S
Sbjct 2017 IELSYEVVDKDSIRSGGPVVVLVQLERE---EEVTGPVIAPLFPQKREEGWWVVIGDAKS 2073
Query 126 NGLNAIKRISVTKQSSTIKLAFEAPETPGKHQFVLFLMCDSYIGADQEHKFEIRVR 181
N L +IKR+++ +Q + +KL F AP T G H + L+ M D+Y+G DQE+KF + V+
Sbjct 2074 NSLISIKRLTL-QQKAKVKLDFVAPAT-GGHNYTLYFMSDAYMGCDQEYKFSVDVK 2127
> dre:799690 fb63a09; wu:fb63a09 (EC:3.6.4.13); K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2134
Score = 154 bits (388), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 78/176 (44%), Positives = 112/176 (63%), Gaps = 6/176 (3%)
Query 6 SALRQLPHFSTDLVKAANAMEVKDVFDFMNMEDGQREKLLASLTQAQLREVAKASNRYPV 65
S LRQLPHF+++L+K V+ +FD M MED R LL L+ Q+ +VA+ NRYP
Sbjct 1954 SYLRQLPHFTSELIKRCTDKGVESIFDIMEMEDEDRTGLL-QLSDVQVADVARFCNRYPN 2012
Query 66 ISLEFELSKTENISPGENIQCTVQLERDLADGDTVGPVYAPLFPKEKEEQWWLVIGQSAS 125
I L +E+ ++I G + VQLER+ + GPV APLFP+++EE WW+VIG S
Sbjct 2013 IELSYEVVDKDDIKSGSPVVVQVQLERE---EEVTGPVIAPLFPQKREEGWWVVIGDPKS 2069
Query 126 NGLNAIKRISVTKQSSTIKLAFEAPETPGKHQFVLFLMCDSYIGADQEHKFEIRVR 181
N L +IKR+++ +Q + +KL F AP G H + L+ M D+Y+G DQE+KF V+
Sbjct 2070 NSLISIKRLTL-QQKAKVKLDFVAP-VVGVHNYTLYFMSDAYMGCDQEYKFSTEVK 2123
> hsa:652147 u5 small nuclear ribonucleoprotein 200 kDa helicase-like
Length=1700
Score = 150 bits (379), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 79/176 (44%), Positives = 113/176 (64%), Gaps = 6/176 (3%)
Query 6 SALRQLPHFSTDLVKAANAMEVKDVFDFMNMEDGQREKLLASLTQAQLREVAKASNRYPV 65
S LR+LP F + L K V+ VFD M MED +R LL LT +Q+ +VA+ NRYP
Sbjct 1522 SYLRRLPPFPSGLFKRCTDKGVESVFDIMEMEDEERNALL-QLTDSQIADVARFCNRYPN 1580
Query 66 ISLEFELSKTENISPGENIQCTVQLERDLADGDTVGPVYAPLFPKEKEEQWWLVIGQSAS 125
I L +E+ ++I G + VQLER+ + GPV APLFP+++EE WW+VIG + S
Sbjct 1581 IELSYEVVDKDSIRSGGPVVVLVQLERE---EEVTGPVIAPLFPQKREEGWWVVIGDAKS 1637
Query 126 NGLNAIKRISVTKQSSTIKLAFEAPETPGKHQFVLFLMCDSYIGADQEHKFEIRVR 181
N L +IKR+++ +Q + +KL F AP T G+H L+ M D+Y+G DQE+KF + V+
Sbjct 1638 NSLISIKRLTL-QQKAKVKLDFVAPATGGRHN-TLYFMSDAYMGCDQEYKFSVDVK 1691
> tpv:TP03_0747 ATP-dependent RNA helicase; K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2249
Score = 139 bits (349), Expect = 7e-33, Method: Compositional matrix adjust.
Identities = 63/175 (36%), Positives = 111/175 (63%), Gaps = 6/175 (3%)
Query 6 SALRQLPHFSTDLVKAANAMEVKDVFDFMNMEDGQREKLLASLTQAQLREVAKASNRYPV 65
S L QLPH + + V AN+M+V D+FDF+ MED R KLL+S ++++ ++A N +
Sbjct 2080 SPLLQLPHSNREFVSKANSMKVNDLFDFIGMEDDDRNKLLSSFNKSEVLDIANFCNSIQI 2139
Query 66 ISLEFELSKTENISPGENIQCTVQLERDLADGDTVGPVYAPLFPKEKEEQWWLVIGQSAS 125
+ +EF+ + +N+ P +++ + + ++ + D + AP FP +K+EQWW+V+G +
Sbjct 2140 LDIEFKFN-NKNVKPSQSVTLLLNITKE-GNNDVIN---APYFPVDKKEQWWIVVGDTKD 2194
Query 126 NGLNAIKRISVTKQSSTIKLAFEAPETPGKHQFVLFLMCDSYIGADQEHKFEIRV 180
N L IKR S+ +++++KL EAP GKH+ L+++ DSY+ D ++K E+ V
Sbjct 2195 NKLYGIKRTSLN-ETNSVKLDIEAPSMKGKHELTLYVVSDSYVSTDYQYKLELNV 2248
> cel:Y46G5A.4 hypothetical protein; K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2145
Score = 135 bits (340), Expect = 8e-32, Method: Compositional matrix adjust.
Identities = 77/174 (44%), Positives = 106/174 (60%), Gaps = 5/174 (2%)
Query 8 LRQLPHFSTDLVKAANAMEVKDVFDFMNMEDGQREKLLASLTQAQLREVAKASNRYPVIS 67
L+QLPH S L++ A A EV VF+ + +E+ R +L + A+L +VA+ N YP I
Sbjct 1959 LKQLPHCSAALLERAKAKEVTSVFELLELENDDRSDIL-QMEGAELADVARFCNHYPSIE 2017
Query 68 LEFELSKTENISPGENIQCTVQLERDLADGDTVGPVYAPLFP-KEKEEQWWLVIGQSASN 126
+ EL + + ++ +N+ V LERD PV APLFP K KEE WWLVIG S SN
Sbjct 2018 VATEL-ENDVVTSNDNLMLAVSLERDNDIDGLAPPVVAPLFPQKRKEEGWWLVIGDSESN 2076
Query 127 GLNAIKRISVTKQSSTIKLAFEAPETPGKHQFVLFLMCDSYIGADQEHKFEIRV 180
L IKR+ + ++SS ++L F AP PG H+F LF + DSY+GADQE +V
Sbjct 2077 ALLTIKRLVINEKSS-VQLDFAAPR-PGHHKFKLFFISDSYLGADQEFDVAFKV 2128
> bbo:BBOV_III000950 17.m07111; sec63 domain containing protein
(EC:3.6.1.-); K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2133
Score = 132 bits (333), Expect = 5e-31, Method: Compositional matrix adjust.
Identities = 66/176 (37%), Positives = 103/176 (58%), Gaps = 5/176 (2%)
Query 6 SALRQLPHFSTDLVKAANAMEVKDVFDFMNMEDGQREKLLASLTQAQLREVAKASNRYPV 65
S L QLP ++ NA V D++D M MED R+ LL+ T Q +A N PV
Sbjct 1961 SCLLQLPGVGPAWIEKCNASGVHDIYDLMGMEDEDRDALLSEFTTQQCAAIANMCNAVPV 2020
Query 66 ISLEFELSKTENISPGENIQCTVQLERDLADGDTVGPVYAPLFPKEKEEQWWLVIGQSAS 125
+++E L E +P E+++ T+Q+ER+ GD VG V+APLFP E+ EQWW+++G S
Sbjct 2021 LNVECSLG-AEQAAPMESVRLTLQIERE---GD-VGTVHAPLFPVERIEQWWILVGDLDS 2075
Query 126 NGLNAIKRISVTKQSSTIKLAFEAPETPGKHQFVLFLMCDSYIGADQEHKFEIRVR 181
+ IKR+++ + + + FEAP G H+ ++++ DSY+G DQ+ + VR
Sbjct 2076 KRVLGIKRVTLLDSVNQVNIDFEAPNKLGSHELSVYVVSDSYVGTDQQQSISLHVR 2131
> pfa:PFD1060w U5 small nuclear ribonucleoprotein-specific protein,
putative; K01509 adenosinetriphosphatase [EC:3.6.1.3]
Length=2874
Score = 128 bits (322), Expect = 9e-30, Method: Composition-based stats.
Identities = 70/175 (40%), Positives = 107/175 (61%), Gaps = 1/175 (0%)
Query 6 SALRQLPHFSTDLVKAANAMEVKDVFDFMNMEDGQREKLLASLTQAQLREVAKASNRYPV 65
S L QLPHF L+K AN +E+ DV+D +N ED R+ LL L + Q E+A N +P+
Sbjct 2699 SNLYQLPHFDEHLIKKANDLEILDVYDLINAEDEPRDILLKHLNEKQRSEIANVCNIFPI 2758
Query 66 ISLEFELSKTENISPGENIQCTVQLERDLADGDTVGPVYAPLFPKEKEEQWWLVIGQSAS 125
I +++E+ ++ E Q + +ERDL D D V ++ P EKEE WW+VIG
Sbjct 2759 IEVQYEIDLDKSYKVNEIAQLNLTIERDLTD-DAVIFAHSLYLPFEKEEMWWIVIGIKKM 2817
Query 126 NGLNAIKRISVTKQSSTIKLAFEAPETPGKHQFVLFLMCDSYIGADQEHKFEIRV 180
N L +IK++S+ K + IK+ FE P+ P + V++++ D Y+G DQE++F+I V
Sbjct 2818 NLLLSIKKLSLLKSVNNIKINFELPDKPNTYDVVIYVINDCYVGCDQEYEFKINV 2872
> sce:YER172C BRR2, PRP44, RSS1, SLT22, SNU246; Brr2p (EC:3.6.1.-);
K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2163
Score = 128 bits (321), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 63/174 (36%), Positives = 110/174 (63%), Gaps = 4/174 (2%)
Query 8 LRQLPHFSTDLVKAANAMEVKDVFDFMNMEDGQREKLLASLTQAQLREVAKASNRYPVIS 67
LRQ+PHF+ +++ + V+ V+D M +ED +R+++L +LT +QL +VA N YP +
Sbjct 1994 LRQIPHFNNKILEKCKEINVETVYDIMALEDEERDEIL-TLTDSQLAQVAAFVNNYPNVE 2052
Query 68 LEFELSKTENISPGENIQCTVQLERDLADGDTVGPVYAPLFPKEKEEQWWLVIGQSASNG 127
L + L+ ++++ G + T+QL RD+ + V + +P +K E WWLV+G+ +
Sbjct 2053 LTYSLNNSDSLISGVKQKITIQLTRDVEPENL--QVTSEKYPFDKLESWWLVLGEVSKKE 2110
Query 128 LNAIKRISVTKQSSTIKLAFEAPETPGKHQFVLFLMCDSYIGADQEHKFEIRVR 181
L AIK++++ K++ +L F+ P T GKH ++ +CDSY+ AD+E FEI V+
Sbjct 2111 LYAIKKVTLNKETQQYELEFDTP-TSGKHNLTIWCVCDSYLDADKELSFEINVK 2163
> pfa:PF13_0102 DnaJ/SEC63 protein, putative; K09540 translocation
protein SEC63
Length=651
Score = 75.9 bits (185), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 51/175 (29%), Positives = 87/175 (49%), Gaps = 11/175 (6%)
Query 6 SALRQLPHFSTDLVKAANA--MEVKDVFDFMNMEDGQREKLLASLTQAQLREVAKASNRY 63
S+L Q+PHF ++V+ + VK+V DF++ +D + K L + Q+ +V N
Sbjct 388 SSLLQIPHFDENIVRHVHKGKFSVKEVLDFVH-QDHENRKGLVDMNPDQILDVKSFCNTI 446
Query 64 PVISL--EFELSKTENISPGENIQCTVQLER-DLADGDTVGPVYAPLFPKEKEEQWWLVI 120
P I + + +I G+ VQ++R +L + + G ++AP FP+ K E+WW++
Sbjct 447 PDIKMTAHIVVEDETHIVKGDVASVYVQIDRSNLKENEAAGYIHAPYFPQPKFEEWWII- 505
Query 121 GQSASNGLNAIKRISVTKQSSTI--KLAFEAPETPGKHQFVLFLMCDSYIGADQE 173
+ N +K + V I KL F + G +F +CDSY G DQ+
Sbjct 506 -ATYKNDDRILKYVHVKNCEKIIEEKLQFLVDKV-GNLSVSVFALCDSYFGCDQK 558
> cpv:cgd5_2980 hypothetical protein
Length=367
Score = 74.7 bits (182), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 53/216 (24%), Positives = 96/216 (44%), Gaps = 63/216 (29%)
Query 27 VKDVFDFMNMEDGQREKLLA---SLTQAQLREVAKASNRYPVISLEFEL----------- 72
V D++D +NM D R++LL SL++ ++ ++A+ N +P+I E+ +
Sbjct 148 VTDIYDLINMNDDDRDELLTEKLSLSEKEISQIAQVCNDFPIIETEYTILGCNDLNEKNK 207
Query 73 ---------SKTEN--------ISPGENIQCTVQLERDLA-----DGDTVG-------PV 103
+ T+N +P + ++ + RD + D D +
Sbjct 208 RRRLDSDNSTNTKNGETQRLFECNPESELTLSIDISRDFSSNEEEDNDLKNQNDIKDQEI 267
Query 104 YAPL------FPKEKEEQWWLVIGQSA-------------SNGLNAIKRISVTKQSSTIK 144
Y + +P EKEE WW+++ + A + + +I+RI + K S+ +
Sbjct 268 YPCIVKNLNYYPLEKEENWWVILIKIAVPNSSKENKDDENEDEILSIRRIQLNKISNQVL 327
Query 145 LAFEAPETPGK-HQFVLFLMCDSYIGADQEHKFEIR 179
L F + E G + L ++CDSYIG DQE F I+
Sbjct 328 LKFNSIEDSGMITNYKLLVICDSYIGCDQEFMFSIK 363
> mmu:77987 Ascc3, ASC1p200, B630009I04Rik, BC023451, D430001L07Rik,
D630041L21, Helic1, RNAH; activating signal cointegrator
1 complex subunit 3; K01529 [EC:3.6.1.-]
Length=2198
Score = 71.6 bits (174), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 29/76 (38%), Positives = 46/76 (60%), Gaps = 0/76 (0%)
Query 105 APLFPKEKEEQWWLVIGQSASNGLNAIKRISVTKQSSTIKLAFEAPETPGKHQFVLFLMC 164
P FPK K+E W+L++G+ L A+KR+ + ++F PETPG++ F L+LM
Sbjct 2105 TPRFPKLKDEGWFLILGEVDKRELMAVKRVGFVRTHHDASISFFTPETPGRYIFTLYLMS 2164
Query 165 DSYIGADQEHKFEIRV 180
D Y+G DQ++ + V
Sbjct 2165 DCYLGLDQQYDIYLNV 2180
> cel:Y54E2A.6 hypothetical protein; K01529 [EC:3.6.1.-]
Length=1798
Score = 69.3 bits (168), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 44/148 (29%), Positives = 69/148 (46%), Gaps = 16/148 (10%)
Query 48 LTQAQLREVAKASNRYPVISLEF--------------ELSKTENISPGENIQCTVQLERD 93
L +AQ REV KA +P+I+++ E K ++ GE + + +ER
Sbjct 1651 LDEAQSREVLKALCNWPIINMKIMQLVDSRGNCVDIDETKKPVKVTAGEVYKLRIVMER- 1709
Query 94 LADGDTVGPVYAPLFPKEKEEQWWLVIGQ-SASNGLNAIKRISVTKQSSTIKLAFEAPET 152
+ G ++ P +PK K+ W +V+G SA LN ST KL AP T
Sbjct 1710 VGPGKNNSSMHLPQWPKPKQAGWIIVVGNVSADMILNTTTVTGSHSTRSTAKLDIRAPAT 1769
Query 153 PGKHQFVLFLMCDSYIGADQEHKFEIRV 180
G H+ + ++ D Y+G DQE+ + V
Sbjct 1770 KGNHELAVLILSDCYLGIDQEYTLRLDV 1797
> hsa:10973 ASCC3, ASC1p200, DJ467N11.1, HELIC1, MGC26074, RNAH,
dJ121G13.4; activating signal cointegrator 1 complex subunit
3; K01529 [EC:3.6.1.-]
Length=2202
Score = 69.3 bits (168), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 27/76 (35%), Positives = 44/76 (57%), Gaps = 0/76 (0%)
Query 105 APLFPKEKEEQWWLVIGQSASNGLNAIKRISVTKQSSTIKLAFEAPETPGKHQFVLFLMC 164
P FPK K+E W+L++G+ L A+KR+ + L+F PE PG++ + L+ M
Sbjct 2104 TPRFPKSKDEGWFLILGEVDKRELIALKRVGYIRNHHVASLSFYTPEIPGRYIYTLYFMS 2163
Query 165 DSYIGADQEHKFEIRV 180
D Y+G DQ++ + V
Sbjct 2164 DCYLGLDQQYDIYLNV 2179
> ath:AT5G61140 DEAD box RNA helicase, putative; K01529 [EC:3.6.1.-]
Length=2146
Score = 66.2 bits (160), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 49/175 (28%), Positives = 82/175 (46%), Gaps = 9/175 (5%)
Query 4 QASALRQLPHFSTDLVKAANAMEVKDVFDFMNMEDGQREKLLASLTQAQLREVAKASNRY 63
Q S+L +P + L+ + A + + +N+ RE L + +++ R+
Sbjct 1974 QDSSLWMIPCMNDLLLGSLTARGIHTLHQLLNLP---RETLQSVTENFPASRLSQDLQRF 2030
Query 64 PVISLEFELSKTENISPGENIQCTVQLERDLADGDTVGPVYAPLFPKEKEEQWWLVIGQS 123
P I + L K + S G+ T+++ + AP FPK K+E WWLV+G +
Sbjct 2031 PRIQMNVRLQKKD--SDGKKKPSTLEIRLEKTSKRNSSRALAPRFPKVKDEAWWLVLGDT 2088
Query 124 ASNGLNAIKRISVTKQSSTIKLAFEAPETPGKHQFV-LFLMCDSYIGADQEHKFE 177
+++ L A+KR+S T + T E P Q L L+ D Y+G +QEH E
Sbjct 2089 STSELFAVKRVSFTGRLIT---RMELPPNITSFQDTKLILVSDCYLGFEQEHSIE 2140
> mmu:140740 Sec63, 5730478J10Rik, AI649014, AW319215; SEC63-like
(S. cerevisiae); K09540 translocation protein SEC63
Length=760
Score = 65.9 bits (159), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 30/80 (37%), Positives = 45/80 (56%), Gaps = 1/80 (1%)
Query 102 PVYAPLFPKEKEEQWWLVIGQSASNGLNAIK-RISVTKQSSTIKLAFEAPETPGKHQFVL 160
PVY+ FP+EK+E WWL I L ++ + K + ++L F AP PG +Q+ +
Sbjct 637 PVYSLYFPEEKQEWWWLYIADRKEQTLISMPYHVCTLKDTEEVELKFPAPGKPGNYQYTV 696
Query 161 FLMCDSYIGADQEHKFEIRV 180
FL DSY+G DQ ++ V
Sbjct 697 FLRSDSYMGLDQIKPLKLEV 716
> hsa:11231 SEC63, ERdj2, PRO2507, SEC63L; SEC63 homolog (S. cerevisiae);
K09540 translocation protein SEC63
Length=760
Score = 65.9 bits (159), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 30/80 (37%), Positives = 45/80 (56%), Gaps = 1/80 (1%)
Query 102 PVYAPLFPKEKEEQWWLVIGQSASNGLNAIK-RISVTKQSSTIKLAFEAPETPGKHQFVL 160
PVY+ FP+EK+E WWL I L ++ + K + ++L F AP PG +Q+ +
Sbjct 637 PVYSLYFPEEKQEWWWLYIADRKEQTLISMPYHVCTLKDTEEVELKFPAPGKPGNYQYTV 696
Query 161 FLMCDSYIGADQEHKFEIRV 180
FL DSY+G DQ ++ V
Sbjct 697 FLRSDSYMGLDQIKPLKLEV 716
> xla:443902 MGC80164 protein; K09540 translocation protein SEC63
Length=755
Score = 64.3 bits (155), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 44/81 (54%), Gaps = 1/81 (1%)
Query 102 PVYAPLFPKEKEEQWWLVIGQSASNGLNAIK-RISVTKQSSTIKLAFEAPETPGKHQFVL 160
PVY+ FP+EK+E WW+ I L ++ + K ++L F AP PG +Q+ +
Sbjct 633 PVYSLYFPEEKQEWWWIYIADRKDQTLISMPYHLCTLKDQEEVELKFPAPNKPGHYQYTV 692
Query 161 FLMCDSYIGADQEHKFEIRVR 181
FL DSY+G DQ ++ V
Sbjct 693 FLRSDSYMGLDQIKPLKLEVH 713
> xla:495416 sec63, MGC132056; SEC63 homolog; K09540 translocation
protein SEC63
Length=754
Score = 64.3 bits (155), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 44/81 (54%), Gaps = 1/81 (1%)
Query 102 PVYAPLFPKEKEEQWWLVIGQSASNGLNAIK-RISVTKQSSTIKLAFEAPETPGKHQFVL 160
PVY+ FP+EK+E WW+ I L ++ + K ++L F AP PG +Q+ +
Sbjct 632 PVYSLYFPEEKQEWWWIYIADRKDQTLISMPYHLCTLKDQEEVELKFPAPNKPGNYQYTV 691
Query 161 FLMCDSYIGADQEHKFEIRVR 181
FL DSY+G DQ ++ V
Sbjct 692 FLRSDSYMGLDQIKPLKLEVH 712
> tgo:TGME49_060630 DnaJ domain-containing protein ; K09540 translocation
protein SEC63
Length=675
Score = 63.2 bits (152), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 49/187 (26%), Positives = 88/187 (47%), Gaps = 14/187 (7%)
Query 4 QASALRQLPHFSTDLVKAA--NAMEVKDVFDFMNMEDGQREKLLASLTQAQLREVAKASN 61
++S L Q+PHF+ + V+ +++ DF+ +D + K L ++ Q ++ +
Sbjct 387 RSSTLLQVPHFTLEAVRHCQRGKHAARELGDFLK-QDPEERKGLVDMSPDQQLDIQAFCH 445
Query 62 RYPVISLEFEL--SKTENISPGENIQCTVQLER-DLADGDTVGPVYAPLFPKEKEEQWWL 118
+ + +E + I G+ C V L R +L +G+ G V+APL P K E+WW+
Sbjct 446 QVSRMKMEATVFVEDEAEIVAGDFATCQVTLTRTNLNEGEAAGAVHAPLLPMAKYEEWWI 505
Query 119 VI---GQSASNG---LNAIKRISVTKQSSTIKLAFEAPETPGKHQFVLFLMCDSYIGADQ 172
+ +SAS G LN ++ S + ++ F GK + +CDSY G D
Sbjct 506 FLVDKTESASTGGRILNFVRSKSAERVVEE-RIQFRVNRV-GKQSVTVLAICDSYAGCDC 563
Query 173 EHKFEIR 179
+ E +
Sbjct 564 TMELEFK 570
> ath:AT1G79940 ATERDJ2A; heat shock protein binding / unfolded
protein binding; K09540 translocation protein SEC63
Length=687
Score = 62.4 bits (150), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 54/221 (24%), Positives = 88/221 (39%), Gaps = 49/221 (22%)
Query 6 SALRQLPHFSTDLVKAANAMEVKDVFDFMNMEDGQREKLL---ASLTQAQLREVAKASNR 62
S QLPHFS +VK +VK D M R +LL A L+ + ++ K
Sbjct 389 SPFLQLPHFSDAVVKKIARKKVKSFQDLQEMRLEDRSELLTQVAGLSATDVEDIEKVLEM 448
Query 63 YPVISLEF--ELSKTENISPGE--NIQCTVQLERDLADGDTVGPVYAPLFPKEKEEQWWL 118
P I+++ E E I G+ +Q V L+R +G +AP FP KEE +W+
Sbjct 449 MPSITVDITCETEGEEGIQEGDIVTLQAWVTLKR--PNGLVGALPHAPYFPFHKEENYWV 506
Query 119 VIGQSASNGLNAIKRISVTKQSSTIKLA-------------------------------- 146
++ S SN + +++S + I A
Sbjct 507 LLADSVSNNVWFSQKVSFLDEGGAITAASKAISESMEGSGAGVKETNDAVREAIEKVKGG 566
Query 147 -------FEAPETPGKHQFVLFLMCDSYIGADQEHKFEIRV 180
+AP G + F +CD++IG D++ +++V
Sbjct 567 SRLVMGKLQAP-AEGTYNLTCFCLCDTWIGCDKKQALKVKV 606
> dre:436861 sec63, zgc:92718; SEC63-like (S. cerevisiae); K09540
translocation protein SEC63
Length=751
Score = 58.9 bits (141), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 27/80 (33%), Positives = 43/80 (53%), Gaps = 1/80 (1%)
Query 102 PVYAPLFPKEKEEQWWLVIGQSASNGLNAI-KRISVTKQSSTIKLAFEAPETPGKHQFVL 160
PVY+ FP+EK+E WWL I L ++ + K + ++L F AP G +Q+ +
Sbjct 630 PVYSLFFPEEKQEWWWLYIADRKEQTLVSMPNHVCTLKDTEEVELKFPAPSKTGNYQYSV 689
Query 161 FLMCDSYIGADQEHKFEIRV 180
L DS++G DQ ++ V
Sbjct 690 ILRSDSFMGLDQIKPLKLEV 709
> dre:100334249 SEC63-like protein-like; K09540 translocation
protein SEC63
Length=751
Score = 58.9 bits (141), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 27/80 (33%), Positives = 43/80 (53%), Gaps = 1/80 (1%)
Query 102 PVYAPLFPKEKEEQWWLVIGQSASNGLNAI-KRISVTKQSSTIKLAFEAPETPGKHQFVL 160
PVY+ FP+EK+E WWL I L ++ + K + ++L F AP G +Q+ +
Sbjct 630 PVYSLFFPEEKQEWWWLYIADRKEQTLVSMPNHVCTLKDTEEVELKFPAPSKTGNYQYSV 689
Query 161 FLMCDSYIGADQEHKFEIRV 180
L DS++G DQ ++ V
Sbjct 690 ILRSDSFMGLDQIKPLKLEV 709
> cpv:cgd7_3880 DNAJ domain protein sec63 ortholog, 4 transmembrane
domains ; K09540 translocation protein SEC63
Length=627
Score = 57.0 bits (136), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 44/173 (25%), Positives = 84/173 (48%), Gaps = 8/173 (4%)
Query 6 SALRQLPHFSTDLVK--AANAMEVKDVFDFMNMEDGQREKLLASLTQAQLREVAKASNRY 63
++ Q+P+ + + V+ V+++ +F+ + R K LA ++Q ++ N
Sbjct 392 ASFLQIPYITENEVQHIKKGKTAVRNLVEFIKQDPANR-KGLAEFNESQKLDIEAFCNLI 450
Query 64 PVISLEFEL--SKTENISPGENIQCTVQLER-DLADGDTVGPVYAPLFPKEKEEQWWLVI 120
I ++ ++ ++I G+ + ++R +L + + GPV++P FP K E+WW+
Sbjct 451 SPIRVDSKVIVDDEQDIVVGDLGTIEINIDRANLKENEACGPVHSPYFPTTKYEEWWIFA 510
Query 121 GQSASNG-LNAIKRISVTKQSSTIKLAFEAPETPGKHQFVLFLMCDSYIGADQ 172
SN + R S ++ +K+ F ETPG L L+ DSY G DQ
Sbjct 511 VTKGSNPQIIGYTRCSSNEKIVDVKIQFLI-ETPGSIDISLHLINDSYEGLDQ 562
> bbo:BBOV_II005390 18.m06448; DnaJ domain containing protein;
K09540 translocation protein SEC63
Length=618
Score = 55.8 bits (133), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 41/141 (29%), Positives = 71/141 (50%), Gaps = 6/141 (4%)
Query 45 LASLTQAQLREVAKASNRYPVISLEFEL--SKTENISPGENIQCTVQLER-DLADGDTV- 100
L +T++QL +V +P I L+ ++ + +I G+ I V + R ++ + TV
Sbjct 432 LVGMTESQLSDVTAFCEYFPQIDLKVDVYVNDANDICVGDVITFEVNITRLNMPENCTVV 491
Query 101 GPVYAPLFPKEKEEQWWLVIGQSASNG-LNAIKRISVTKQSSTIKLAFEAPETPGKHQFV 159
GPV+AP FP K E+W ++I ++ + A + + T K++ A E G H +
Sbjct 492 GPVHAPHFPWVKYEEWLVMINYGENDDKILAFSTCTSRGRVITEKISVLA-ENVGMHSVL 550
Query 160 LFLMCDSYIGADQEHKFEIRV 180
+ M DSY G D+ + E V
Sbjct 551 VTAMSDSYFGCDKSTRVEFMV 571
> tpv:TP02_0490 hypothetical protein; K09540 translocation protein
SEC63
Length=659
Score = 52.8 bits (125), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 34/122 (27%), Positives = 68/122 (55%), Gaps = 6/122 (4%)
Query 4 QASALRQLPHFSTDLVKAANAMEV--KDVFDFMNMEDGQREKLLASLTQAQLREVAKASN 61
++ + Q+PHF+ + + K + ++ D + +K L +LT+ Q ++V +
Sbjct 390 RSESFYQIPHFTEYEINHVGRGKASSKSIEQYVKT-DFKYKKGLNNLTEEQKQDVEEFCK 448
Query 62 RYPVISLEFEL--SKTENISPGENIQCTVQLERD-LADGDTVGPVYAPLFPKEKEEQWWL 118
+P +SLE ++ + I G+ + V+L R+ L D + +GPV+APLFP K EQ+++
Sbjct 449 YFPNVSLEVKVYVEDEDEIYEGDLVTVEVRLRRNNLKDKELIGPVHAPLFPYVKYEQYYV 508
Query 119 VI 120
++
Sbjct 509 LL 510
> ath:AT3G28870 hypothetical protein
Length=355
Score = 50.1 bits (118), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 17/38 (44%), Positives = 25/38 (65%), Gaps = 0/38 (0%)
Query 141 STIKLAFEAPETPGKHQFVLFLMCDSYIGADQEHKFEI 178
+ +KL F P PG+ + L+ MCDSY+G DQE+ F +
Sbjct 305 AKVKLDFTVPSEPGEKSYTLYFMCDSYLGCDQEYSFSV 342
> dre:100334981 hypothetical protein LOC100334981
Length=127
Score = 49.7 bits (117), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 22/51 (43%), Positives = 32/51 (62%), Gaps = 0/51 (0%)
Query 105 APLFPKEKEEQWWLVIGQSASNGLNAIKRISVTKQSSTIKLAFEAPETPGK 155
AP FPK K+E W+LV+G+ L A+KR+ + S++ +AF PE GK
Sbjct 77 APRFPKPKDEGWFLVLGEVEKKELLAVKRVGFVRNHSSVSVAFYTPEKTGK 127
> ath:AT4G21180 ATERDJ2B; heat shock protein binding / unfolded
protein binding
Length=661
Score = 49.7 bits (117), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 53/215 (24%), Positives = 92/215 (42%), Gaps = 48/215 (22%)
Query 10 QLPHFSTDLVKAANAMEVKDVFDFMNMEDGQREKLL---ASLTQAQLREVAKASNRYPV- 65
QLPHF+ + K+ A++VK F + +R KLL SL++ ++++ K P
Sbjct 390 QLPHFNESIAKSI-ALQVKSFQKFQELSLAERSKLLREVVSLSETDVQDIEKVLEMIPSL 448
Query 66 -ISLEFELSKTENISPGE--NIQCTVQLERDLADGDTVGPVYAPLFPKEKEEQWWLVIGQ 122
I++ + E I G+ +Q + L+R +G ++P FP KEE +W+++
Sbjct 449 KINVTCKTEGEEGIQEGDIMTVQAWITLKR--PNGLIGAIPHSPYFPFHKEENFWVLLAD 506
Query 123 S--------------------ASNGL---------------NAIKRISVTKQSSTIKLAF 147
S ASN + +A+K +V K S +L
Sbjct 507 SNHVWFFQKVKFMDEAGAIAAASNTITETMEPLGASVKETNDAVKE-AVEKVKSGSRLVM 565
Query 148 EAPETPGKHQFVLFLMC--DSYIGADQEHKFEIRV 180
PG+ + L C D++IG DQ+ ++ V
Sbjct 566 GRLLAPGEGTYNLTCFCLSDTWIGCDQKTSLKVEV 600
> cel:Y63D3A.6 dnj-29; DNaJ domain (prokaryotic heat shock protein)
family member (dnj-29); K09540 translocation protein SEC63
Length=752
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 25/84 (29%), Positives = 40/84 (47%), Gaps = 5/84 (5%)
Query 103 VYAPLFPKEKEEQWWLVIG-----QSASNGLNAIKRISVTKQSSTIKLAFEAPETPGKHQ 157
V+AP FP EK E WW+ + + + L + + TI + F AP G +
Sbjct 632 VHAPFFPTEKFEWWWITVAYVDKKEKSRQLLTFPQLVKTLIDEQTIDIRFAAPPHKGIYT 691
Query 158 FVLFLMCDSYIGADQEHKFEIRVR 181
+ L + DSY+ A+ F+I V+
Sbjct 692 YNLSVKSDSYMDAEYSVDFKIDVK 715
> xla:379332 MGC52970; similar to RU2S
Length=486
Score = 32.7 bits (73), Expect = 0.78, Method: Compositional matrix adjust.
Identities = 18/40 (45%), Positives = 25/40 (62%), Gaps = 5/40 (12%)
Query 83 NIQ--CTVQLERDLADGDTVGPVYAPLFPKEKEEQWWLVI 120
N+Q CTV L +A+GDT+ P L P++ EQW LV+
Sbjct 140 NVQEPCTVFL---VANGDTLNPFIRLLIPRKTLEQWELVL 176
> dre:100331964 ubiquitin specific peptidase 29-like; K11850 ubiquitin
carboxyl-terminal hydrolase 26/29/37 [EC:3.1.2.15]
Length=935
Score = 32.0 bits (71), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 28/49 (57%), Gaps = 6/49 (12%)
Query 118 LVIGQSASNGLNAIKRISVTKQSSTIKLAFEAPETPGKHQFVLFLMCDS 166
L +G SA N L+ +IS + SST++ A + PE+ G ++CDS
Sbjct 588 LSLGWSAQNALSRTLKISQSVNSSTLRRASQRPESSGS------VLCDS 630
> dre:768201 usp37, MGC153999, wu:fi15b04, wu:fi38d03, zgc:152882,
zgc:153999; ubiquitin specific peptidase 37 (EC:3.1.2.15);
K11850 ubiquitin carboxyl-terminal hydrolase 26/29/37 [EC:3.1.2.15]
Length=938
Score = 31.6 bits (70), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 28/49 (57%), Gaps = 6/49 (12%)
Query 118 LVIGQSASNGLNAIKRISVTKQSSTIKLAFEAPETPGKHQFVLFLMCDS 166
L +G SA N L+ +IS + SST++ A + PE+ G ++CDS
Sbjct 588 LSLGWSAQNALSRTLKISQSVNSSTLRKASQRPESSGS------VLCDS 630
> pfa:PF14_0370 DEAD/DEAH box helicase, putative; K01509 adenosinetriphosphatase
[EC:3.6.1.3]
Length=2472
Score = 31.2 bits (69), Expect = 2.0, Method: Composition-based stats.
Identities = 18/61 (29%), Positives = 32/61 (52%), Gaps = 3/61 (4%)
Query 115 QWWLVIGQSASNGLNAIKRISVT--KQSSTIKLAFEAPETPGKHQFVLFLMCDSYIGADQ 172
QW+ ++ + + +IKR + K+ S I E E GK+ FV+++ D+Y G +
Sbjct 2405 QWFAILHDTEQDESISIKRFNNNNLKKVSVISFTLEDMEK-GKYNFVIYIHNDTYYGIEH 2463
Query 173 E 173
E
Sbjct 2464 E 2464
> mmu:215654 Cdh12, Cdhb, MGC92959; cadherin 12; K06804 cadherin
12, type 2, N-cadherin 2
Length=794
Score = 29.3 bits (64), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 13/32 (40%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 64 PVISLEFELSKTENISPGENIQCTVQLERDLA 95
P IS+ +E + EN PG+ IQ ++RDL+
Sbjct 487 PEISVPYETAVCENAKPGQIIQIVGAVDRDLS 518
> cel:F32B5.7 hypothetical protein
Length=792
Score = 28.9 bits (63), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 20/76 (26%), Positives = 38/76 (50%), Gaps = 1/76 (1%)
Query 20 KAANAMEVK-DVFDFMNMEDGQREKLLASLTQAQLREVAKASNRYPVISLEFELSKTENI 78
+AA + V+ ++FD M ++ KL+ T QL + + + + V L+ + SK
Sbjct 404 RAAKDLTVELEIFDKMFLQRKDLIKLIGKATLDQLLPPSNSKSAHNVAELQLKASKDSEF 463
Query 79 SPGENIQCTVQLERDL 94
+ N+ +V L R+L
Sbjct 464 TAKVNVTTSVNLNREL 479
> dre:567704 cdh7; cadherin 7, type 2; K06799 cadherin 7, type
2
Length=787
Score = 28.9 bits (63), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 19/76 (25%), Positives = 38/76 (50%), Gaps = 6/76 (7%)
Query 18 LVKAANAMEVKDVFDFMNMEDGQREKLLASLTQAQLREVAKASNRYPVISLEFELSKTEN 77
L + ANA+ + +M+ Q K +A +T + + A PV ++E+E E+
Sbjct 437 LDREANAVHNITILAIESMDPSQVGKGVALITVMDINDNA------PVFAIEYETFLCES 490
Query 78 ISPGENIQCTVQLERD 93
+ PG+ I+ +++D
Sbjct 491 VGPGQVIETISAVDKD 506
Lambda K H
0.316 0.131 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4924747408
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40