bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0529_orf2
Length=204
Score E
Sequences producing significant alignments: (Bits) Value
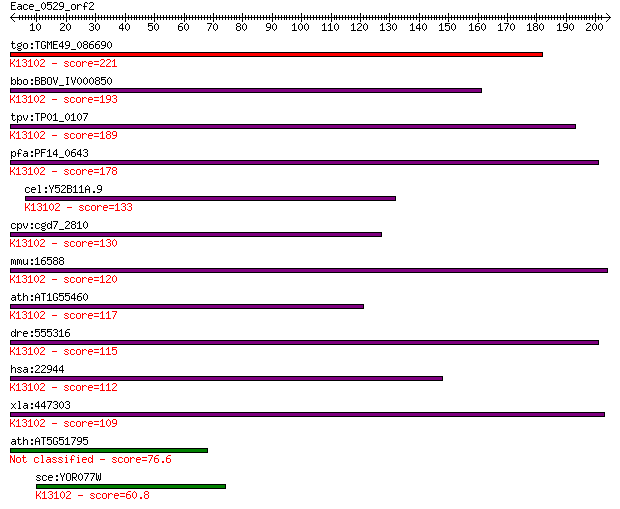
tgo:TGME49_086690 hypothetical protein ; K13102 DNA/RNA-bindin... 221 1e-57
bbo:BBOV_IV000850 21.m02949; hypothetical protein; K13102 DNA/... 193 3e-49
tpv:TP01_0107 hypothetical protein; K13102 DNA/RNA-binding pro... 189 8e-48
pfa:PF14_0643 conserved Plasmodium protein, unknown function; ... 178 1e-44
cel:Y52B11A.9 hypothetical protein; K13102 DNA/RNA-binding pro... 133 4e-31
cpv:cgd7_2810 ortholog of kin17 (RNA metabolism proteins): a f... 130 4e-30
mmu:16588 Kin, Kin17; antigenic determinant of rec-A protein; ... 120 4e-27
ath:AT1G55460 Kin17 DNA-binding protein-related; K13102 DNA/RN... 117 3e-26
dre:555316 kin, MGC123169, fc49f10, si:ch211-235a22.3, wu:fc49... 115 7e-26
hsa:22944 KIN, BTCD, KIN17; KIN, antigenic determinant of recA... 112 1e-24
xla:447303 kin, MGC81626; KIN, antigenic determinant of recA p... 109 6e-24
ath:AT5G51795 Kin17 DNA-binding protein-related 76.6 5e-14
sce:YOR077W RTS2; Basic zinc-finger protein, similar to human ... 60.8 3e-09
> tgo:TGME49_086690 hypothetical protein ; K13102 DNA/RNA-binding
protein KIN17
Length=291
Score = 221 bits (563), Expect = 1e-57, Method: Compositional matrix adjust.
Identities = 113/195 (57%), Positives = 143/195 (73%), Gaps = 14/195 (7%)
Query 1 QRPGKFMDEFSREFEEEFMKLMRTRYCRTRVLANSVYNNVVCDRHHIHMNSTIWVTLSEF 60
Q P KFMD+FS+EFE EFM+LMRTRYCRTRVLANSVYNN+V DR HIHMNSTIWVTLSEF
Sbjct 59 QNPTKFMDDFSQEFEREFMRLMRTRYCRTRVLANSVYNNIVADRQHIHMNSTIWVTLSEF 118
Query 61 VQYLGATQKCKIEHTPKGWYVEYIDHEELAKKEEEAKRRKIEKSQEDARMEQIQAMVEES 120
VQYLG+T KCKIE TPKGWYVEYIDH+E + EE+ KRRK+E SQE+A +++ +VEE+
Sbjct 119 VQYLGSTNKCKIELTPKGWYVEYIDHDERRRLEEQEKRRKVELSQEEAEARRLEELVEEA 178
Query 121 RRRGGFQEPEYTPLQREEGHTVALTFNKTQPE----------SKERKVNALQQLHEQLKQ 170
++RGG+++ EYTPL R G + +P S ER NAL+ L +LK+
Sbjct 179 KKRGGYRDAEYTPLLRRNGDEKIVLKGLARPSEKSNEMSDGHSGERPANALKLLEAELKR 238
Query 171 ER----LSKSGSQAE 181
+R ++KS + +E
Sbjct 239 KRRFAEMAKSSASSE 253
> bbo:BBOV_IV000850 21.m02949; hypothetical protein; K13102 DNA/RNA-binding
protein KIN17
Length=437
Score = 193 bits (491), Expect = 3e-49, Method: Compositional matrix adjust.
Identities = 86/160 (53%), Positives = 113/160 (70%), Gaps = 0/160 (0%)
Query 1 QRPGKFMDEFSREFEEEFMKLMRTRYCRTRVLANSVYNNVVCDRHHIHMNSTIWVTLSEF 60
Q G+FMD FSR FE EFMKLMRTRYC+T++LANSVY V+ D+ H+HMN+T+WVTLSEF
Sbjct 59 QNAGRFMDGFSRAFEHEFMKLMRTRYCKTKILANSVYQEVISDKEHVHMNATVWVTLSEF 118
Query 61 VQYLGATQKCKIEHTPKGWYVEYIDHEELAKKEEEAKRRKIEKSQEDARMEQIQAMVEES 120
V YLG + KCK+E +P+GW +EYID +++ + ++ A RRK E S E IQ MVEE+
Sbjct 119 VLYLGRSGKCKVEDSPRGWMIEYIDQDQIRRDQDAASRRKREISLEQRHQRLIQKMVEEA 178
Query 121 RRRGGFQEPEYTPLQREEGHTVALTFNKTQPESKERKVNA 160
R RGGFQEPEYTPL ++ + N + S+ N+
Sbjct 179 RARGGFQEPEYTPLMKDSDEKLCFDTNLVKSNSEPAPKNS 218
> tpv:TP01_0107 hypothetical protein; K13102 DNA/RNA-binding protein
KIN17
Length=378
Score = 189 bits (479), Expect = 8e-48, Method: Compositional matrix adjust.
Identities = 93/199 (46%), Positives = 134/199 (67%), Gaps = 8/199 (4%)
Query 1 QRPGKFMDEFSREFEEEFMKLMRTRYCRTRVLANSVYNNVVCDRHHIHMNSTIWVTLSEF 60
Q G+FMD FSR FE EFMKLMRTRYC+T++LANSVY ++ D+ H+HMN+TIWVTLSEF
Sbjct 59 QNSGRFMDGFSRTFENEFMKLMRTRYCKTKILANSVYQELISDKQHVHMNATIWVTLSEF 118
Query 61 VQYLGATQKCKIEHTPKGWYVEYIDHEELAKKEEEAKRRKIEKSQEDARMEQIQAMVEES 120
V YLG + KCK+EHTPKGWY+EYID E+L ++E+E + K E + E IQ +VEE+
Sbjct 119 VLYLGRSGKCKVEHTPKGWYIEYIDQEKLKREEDERAKMKREITLEQMHQRLIQKLVEEA 178
Query 121 RRRGGFQ-EPEYTPLQREEGHTVALTFNKTQPESKE--RKVNALQQLHEQ----LKQERL 173
+ G F+ E EYTPL ++ + + N T S+E + +N +Q + + + ++
Sbjct 179 KESGAFEHEEEYTPLMKDPDQKIVINTNLTTTNSEESNKGLNVFKQFYRKNKSTIGKQEP 238
Query 174 SKSGSQAEKRKLSAMEALV 192
++ KRK SA+E+++
Sbjct 239 ETPKTEGSKRK-SALESIM 256
> pfa:PF14_0643 conserved Plasmodium protein, unknown function;
K13102 DNA/RNA-binding protein KIN17
Length=442
Score = 178 bits (452), Expect = 1e-44, Method: Compositional matrix adjust.
Identities = 90/216 (41%), Positives = 143/216 (66%), Gaps = 16/216 (7%)
Query 1 QRPGKFMDEFSREFEEEFMKLMRTRYCRTRVLANSVYNNVVCDRHHIHMNSTIWVTLSEF 60
Q KFMD++S FE+EFM+LM+T+YCR R+LAN+VY N++ D+ HIHMN+T+WVTL++F
Sbjct 59 QDANKFMDDYSAMFEKEFMRLMKTKYCRARILANTVYTNMISDKGHIHMNATVWVTLTDF 118
Query 61 VQYLGATQKCKIEHTPKGWYVEYIDHEELAKKEEEAKRRKIEKSQEDARMEQIQAMVEES 120
V YLG T KCKIE T +GWY+EYID E++ +++ +R+KIE S E+ + ++I ++E+
Sbjct 119 VLYLGKTGKCKIEQTERGWYLEYIDREKIEREKAYQERKKIEYSYEELKEKKINEAIQEA 178
Query 121 RRRGGFQEPEYTPLQREEGHTVALTFNKTQPES---KERKVNALQQLHEQ-LKQERLSKS 176
+++G F E EYT L+++ + ++ KT ++ K+ K N L + LKQ+ +K
Sbjct 179 KKKGTFIESEYTALEKKNDEKIIISSIKTNNDTNSIKDLKPNVNIFLDKNLLKQDSQNKK 238
Query 177 GSQAE------------KRKLSAMEALVLEAEQRKQ 200
S + KR LS ++ L+LE E++K+
Sbjct 239 KSIIDDKNINHRSNTKNKRPLSELDLLILENEEKKK 274
> cel:Y52B11A.9 hypothetical protein; K13102 DNA/RNA-binding protein
KIN17
Length=404
Score = 133 bits (335), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 58/126 (46%), Positives = 86/126 (68%), Gaps = 0/126 (0%)
Query 6 FMDEFSREFEEEFMKLMRTRYCRTRVLANSVYNNVVCDRHHIHMNSTIWVTLSEFVQYLG 65
++ +FS +FE+ FM+L+RT Y RV AN VYN + D+ H+HMNST+W +L+ FVQYLG
Sbjct 64 YLRQFSNDFEKNFMQLLRTSYGTKRVRANEVYNAFIKDKGHVHMNSTVWHSLTGFVQYLG 123
Query 66 ATQKCKIEHTPKGWYVEYIDHEELAKKEEEAKRRKIEKSQEDARMEQIQAMVEESRRRGG 125
++ KCKI+ KGWY+ YID E L +KEE+ ++++ EK E+ M+ + MV+ + G
Sbjct 124 SSGKCKIDEGDKGWYIAYIDQEALIRKEEDQRKQQQEKDDEERHMQIMDGMVQRGKELAG 183
Query 126 FQEPEY 131
E EY
Sbjct 184 DDEHEY 189
> cpv:cgd7_2810 ortholog of kin17 (RNA metabolism proteins): a
family of highly conserved zinc binding C-rich proteins with
KOW domains ; K13102 DNA/RNA-binding protein KIN17
Length=265
Score = 130 bits (327), Expect = 4e-30, Method: Compositional matrix adjust.
Identities = 57/129 (44%), Positives = 91/129 (70%), Gaps = 3/129 (2%)
Query 1 QRPGKFMDEFSREFEEEFMKLMRTRYCRTRVLANSVYNNVVCDRHHIHMNSTIWVTLSEF 60
Q K ++++S++FE FM+LM+TRY +++VLAN+VYN+++ DR+HIHMNSTIW TL+EF
Sbjct 59 QNSKKILNDYSKQFELAFMRLMKTRYSKSKVLANTVYNDLIHDRNHIHMNSTIWTTLTEF 118
Query 61 VQYLGATQKCKIEHTPKGWYVEYIDHEELAKKEEEAKRRKIEKSQEDARMEQIQAMVE-- 118
+ YL T KC+IE++ KGWY++YI HE K ++ + K ++++ R +I +V+
Sbjct 119 IHYLSNTNKCEIEYSDKGWYIQYIGHEGEKKNKDLDSKNKFRLTEDNRRERRIGMVVKSN 178
Query 119 -ESRRRGGF 126
S +R F
Sbjct 179 SSSNQRASF 187
> mmu:16588 Kin, Kin17; antigenic determinant of rec-A protein;
K13102 DNA/RNA-binding protein KIN17
Length=391
Score = 120 bits (301), Expect = 4e-27, Method: Compositional matrix adjust.
Identities = 81/222 (36%), Positives = 122/222 (54%), Gaps = 26/222 (11%)
Query 1 QRPGKFMDEFSREFEEEFMKLMRTRYCRTRVLANSVYNNVVCDRHHIHMNSTIWVTLSEF 60
+ P +FMD FS EF +F++L+R R+ RV N VYN + R HIHMN+T W TL++F
Sbjct 59 ENPQQFMDYFSEEFRNDFLELLRRRFGTKRVHNNIVYNEYISHREHIHMNATQWETLTDF 118
Query 61 VQYLGATQKCKIEHTPKGWYVEYIDHEELAKKEEEAKRRKIE----KSQEDARMEQIQAM 116
++LG CK++ TPKGWY++YID ++ E RR++E K Q+ E+
Sbjct 119 TKWLGREGLCKVDETPKGWYIQYID------RDPETIRRQLELEKKKKQDLDDEEKTAKF 172
Query 117 VEESRRRG--GFQE--PEYTPLQRE-EGHTVALTFNK--------TQPESKERKVNALQQ 163
+EE RRG G ++ P +T L RE E V NK T +S +AL+
Sbjct 173 IEEQVRRGLEGKEQETPVFTELSRENEEEKVTFNLNKGAGGSAGATTSKSSSLGPSALKL 232
Query 164 LHEQLKQER--LSKSGSQAEKRKLSAMEALVLEAEQRKQKAA 203
L +R S+S +Q K+K SA++ ++E E+ K++ A
Sbjct 233 LGSAASGKRKESSQSSAQPAKKKKSALDE-IMELEEEKKRTA 273
> ath:AT1G55460 Kin17 DNA-binding protein-related; K13102 DNA/RNA-binding
protein KIN17
Length=411
Score = 117 bits (293), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 55/122 (45%), Positives = 82/122 (67%), Gaps = 2/122 (1%)
Query 1 QRPGKFMDEFSREFEEEFMKLMRTRYCRTRVLANSVYNNVVCDRHHIHMNSTIWVTLSEF 60
Q P + +D +S EFE+ F+ LMR + +R+ A VYN + DRHH+HMNST W TL+EF
Sbjct 59 QNPTRVVDGYSEEFEQTFLDLMRRSHRFSRIAATVVYNEYINDRHHVHMNSTEWATLTEF 118
Query 61 VQYLGATQKCKIEHTPKGWYVEYIDH--EELAKKEEEAKRRKIEKSQEDARMEQIQAMVE 118
+++LG T KCK+E TPKGW++ YID E L K+ + KR K + ++E+ + +IQ +E
Sbjct 119 IKHLGKTGKCKVEETPKGWFITYIDRDSETLFKERLKNKRVKSDLAEEEKQEREIQRQIE 178
Query 119 ES 120
+
Sbjct 179 RA 180
> dre:555316 kin, MGC123169, fc49f10, si:ch211-235a22.3, wu:fc49f10,
zgc:123169; KIN, antigenic determinant of recA protein
homolog (mouse); K13102 DNA/RNA-binding protein KIN17
Length=385
Score = 115 bits (289), Expect = 7e-26, Method: Compositional matrix adjust.
Identities = 74/212 (34%), Positives = 122/212 (57%), Gaps = 17/212 (8%)
Query 1 QRPGKFMDEFSREFEEEFMKLMRTRYCRTRVLANSVYNNVVCDRHHIHMNSTIWVTLSEF 60
+ P +F+ FS EF+ +F++L+R R+ RV N VYN + R H+HMN+T W TL++F
Sbjct 59 ENPNQFLSYFSDEFKTDFLELLRRRFGTKRVHNNIVYNEYISHREHVHMNATQWETLTDF 118
Query 61 VQYLGATQKCKIEHTPKGWYVEYIDHE-ELAKKEEEAKRRKIEKSQEDARMEQIQAMVEE 119
++LG CK++ PKGWYV+YID + E +++EE +R+K + ++ R + +EE
Sbjct 119 TKWLGREGFCKVDEAPKGWYVQYIDRDPETIRRQEELERKKKQDLDDEERSAK---FIEE 175
Query 120 SRRRG-----GFQEPEYTPLQRE-EGHTVALTF-----NKTQPESKERKVNALQQLHEQL 168
RRG +EP +T LQR+ E VA ++P S +AL+ +
Sbjct 176 QVRRGREGREPTEEPVFTELQRKSEDEKVAFNLEIAAAGPSKP-STTLGPSALKTT-ASV 233
Query 169 KQERLSKSGSQAEKRKLSAMEALVLEAEQRKQ 200
K++ S SG +K+K SA++ ++ EQ+K+
Sbjct 234 KRKESSLSGEFKDKKKKSALDEIIEMEEQKKK 265
> hsa:22944 KIN, BTCD, KIN17; KIN, antigenic determinant of recA
protein homolog (mouse); K13102 DNA/RNA-binding protein KIN17
Length=393
Score = 112 bits (280), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 63/155 (40%), Positives = 91/155 (58%), Gaps = 15/155 (9%)
Query 1 QRPGKFMDEFSREFEEEFMKLMRTRYCRTRVLANSVYNNVVCDRHHIHMNSTIWVTLSEF 60
+ P +FMD FS EF +F++L+R R+ RV N VYN + R HIHMN+T W TL++F
Sbjct 59 ENPQQFMDYFSEEFRNDFLELLRRRFGTKRVHNNIVYNEYISHREHIHMNATQWETLTDF 118
Query 61 VQYLGATQKCKIEHTPKGWYVEYIDHEELAKKEEEAKRRKIE----KSQEDARMEQIQAM 116
++LG CK++ TPKGWY++YID ++ E RR++E K Q+ E+
Sbjct 119 TKWLGREGLCKVDETPKGWYIQYID------RDPETIRRQLELEKKKKQDLDDEEKTAKF 172
Query 117 VEESRRRG--GFQE--PEYTPLQREEGHTVALTFN 147
+EE RRG G ++ P +T L RE +TFN
Sbjct 173 IEEQVRRGLEGKEQEVPTFTELSRENDEE-KVTFN 206
> xla:447303 kin, MGC81626; KIN, antigenic determinant of recA
protein homolog; K13102 DNA/RNA-binding protein KIN17
Length=387
Score = 109 bits (273), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 72/213 (33%), Positives = 119/213 (55%), Gaps = 14/213 (6%)
Query 1 QRPGKFMDEFSREFEEEFMKLMRTRYCRTRVLANSVYNNVVCDRHHIHMNSTIWVTLSEF 60
+ P + MD FS EF EF++L++ R+ RV N VYN + R H+HMN+T W TL++F
Sbjct 59 ENPQQIMDSFSDEFRTEFLELLKRRFGTKRVHNNIVYNEYIGHREHVHMNATQWETLTDF 118
Query 61 VQYLGATQKCKIEHTPKGWYVEYIDHEELAKKEEEAKRRKIEKSQEDARMEQIQAMVEES 120
++LG CK++ TPKGWY++YID + ++++ + +K ++ +D E+ +E+
Sbjct 119 TKWLGREGYCKVDETPKGWYIQYIDRDPETIRKQQEQEKKKKQDLDDE--ERTSKFIEDQ 176
Query 121 RRRG--GFQE--PEYTPLQRE-EGHTVALTFNK-----TQPESKERKVNALQQLHEQLKQ 170
++G G ++ P +T L R+ E VA NK S NAL+
Sbjct 177 VKKGLEGVEQNTPTFTELSRQNEEEKVAFNLNKGAGTSGTTASSSHASNALKSAAVMGSV 236
Query 171 ERLSKSGSQA-EKRKLSAMEALVLEAEQRKQKA 202
+R S QA EK+K SA++ ++E E++K+K
Sbjct 237 KRKDTSQGQAKEKKKKSALDE-IMEMEEQKKKT 268
> ath:AT5G51795 Kin17 DNA-binding protein-related
Length=347
Score = 76.6 bits (187), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 33/67 (49%), Positives = 47/67 (70%), Gaps = 0/67 (0%)
Query 1 QRPGKFMDEFSREFEEEFMKLMRTRYCRTRVLANSVYNNVVCDRHHIHMNSTIWVTLSEF 60
Q P + ++ +S+EFE+ F+ LMR + +RV A VYN + DR H+HMNST W TL+EF
Sbjct 75 QNPTRVVNGYSQEFEQTFLDLMRRSHRFSRVAATVVYNEYINDRLHVHMNSTKWATLTEF 134
Query 61 VQYLGAT 67
++YLG T
Sbjct 135 IKYLGKT 141
> sce:YOR077W RTS2; Basic zinc-finger protein, similar to human
and mouse Kin17 proteins which are chromatin-associated proteins
involved in UV response and DNA replication; K13102 DNA/RNA-binding
protein KIN17
Length=232
Score = 60.8 bits (146), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 25/64 (39%), Positives = 44/64 (68%), Gaps = 0/64 (0%)
Query 10 FSREFEEEFMKLMRTRYCRTRVLANSVYNNVVCDRHHIHMNSTIWVTLSEFVQYLGATQK 69
++ +FE+ F++L++ R+ + AN VYN V DR H+HMN+T+ +L++FV+YLG K
Sbjct 63 YNIQFEKGFLQLLKQRHGEKWIDANKVYNEYVQDRDHVHMNATMHRSLTQFVRYLGRAGK 122
Query 70 CKIE 73
++
Sbjct 123 VDVD 126
Lambda K H
0.314 0.127 0.354
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6253620652
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40