bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0571_orf1
Length=184
Score E
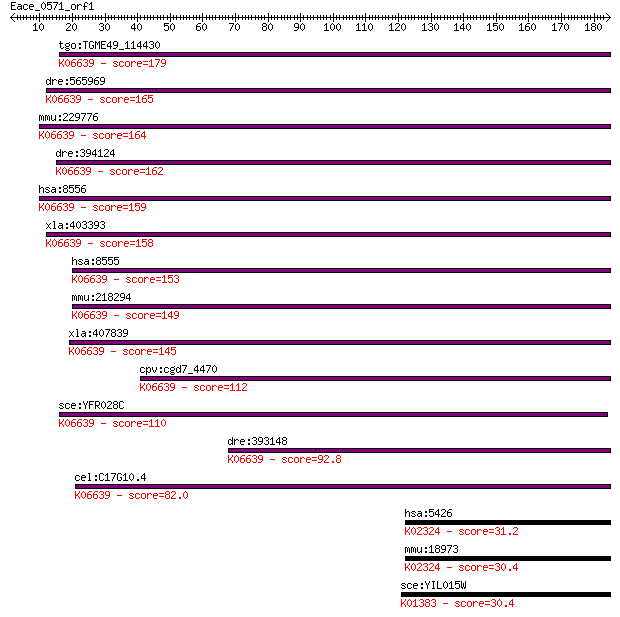
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_114430 dual specificity protein phosphatase CDC14A,... 179 4e-45
dre:565969 cdc14ab, si:dkey-168j9.1; CDC14 cell division cycle... 165 8e-41
mmu:229776 Cdc14a, A830059A17Rik, CDC14A2, CDC14a1, Cdc14; CDC... 164 2e-40
dre:394124 cdc14aa, CDC14A, MGC63654, zgc:63654; CDC14 cell di... 162 5e-40
hsa:8556 CDC14A, cdc14, hCDC14; CDC14 cell division cycle 14 h... 159 4e-39
xla:403393 cdc14a, xcdc14a; CDC14 cell division cycle 14 homol... 158 8e-39
hsa:8555 CDC14B, CDC14B3, Cdc14B1, Cdc14B2, hCDC14B; CDC14 cel... 153 3e-37
mmu:218294 Cdc14b, 2810432N10Rik, A530086E13Rik, AA472821, CDC... 149 4e-36
xla:407839 cdc14b, MGC81657, cdc14beta, xcdc14b; CDC14 cell di... 145 6e-35
cpv:cgd7_4470 CDC14 phosphatase ; K06639 cell division cycle 1... 112 1e-24
sce:YFR028C CDC14, OAF3; Cdc14p (EC:3.1.3.48); K06639 cell div... 110 3e-24
dre:393148 cdc14b, MGC55844, cdc14a, zgc:55844; CDC14 cell div... 92.8 6e-19
cel:C17G10.4 cdc-14; Cell Division Cycle related family member... 82.0 9e-16
hsa:5426 POLE, DKFZp434F222, FLJ21434, POLE1; polymerase (DNA ... 31.2 2.1
mmu:18973 Pole; polymerase (DNA directed), epsilon (EC:2.7.7.7... 30.4 3.4
sce:YIL015W BAR1, SST1; Aspartyl protease secreted into the pe... 30.4 3.6
> tgo:TGME49_114430 dual specificity protein phosphatase CDC14A,
putative (EC:3.1.3.16); K06639 cell division cycle 14 [EC:3.1.3.48]
Length=479
Score = 179 bits (454), Expect = 4e-45, Method: Compositional matrix adjust.
Identities = 80/169 (47%), Positives = 116/169 (68%), Gaps = 0/169 (0%)
Query 16 LREAVEVLQGRLYFRSVFSPPRDTPTIHFFSTDKLFVYEPFFLDFGPLNLSCIYRYCMLL 75
LR+AV+++ GR +F S+ P DT +IHFF TD+ F YEPFF DFGPL+L CIY+Y LL
Sbjct 16 LRDAVQIIPGRFFFLSLTGTPWDTQSIHFFCTDQTFQYEPFFADFGPLSLGCIYKYAKLL 75
Query 76 KAKLASDLLQRKVLVHYTTYSQQKVTNAVLLLGSFLVAAMGKTPEEAMKPFARLNSHLKP 135
++KL ++ +L+HY+ +K NA LL+G+ + G + +EA +PF ++ P
Sbjct 76 ESKLKEAEERQHILIHYSAIHPEKRANAALLIGAAQMLLFGMSAQEAYRPFLNISPRFVP 135
Query 136 YRDATYSACDFRLPVLDCLRGLAYAMQLGWFSLSSFNQAAYDHYEKLEN 184
+RDAT C+F+L +LDCL+GL +AM+LGWF +FN YD+YEKL+N
Sbjct 136 FRDATCGPCNFKLTILDCLKGLEFAMKLGWFDYKTFNVDEYDYYEKLKN 184
> dre:565969 cdc14ab, si:dkey-168j9.1; CDC14 cell division cycle
14 homolog A, b; K06639 cell division cycle 14 [EC:3.1.3.48]
Length=510
Score = 165 bits (417), Expect = 8e-41, Method: Compositional matrix adjust.
Identities = 79/174 (45%), Positives = 115/174 (66%), Gaps = 1/174 (0%)
Query 12 DRGQLREAVEVLQGRLYFRSVFSPPRDTPTIHFFSTDKLFVYEPFFLDFGPLNLSCIYRY 71
D L A E ++ RLYF ++ S P+ T H+FSTD+ FVYE F+ DFGPLNL+ +YRY
Sbjct 20 DDNDLLGASEFIKDRLYFATLRSKPKSTANTHYFSTDEEFVYENFYADFGPLNLAMLYRY 79
Query 72 CMLLKAKLASDLLQRKVLVHYTTYSQQKVTNAVLLLGSFLVAAMGKTPEEAMKP-FARLN 130
C L KL S L RK +VHYT++ Q+K NA +L+G++ V + KTPEEA + + N
Sbjct 80 CCKLNKKLKSFTLTRKRIVHYTSFDQRKRANAAVLIGAYAVIYLKKTPEEAYRALISGSN 139
Query 131 SHLKPYRDATYSACDFRLPVLDCLRGLAYAMQLGWFSLSSFNQAAYDHYEKLEN 184
+ P+RDA++ C + L VLDCL+G+ A+Q G+ + +F+ Y+HYE++EN
Sbjct 140 ASYLPFRDASFGNCTYNLTVLDCLQGIRKALQHGFLNFETFDVNEYEHYERVEN 193
> mmu:229776 Cdc14a, A830059A17Rik, CDC14A2, CDC14a1, Cdc14; CDC14
cell division cycle 14 homolog A (S. cerevisiae) (EC:3.1.3.16
3.1.3.48); K06639 cell division cycle 14 [EC:3.1.3.48]
Length=603
Score = 164 bits (414), Expect = 2e-40, Method: Composition-based stats.
Identities = 78/176 (44%), Positives = 114/176 (64%), Gaps = 1/176 (0%)
Query 10 AEDRGQLREAVEVLQGRLYFRSVFSPPRDTPTIHFFSTDKLFVYEPFFLDFGPLNLSCIY 69
A + G+L A E ++ RLYF ++ + P+ T IH+FS D+ VYE F+ DFGPLNL+ +Y
Sbjct 2 AAESGELIGACEFMKDRLYFATLRNRPKSTINIHYFSIDEELVYENFYADFGPLNLAMVY 61
Query 70 RYCMLLKAKLASDLLQRKVLVHYTTYSQQKVTNAVLLLGSFLVAAMGKTPEEAMKP-FAR 128
RYC L KL S L RK +VHYT++ Q+K NA L+G++ V + KTPEEA + +
Sbjct 62 RYCCKLNKKLKSYSLSRKKIVHYTSFDQRKRANAAFLIGAYAVIYLKKTPEEAYRALLSG 121
Query 129 LNSHLKPYRDATYSACDFRLPVLDCLRGLAYAMQLGWFSLSSFNQAAYDHYEKLEN 184
N P+RDA++ C + L VLDCL+G+ +Q G+F +F+ Y+HYE++EN
Sbjct 122 SNPPYLPFRDASFGNCTYNLTVLDCLQGIRKGLQHGFFDFETFDAEEYEHYERVEN 177
> dre:394124 cdc14aa, CDC14A, MGC63654, zgc:63654; CDC14 cell
division cycle 14 homolog A, a; K06639 cell division cycle 14
[EC:3.1.3.48]
Length=592
Score = 162 bits (410), Expect = 5e-40, Method: Compositional matrix adjust.
Identities = 75/171 (43%), Positives = 111/171 (64%), Gaps = 1/171 (0%)
Query 15 QLREAVEVLQGRLYFRSVFSPPRDTPTIHFFSTDKLFVYEPFFLDFGPLNLSCIYRYCML 74
+LR+A E ++ RLYF ++ S P+ T HFF TD ++YE F+ DFGPLNL+ +YRYC
Sbjct 5 ELRDASEFIKDRLYFATLRSKPKSTANTHFFCTDDEYIYENFYADFGPLNLAMLYRYCCK 64
Query 75 LKAKLASDLLQRKVLVHYTTYSQQKVTNAVLLLGSFLVAAMGKTPEEAMKP-FARLNSHL 133
L KL S L RK ++HYT+Y Q+K NA L+G++ V + +TPEE + + N
Sbjct 65 LNKKLKSFTLSRKRIIHYTSYDQRKRANAAFLIGAYAVIYLKRTPEEVYRALISGTNVSY 124
Query 134 KPYRDATYSACDFRLPVLDCLRGLAYAMQLGWFSLSSFNQAAYDHYEKLEN 184
P+RDA + C + L +LDCL+G+ A+Q G+F +F+ Y+HYE++EN
Sbjct 125 LPFRDAAFGNCTYDLTILDCLQGIRKALQHGFFDFENFDVEEYEHYERVEN 175
> hsa:8556 CDC14A, cdc14, hCDC14; CDC14 cell division cycle 14
homolog A (S. cerevisiae) (EC:3.1.3.16 3.1.3.48); K06639 cell
division cycle 14 [EC:3.1.3.48]
Length=594
Score = 159 bits (403), Expect = 4e-39, Method: Compositional matrix adjust.
Identities = 76/176 (43%), Positives = 112/176 (63%), Gaps = 1/176 (0%)
Query 10 AEDRGQLREAVEVLQGRLYFRSVFSPPRDTPTIHFFSTDKLFVYEPFFLDFGPLNLSCIY 69
A + G+L A E ++ RLYF ++ + P+ T H+FS D+ VYE F+ DFGPLNL+ +Y
Sbjct 2 AAESGELIGACEFMKDRLYFATLRNRPKSTVNTHYFSIDEELVYENFYADFGPLNLAMVY 61
Query 70 RYCMLLKAKLASDLLQRKVLVHYTTYSQQKVTNAVLLLGSFLVAAMGKTPEEAMKP-FAR 128
RYC L KL S L RK +VHYT + Q+K NA L+G++ V + KTPEEA + +
Sbjct 62 RYCCKLNKKLKSYSLSRKKIVHYTCFDQRKRANAAFLIGAYAVIYLKKTPEEAYRALLSG 121
Query 129 LNSHLKPYRDATYSACDFRLPVLDCLRGLAYAMQLGWFSLSSFNQAAYDHYEKLEN 184
N P+RDA++ C + L +LDCL+G+ +Q G+F +F+ Y+HYE++EN
Sbjct 122 SNPPYLPFRDASFGNCTYNLTILDCLQGIRKGLQHGFFDFETFDVDEYEHYERVEN 177
> xla:403393 cdc14a, xcdc14a; CDC14 cell division cycle 14 homolog
A; K06639 cell division cycle 14 [EC:3.1.3.48]
Length=576
Score = 158 bits (400), Expect = 8e-39, Method: Compositional matrix adjust.
Identities = 74/174 (42%), Positives = 114/174 (65%), Gaps = 1/174 (0%)
Query 12 DRGQLREAVEVLQGRLYFRSVFSPPRDTPTIHFFSTDKLFVYEPFFLDFGPLNLSCIYRY 71
D +L A EV++ RLYF + + P+ T H+F TD+ FVYE F+ DFGPLNL+ +YRY
Sbjct 2 DNQELISASEVIKDRLYFAILRNKPKSTLNTHYFCTDEEFVYENFYADFGPLNLAQLYRY 61
Query 72 CMLLKAKLASDLLQRKVLVHYTTYSQQKVTNAVLLLGSFLVAAMGKTPEEAMKP-FARLN 130
C L KL S L RK +VHYT++ Q+K +NA L+ ++ V + K+PEE + + N
Sbjct 62 CCKLNKKLKSFSLSRKKIVHYTSFEQRKRSNAGFLISAYAVIYLKKSPEEVYRALLSGSN 121
Query 131 SHLKPYRDATYSACDFRLPVLDCLRGLAYAMQLGWFSLSSFNQAAYDHYEKLEN 184
+ P+RDA++ +C + L +LDCL+G+ A+ G+F+ +F+ Y+HYE++EN
Sbjct 122 AQYLPFRDASFGSCTYNLTILDCLQGIRKALHFGFFNFENFDADEYEHYERVEN 175
> hsa:8555 CDC14B, CDC14B3, Cdc14B1, Cdc14B2, hCDC14B; CDC14 cell
division cycle 14 homolog B (S. cerevisiae) (EC:3.1.3.16
3.1.3.48); K06639 cell division cycle 14 [EC:3.1.3.48]
Length=461
Score = 153 bits (387), Expect = 3e-37, Method: Compositional matrix adjust.
Identities = 70/165 (42%), Positives = 102/165 (61%), Gaps = 0/165 (0%)
Query 20 VEVLQGRLYFRSVFSPPRDTPTIHFFSTDKLFVYEPFFLDFGPLNLSCIYRYCMLLKAKL 79
EV++ RL F ++S P+ +H+FS D YE F+ DFGPLNL+ +YRYC + KL
Sbjct 12 AEVIKDRLCFAILYSRPKSASNVHYFSIDNELEYENFYADFGPLNLAMVYRYCCKINKKL 71
Query 80 ASDLLQRKVLVHYTTYSQQKVTNAVLLLGSFLVAAMGKTPEEAMKPFARLNSHLKPYRDA 139
S + RK +VH+T Q+K NA L+G ++V +G+TPEEA + + P+RDA
Sbjct 72 KSITMLRKKIVHFTGSDQRKQANAAFLVGCYMVIYLGRTPEEAYRILIFGETSYIPFRDA 131
Query 140 TYSACDFRLPVLDCLRGLAYAMQLGWFSLSSFNQAAYDHYEKLEN 184
Y +C+F + +LDC + AMQ G+ + +SFN Y+HYEK EN
Sbjct 132 AYGSCNFYITLLDCFHAVKKAMQYGFLNFNSFNLDEYEHYEKAEN 176
> mmu:218294 Cdc14b, 2810432N10Rik, A530086E13Rik, AA472821, CDC14B3,
Cdc14B1; CDC14 cell division cycle 14 homolog B (S.
cerevisiae) (EC:3.1.3.16 3.1.3.48); K06639 cell division cycle
14 [EC:3.1.3.48]
Length=448
Score = 149 bits (377), Expect = 4e-36, Method: Compositional matrix adjust.
Identities = 69/165 (41%), Positives = 102/165 (61%), Gaps = 0/165 (0%)
Query 20 VEVLQGRLYFRSVFSPPRDTPTIHFFSTDKLFVYEPFFLDFGPLNLSCIYRYCMLLKAKL 79
EV++ RL F ++S P+ H+FS D YE F+ DFGPLNL+ +YRYC + KL
Sbjct 12 AEVIRDRLCFAILYSRPKSATNEHYFSIDNELEYENFYADFGPLNLAMVYRYCCKINKKL 71
Query 80 ASDLLQRKVLVHYTTYSQQKVTNAVLLLGSFLVAAMGKTPEEAMKPFARLNSHLKPYRDA 139
S + RK ++H+T Q+K NA L+G ++V +G+TPE+A + ++ P+RDA
Sbjct 72 KSITMLRKKIIHFTGTDQRKQANAAFLVGCYMVIYLGRTPEDAYRTLIFGDTAYIPFRDA 131
Query 140 TYSACDFRLPVLDCLRGLAYAMQLGWFSLSSFNQAAYDHYEKLEN 184
Y +C F + +LDC + AMQ G+F+ +SFN Y+HYEK EN
Sbjct 132 AYGSCSFYITLLDCFHAVKKAMQYGFFNFNSFNLDEYEHYEKAEN 176
> xla:407839 cdc14b, MGC81657, cdc14beta, xcdc14b; CDC14 cell
division cycle 14 homolog B; K06639 cell division cycle 14 [EC:3.1.3.48]
Length=452
Score = 145 bits (367), Expect = 6e-35, Method: Compositional matrix adjust.
Identities = 71/166 (42%), Positives = 99/166 (59%), Gaps = 0/166 (0%)
Query 19 AVEVLQGRLYFRSVFSPPRDTPTIHFFSTDKLFVYEPFFLDFGPLNLSCIYRYCMLLKAK 78
AVE+++ RLYF + S P+ T H+FS D VYE F+ DFGPLNL+ +YRYCM L K
Sbjct 11 AVEIVKDRLYFAILCSRPKTTYGTHYFSIDDELVYENFYADFGPLNLAMLYRYCMKLNKK 70
Query 79 LASDLLQRKVLVHYTTYSQQKVTNAVLLLGSFLVAAMGKTPEEAMKPFARLNSHLKPYRD 138
+ S L +K +VHYT +K NA LLGSF + + K P E + N++ P+RD
Sbjct 71 IKSFSLTKKKIVHYTCGDDKKQANAAFLLGSFAIIYLNKQPLELYRLLQAGNTNYLPFRD 130
Query 139 ATYSACDFRLPVLDCLRGLAYAMQLGWFSLSSFNQAAYDHYEKLEN 184
A++ C F L +LDC + A+Q +F +F+ Y HYE+ EN
Sbjct 131 ASFGTCSFHLTLLDCFNAVHKALQYDFFDFKTFDVEEYQHYERAEN 176
> cpv:cgd7_4470 CDC14 phosphatase ; K06639 cell division cycle
14 [EC:3.1.3.48]
Length=453
Score = 112 bits (279), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 61/145 (42%), Positives = 81/145 (55%), Gaps = 3/145 (2%)
Query 41 TIHFFSTDKLFVYEPFFLDFGPLNLSCIYRYCMLLKAKLASDLLQRKVLVHYTT-YSQQK 99
T ++F D VYEPFF DFGPLNL CIY+YC + L + +V T+ Y +K
Sbjct 41 TNYYFCIDDELVYEPFFQDFGPLNLRCIYKYCKKVDNLLRTVESTGGYIVQCTSVYDMKK 100
Query 100 VTNAVLLLGSFLVAAMGKTPEEAMKPFARLNSHLKPYRDATYSACDFRLPVLDCLRGLAY 159
TN+ L +++ GKTP ++ F + L +RDATY C + L V DCL GL Y
Sbjct 101 RTNSAFLACCYMMIRQGKTPGSSLALFNSV--PLLSFRDATYGPCSYSLTVGDCLLGLYY 158
Query 160 AMQLGWFSLSSFNQAAYDHYEKLEN 184
AM L WF +F+ Y YEK+EN
Sbjct 159 AMLLKWFDYETFDINEYSTYEKVEN 183
> sce:YFR028C CDC14, OAF3; Cdc14p (EC:3.1.3.48); K06639 cell division
cycle 14 [EC:3.1.3.48]
Length=551
Score = 110 bits (274), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 53/168 (31%), Positives = 90/168 (53%), Gaps = 0/168 (0%)
Query 16 LREAVEVLQGRLYFRSVFSPPRDTPTIHFFSTDKLFVYEPFFLDFGPLNLSCIYRYCMLL 75
L +E L+GR+Y + P DT + FF+ + Y F LDFGP+N+ +YR+ ++
Sbjct 7 LDNTIEFLRGRVYLGAYDYTPEDTDELVFFTVEDAIFYNSFHLDFGPMNIGHLYRFAVIF 66
Query 76 KAKLASDLLQRKVLVHYTTYSQQKVTNAVLLLGSFLVAAMGKTPEEAMKPFARLNSHLKP 135
L K +V Y++ S ++ NA +L +++ TP + ++P A+++ P
Sbjct 67 HEILNDPENANKAVVFYSSASTRQRANAACMLCCYMILVQAWTPHQVLQPLAQVDPPFMP 126
Query 136 YRDATYSACDFRLPVLDCLRGLAYAMQLGWFSLSSFNQAAYDHYEKLE 183
+RDA YS DF + + D + G+ A + G L SFN +Y+ YE +E
Sbjct 127 FRDAGYSNADFEITIQDVVYGVWRAKEKGLIDLHSFNLESYEKYEHVE 174
> dre:393148 cdc14b, MGC55844, cdc14a, zgc:55844; CDC14 cell division
cycle 14 homolog B; K06639 cell division cycle 14 [EC:3.1.3.48]
Length=404
Score = 92.8 bits (229), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 50/117 (42%), Positives = 70/117 (59%), Gaps = 0/117 (0%)
Query 68 IYRYCMLLKAKLASDLLQRKVLVHYTTYSQQKVTNAVLLLGSFLVAAMGKTPEEAMKPFA 127
+YR+C LK KL S +K +V YT ++K NA L+GS+ V + KTPEEA
Sbjct 2 LYRFCCKLKKKLKSCAHAKKKIVFYTYGDRKKQANAAYLIGSYAVMHLQKTPEEAYSLLV 61
Query 128 RLNSHLKPYRDATYSACDFRLPVLDCLRGLAYAMQLGWFSLSSFNQAAYDHYEKLEN 184
N+ P+RDA++ AC + L +LDCLR + A+Q GW + FN Y+HYE+ EN
Sbjct 62 SQNAPYLPFRDASFGACMYNLNILDCLRAVHKALQFGWLDFTQFNVEEYEHYERAEN 118
> cel:C17G10.4 cdc-14; Cell Division Cycle related family member
(cdc-14); K06639 cell division cycle 14 [EC:3.1.3.48]
Length=1063
Score = 82.0 bits (201), Expect = 9e-16, Method: Composition-based stats.
Identities = 52/177 (29%), Positives = 89/177 (50%), Gaps = 14/177 (7%)
Query 21 EVLQGRLYFRSVFSPP------RDTPTIHFFSTDKLFVYEPFFLDFGPLNLSCIYRYCML 74
E+L RLYF +P + F + + F YEPF+ DFGP NLS +YR C+
Sbjct 19 ELLPNRLYFGCFPNPDAIDKSDKSVKKTCFININNKFHYEPFYEDFGPWNLSVLYRLCVQ 78
Query 75 LKAKLASDLLQRKVLVHYTT------YSQQKVTNAVLLLGSFLVAAMGKTPEEA-MKPFA 127
+ L + + + +V + Y + +V N +LG++L+ G + ++A +K +
Sbjct 79 VGKLLEVEEKRSRRVVLFCQDDGTGQYDKIRV-NTAYVLGAYLIIYQGFSADDAYLKVSS 137
Query 128 RLNSHLKPYRDATYSACDFRLPVLDCLRGLAYAMQLGWFSLSSFNQAAYDHYEKLEN 184
+RDA+ + + L + D LRG+ A++ GW S F+ Y+ YE++EN
Sbjct 138 GETVKFVGFRDASMGSPQYLLHLHDVLRGIEKALKFGWLDFSDFDYEEYEFYERVEN 194
> hsa:5426 POLE, DKFZp434F222, FLJ21434, POLE1; polymerase (DNA
directed), epsilon (EC:2.7.7.7); K02324 DNA polymerase epsilon
subunit 1 [EC:2.7.7.7]
Length=2286
Score = 31.2 bits (69), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 20/66 (30%), Positives = 31/66 (46%), Gaps = 9/66 (13%)
Query 122 AMKPFARLNSHLKP---YRDATYSACDFRLPVLDCLRGLAYAMQLGWFSLSSFNQAAYDH 178
AM P L + L+P +AT +ACDF P +C R +A W F A+
Sbjct 629 AMYPNIILTNRLQPSAMVDEATCAACDFNKPGANCQRKMA------WQWRGEFMPASRSE 682
Query 179 YEKLEN 184
Y ++++
Sbjct 683 YHRIQH 688
> mmu:18973 Pole; polymerase (DNA directed), epsilon (EC:2.7.7.7);
K02324 DNA polymerase epsilon subunit 1 [EC:2.7.7.7]
Length=2283
Score = 30.4 bits (67), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 20/66 (30%), Positives = 30/66 (45%), Gaps = 9/66 (13%)
Query 122 AMKPFARLNSHLKP---YRDATYSACDFRLPVLDCLRGLAYAMQLGWFSLSSFNQAAYDH 178
AM P L + L+P +AT +ACDF P C R +A W F A+
Sbjct 629 AMYPNIILTNRLQPSAIVDEATCAACDFNKPGASCQRKMA------WQWRGEFMPASRSE 682
Query 179 YEKLEN 184
Y ++++
Sbjct 683 YHRIQH 688
> sce:YIL015W BAR1, SST1; Aspartyl protease secreted into the
periplasmic space of mating type a cells, helps cells find mating
partners, cleaves and inactivates alpha factor allowing
cells to recover from alpha-factor-induced cell cycle arrest
(EC:3.4.23.35); K01383 barrierpepsin [EC:3.4.23.35]
Length=587
Score = 30.4 bits (67), Expect = 3.6, Method: Composition-based stats.
Identities = 18/64 (28%), Positives = 31/64 (48%), Gaps = 13/64 (20%)
Query 121 EAMKPFARLNSHLKPYRDATYSACDFRLPVLDCLRGLAYAMQLGWFSLSSFNQAAYDHYE 180
++ PF NS+ Y +ATY+ + + P +DC S+S++N+ Y+
Sbjct 73 DSSNPFCLPNSNTSSYSNATYNGEEVK-PSIDCR------------SMSTYNEHRSSTYQ 119
Query 181 KLEN 184
LEN
Sbjct 120 YLEN 123
Lambda K H
0.326 0.138 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5041515336
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40