bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0572_orf1
Length=135
Score E
Sequences producing significant alignments: (Bits) Value
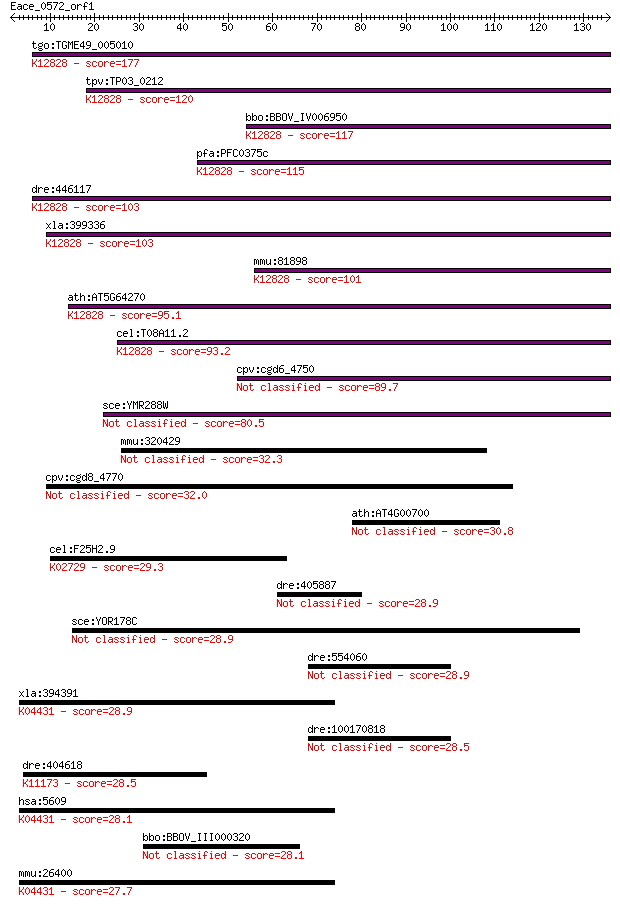
tgo:TGME49_005010 splicing factor 3B subunit 1, putative (EC:5... 177 6e-45
tpv:TP03_0212 splicing factor 3B subunit 1; K12828 splicing fa... 120 2e-27
bbo:BBOV_IV006950 23.m06336; splicing factor; K12828 splicing ... 117 8e-27
pfa:PFC0375c U2 snRNP spliceosome subunit, putative; K12828 sp... 115 5e-26
dre:446117 sf3b1, wu:fb99f09; splicing factor 3b, subunit 1; K... 103 1e-22
xla:399336 sf3b1, MGC115390, hsh155, prp10, prpf10, sap155; sp... 103 2e-22
mmu:81898 Sf3b1, 155kDa, 2810001M05Rik, AA409119, Prp10, SAP15... 101 6e-22
ath:AT5G64270 splicing factor, putative; K12828 splicing facto... 95.1 6e-20
cel:T08A11.2 hypothetical protein; K12828 splicing factor 3B s... 93.2 2e-19
cpv:cgd6_4750 splicing factor 3B subunit1-like HEAT repeat con... 89.7 2e-18
sce:YMR288W HSH155; Hsh155p 80.5 1e-15
mmu:320429 Trank1, A230061D21Rik, C030048J01Rik, Gm187, Lba1; ... 32.3 0.45
cpv:cgd8_4770 hypothetical protein 32.0 0.59
ath:AT4G00700 C2 domain-containing protein 30.8 1.3
cel:F25H2.9 pas-5; Proteasome Alpha Subunit family member (pas... 29.3 4.0
dre:405887 nf2b, MGC123124, NF2a, nf2; neurofibromin 2b (merlin) 28.9 4.7
sce:YOR178C GAC1; Gac1p 28.9 4.8
dre:554060 pcdh2g13; protocadherin 2 gamma 13 28.9
xla:394391 map2k7, MKK7, map2k7-A; mitogen-activated protein k... 28.9 5.3
dre:100170818 zgc:193751 28.5 5.8
dre:404618 adhfe1, MGC77479, zgc:77479; alcohol dehydrogenase,... 28.5 6.2
hsa:5609 MAP2K7, Jnkk2, MAPKK7, MKK7, PRKMK7; mitogen-activate... 28.1 8.2
bbo:BBOV_III000320 17.m10445; hypothetical protein 28.1 8.9
mmu:26400 Map2k7, 5930412N11Rik, Jnkk2, MKK7, Prkmk7, sek2; mi... 27.7 9.4
> tgo:TGME49_005010 splicing factor 3B subunit 1, putative (EC:5.5.1.4);
K12828 splicing factor 3B subunit 1
Length=1386
Score = 177 bits (450), Expect = 6e-45, Method: Compositional matrix adjust.
Identities = 92/132 (69%), Positives = 107/132 (81%), Gaps = 3/132 (2%)
Query 6 TPGVSSAATPLGVTPMYVMPEEGGI--SVKAVDPTGSLLDPQGLGEASGGVQMKAEDFHF 63
TPGV++ TP+GVTP+Y MPEEG I ++ + G + Q L GGV MKAED+ F
Sbjct 497 TPGVAATPTPIGVTPLYTMPEEGVIPGGMEGLMTMGGGIGGQ-LTAEGGGVTMKAEDYQF 555
Query 64 FSKLFEEKSEDQMTQEEVKEKKIQLLLLKIKNGTPPLRRSALRILTDKAKEFGAAALFNQ 123
F+KLFE+K ED MTQEEVKE+KIQLLLLKIKNGTPPLRRSALR LT+K+KEFGAAALFNQ
Sbjct 556 FAKLFEDKDEDAMTQEEVKERKIQLLLLKIKNGTPPLRRSALRTLTEKSKEFGAAALFNQ 615
Query 124 ILPLMMQTTLED 135
ILPLMMQ+TLED
Sbjct 616 ILPLMMQSTLED 627
> tpv:TP03_0212 splicing factor 3B subunit 1; K12828 splicing
factor 3B subunit 1
Length=1107
Score = 120 bits (300), Expect = 2e-27, Method: Composition-based stats.
Identities = 64/118 (54%), Positives = 83/118 (70%), Gaps = 10/118 (8%)
Query 18 VTPMYVMPEEGGISVKAVDPTGSLLDPQGLGEASGGVQMKAEDFHFFSKLFEEKSEDQMT 77
VTP + +PE+ K D G+ P L + V++KAED HFFSKLF++ +ED +T
Sbjct 230 VTPHFTIPED---VRKPYDIPGT---PSILQD----VEIKAEDQHFFSKLFDDSTEDDLT 279
Query 78 QEEVKEKKIQLLLLKIKNGTPPLRRSALRILTDKAKEFGAAALFNQILPLMMQTTLED 135
+E+ E++I LLLK+KNGTPP RR ALR+L KAKEFG LFNQILPLMMQ+TL+D
Sbjct 280 SDEITERRILALLLKVKNGTPPHRRQALRLLASKAKEFGPGPLFNQILPLMMQSTLQD 337
> bbo:BBOV_IV006950 23.m06336; splicing factor; K12828 splicing
factor 3B subunit 1
Length=1147
Score = 117 bits (294), Expect = 8e-27, Method: Compositional matrix adjust.
Identities = 56/82 (68%), Positives = 69/82 (84%), Gaps = 0/82 (0%)
Query 54 VQMKAEDFHFFSKLFEEKSEDQMTQEEVKEKKIQLLLLKIKNGTPPLRRSALRILTDKAK 113
V++KAED FF KLF++K+E+ +T +E+ E++I LLLKIKNGTPPLRR ALR+LT KA+
Sbjct 302 VEVKAEDQGFFGKLFDDKTEEDLTADEITERRILALLLKIKNGTPPLRRQALRLLTQKAR 361
Query 114 EFGAAALFNQILPLMMQTTLED 135
EFG LFNQILPLMMQTTLED
Sbjct 362 EFGPGPLFNQILPLMMQTTLED 383
> pfa:PFC0375c U2 snRNP spliceosome subunit, putative; K12828
splicing factor 3B subunit 1
Length=1386
Score = 115 bits (287), Expect = 5e-26, Method: Composition-based stats.
Identities = 55/93 (59%), Positives = 72/93 (77%), Gaps = 1/93 (1%)
Query 43 DPQGLGEASGGVQMKAEDFHFFSKLFEEKSEDQMTQEEVKEKKIQLLLLKIKNGTPPLRR 102
+PQ L E +Q+K ED+ +FSKLFE +E++++QEE+KE+K +LLLKIKNGTP +RR
Sbjct 536 NPQLLNELKY-IQLKNEDYIYFSKLFESINEEELSQEELKERKFMILLLKIKNGTPSIRR 594
Query 103 SALRILTDKAKEFGAAALFNQILPLMMQTTLED 135
+ALR +T+K KE G LFN ILPLMMQ TLED
Sbjct 595 TALRTITEKVKELGPETLFNLILPLMMQNTLED 627
> dre:446117 sf3b1, wu:fb99f09; splicing factor 3b, subunit 1;
K12828 splicing factor 3B subunit 1
Length=1315
Score = 103 bits (258), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 61/133 (45%), Positives = 80/133 (60%), Gaps = 15/133 (11%)
Query 6 TPGVSSAATPL---GVTPMYVMPEEGGISVKAVDPTGSLLDPQGLGEASGGVQMKAEDFH 62
TP AATP G+T ++ E+ + P+G+L +K +D
Sbjct 437 TPARKLAATPTPIGGMTGFHMQTEDRSMKQVNDQPSGNL------------PFLKPDDIQ 484
Query 63 FFSKLFEEKSEDQMTQEEVKEKKIQLLLLKIKNGTPPLRRSALRILTDKAKEFGAAALFN 122
+F KL E E ++ EE KE+KI LLLKIKNGTPP+R++ALR +TDKA+EFGA LFN
Sbjct 485 YFDKLLVEVDESTLSPEEQKERKIMKLLLKIKNGTPPMRKAALRQITDKAREFGAGPLFN 544
Query 123 QILPLMMQTTLED 135
QILPL+M TLED
Sbjct 545 QILPLLMSPTLED 557
> xla:399336 sf3b1, MGC115390, hsh155, prp10, prpf10, sap155;
splicing factor 3b, subunit 1, 155kDa; K12828 splicing factor
3B subunit 1
Length=1307
Score = 103 bits (256), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 61/129 (47%), Positives = 82/129 (63%), Gaps = 15/129 (11%)
Query 9 VSSAATPLGVTPMYVMPEEGGISVKAVD--PTGSLLDPQGLGEASGGVQMKAEDFHFFSK 66
+++ TPLG + MP E S+K+V P+G+L +K +D +F K
Sbjct 434 LTATPTPLGGLTGFHMPTEDR-SMKSVSDQPSGNL------------PFLKPDDIQYFDK 480
Query 67 LFEEKSEDQMTQEEVKEKKIQLLLLKIKNGTPPLRRSALRILTDKAKEFGAAALFNQILP 126
L + E ++ EE KE+KI LLLKIKNGTPP+R++ALR +TDKA+EFGA LFNQILP
Sbjct 481 LLVDVDESTLSPEEQKERKIMKLLLKIKNGTPPMRKAALRQITDKAREFGAGPLFNQILP 540
Query 127 LMMQTTLED 135
L+M TLED
Sbjct 541 LLMSPTLED 549
> mmu:81898 Sf3b1, 155kDa, 2810001M05Rik, AA409119, Prp10, SAP155,
SF3b155, TA-8, Targ4; splicing factor 3b, subunit 1; K12828
splicing factor 3B subunit 1
Length=1304
Score = 101 bits (252), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 48/80 (60%), Positives = 61/80 (76%), Gaps = 0/80 (0%)
Query 56 MKAEDFHFFSKLFEEKSEDQMTQEEVKEKKIQLLLLKIKNGTPPLRRSALRILTDKAKEF 115
+K +D +F KL + E ++ EE KE+KI LLLKIKNGTPP+R++ALR +TDKA+EF
Sbjct 467 LKPDDIQYFDKLLVDVDESTLSPEEQKERKIMKLLLKIKNGTPPMRKAALRQITDKAREF 526
Query 116 GAAALFNQILPLMMQTTLED 135
GA LFNQILPL+M TLED
Sbjct 527 GAGPLFNQILPLLMSPTLED 546
> ath:AT5G64270 splicing factor, putative; K12828 splicing factor
3B subunit 1
Length=1269
Score = 95.1 bits (235), Expect = 6e-20, Method: Compositional matrix adjust.
Identities = 63/123 (51%), Positives = 81/123 (65%), Gaps = 13/123 (10%)
Query 14 TPLGVTPMYVMPEEGGISVKAVDPTGSLLDPQGLGEASGGVQ-MKAEDFHFFSKLFEEKS 72
TP+ TP YV+PEE V P E GG+ MK ED+ +F L E++
Sbjct 400 TPMA-TPGYVIPEENRGQQYDVPP-----------EVPGGLPFMKPEDYQYFGSLLNEEN 447
Query 73 EDQMTQEEVKEKKIQLLLLKIKNGTPPLRRSALRILTDKAKEFGAAALFNQILPLMMQTT 132
E++++ EE KE+KI LLLK+KNGTPP R++ALR LTDKA+E GA LFN+ILPL+MQ T
Sbjct 448 EEELSPEEQKERKIMKLLLKVKNGTPPQRKTALRQLTDKARELGAGPLFNKILPLLMQPT 507
Query 133 LED 135
LED
Sbjct 508 LED 510
> cel:T08A11.2 hypothetical protein; K12828 splicing factor 3B
subunit 1
Length=1322
Score = 93.2 bits (230), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 51/111 (45%), Positives = 72/111 (64%), Gaps = 5/111 (4%)
Query 25 PEEGGISVKAVDPTGSLLDPQGLGEASGGVQMKAEDFHFFSKLFEEKSEDQMTQEEVKEK 84
P+ GI K V G L+D Q + + +K +D +F KL + E Q+T+EE E+
Sbjct 459 PDRDGIGEKGV---GGLVDTQP--KNAELPPLKPDDMQYFDKLLMDVDESQLTKEEKNER 513
Query 85 KIQLLLLKIKNGTPPLRRSALRILTDKAKEFGAAALFNQILPLMMQTTLED 135
+I LLKIKNGTPP+R+S LR +T+ A+++GA LFNQILPL+M +LED
Sbjct 514 EIMEHLLKIKNGTPPMRKSGLRKITENARKYGAGPLFNQILPLLMSPSLED 564
> cpv:cgd6_4750 splicing factor 3B subunit1-like HEAT repeat containing
protein
Length=1031
Score = 89.7 bits (221), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 40/84 (47%), Positives = 59/84 (70%), Gaps = 0/84 (0%)
Query 52 GGVQMKAEDFHFFSKLFEEKSEDQMTQEEVKEKKIQLLLLKIKNGTPPLRRSALRILTDK 111
G + ++ EDFHFF KLF S+D ++ EEV E+ + LLLKIKNG P LRR A++ + +
Sbjct 189 GELSLRIEDFHFFGKLFSNISDDDLSPEEVNERLVLTLLLKIKNGAPILRRKAMKQIVET 248
Query 112 AKEFGAAALFNQILPLMMQTTLED 135
A++ G + N +LPL+MQ+TLE+
Sbjct 249 ARDHGPGIILNHLLPLLMQSTLEE 272
> sce:YMR288W HSH155; Hsh155p
Length=971
Score = 80.5 bits (197), Expect = 1e-15, Method: Composition-based stats.
Identities = 41/114 (35%), Positives = 72/114 (63%), Gaps = 4/114 (3%)
Query 22 YVMPEEGGISVKAVDPTGSLLDPQGLGEASGGVQMKAEDFHFFSKLFEEKSEDQMTQEEV 81
Y P+E +VK + +L++ +G+ + + K D +F+ + +K D++ ++E
Sbjct 106 YEPPDESSTAVKE-NSDSALVNVEGIHDL---MFFKPSDHKYFADVISKKPIDELNKDEK 161
Query 82 KEKKIQLLLLKIKNGTPPLRRSALRILTDKAKEFGAAALFNQILPLMMQTTLED 135
KE+ + +LLLKIKNG RR+++RILTDKA FG +FN++LP+++ +LED
Sbjct 162 KERTLSMLLLKIKNGNTASRRTSMRILTDKAVTFGPEMIFNRLLPILLDRSLED 215
> mmu:320429 Trank1, A230061D21Rik, C030048J01Rik, Gm187, Lba1;
tetratricopeptide repeat and ankyrin repeat containing 1
Length=2999
Score = 32.3 bits (72), Expect = 0.45, Method: Composition-based stats.
Identities = 24/85 (28%), Positives = 43/85 (50%), Gaps = 3/85 (3%)
Query 26 EEGGISVKAVDPTGSLLDPQGLGEASGGVQMKA-EDFHFFSKLFEE--KSEDQMTQEEVK 82
EEG + V+ ++ + + G E S +Q K E S L EE + + EEV
Sbjct 2879 EEGKVVVQGIEQSVWIRSHLGSKEQSHMLQRKVQEHIRRVSDLVEELYRRKAWAEAEEVM 2938
Query 83 EKKIQLLLLKIKNGTPPLRRSALRI 107
+++++L L +KN L+++ LR+
Sbjct 2939 TQQVKILTLSVKNAQEWLKKTELRL 2963
> cpv:cgd8_4770 hypothetical protein
Length=1965
Score = 32.0 bits (71), Expect = 0.59, Method: Composition-based stats.
Identities = 30/117 (25%), Positives = 53/117 (45%), Gaps = 16/117 (13%)
Query 9 VSSAATPLGVTPMYVMPEEGGISVKAVDPTGSLLDPQGLGEASGGVQ----MKAEDFHFF 64
++S TP+ Y+ E GGIS+ + L+P GL +S G ++ + +
Sbjct 1047 ITSICTPIIEPKYYIAKEGGGISISSTFS----LNPSGLSTSSQGGNDSNFTQSSNSYLI 1102
Query 65 S----KLFEEKSEDQMTQ----EEVKEKKIQLLLLKIKNGTPPLRRSALRILTDKAK 113
S K+ EE S++ ++ V +K I + L ++N + RS I + K K
Sbjct 1103 SCELIKMEEEDSKEGNSEMILVASVSQKPILVYRLDLRNNNSNISRSGDFIFSRKWK 1159
> ath:AT4G00700 C2 domain-containing protein
Length=1006
Score = 30.8 bits (68), Expect = 1.3, Method: Composition-based stats.
Identities = 12/33 (36%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 78 QEEVKEKKIQLLLLKIKNGTPPLRRSALRILTD 110
QE +K + I ++++++ PPLRR + LTD
Sbjct 757 QEALKMQAINIIIVRLGRSEPPLRREVVDYLTD 789
> cel:F25H2.9 pas-5; Proteasome Alpha Subunit family member (pas-5);
K02729 20S proteasome subunit alpha 5 [EC:3.4.25.1]
Length=248
Score = 29.3 bits (64), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 15/56 (26%), Positives = 30/56 (53%), Gaps = 3/56 (5%)
Query 10 SSAATPLGVTPMYVMPEEGGISVKAVDPTGSLLD--PQGLGEASGGVQMK-AEDFH 62
+S + P GV ++ ++ G + +DP+G+ +D + +G AS G + E +H
Sbjct 130 ASMSRPFGVAMLFAGVDQEGAKLFHLDPSGTFIDCKAKSIGAASDGAEQNLKEQYH 185
> dre:405887 nf2b, MGC123124, NF2a, nf2; neurofibromin 2b (merlin)
Length=586
Score = 28.9 bits (63), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 11/19 (57%), Positives = 14/19 (73%), Gaps = 0/19 (0%)
Query 61 FHFFSKLFEEKSEDQMTQE 79
FHF +K F EK ED++ QE
Sbjct 86 FHFLAKFFPEKVEDELVQE 104
> sce:YOR178C GAC1; Gac1p
Length=793
Score = 28.9 bits (63), Expect = 4.8, Method: Composition-based stats.
Identities = 31/123 (25%), Positives = 50/123 (40%), Gaps = 17/123 (13%)
Query 15 PLGVTPMYVMPEEGGISVKAVDPTGSLLDPQGLGEASGGVQMKAEDFHFFSKLFEEKSED 74
P T + P+E + +PT S PQ + +ED +++ +
Sbjct 380 PASQTDAAMSPKEMKARFVSSNPTLSRFLPQS--------RKFSEDTDYYNTSPLKHLYH 431
Query 75 QMTQEEVKEKKIQLLLLKIKNGTPPLRRSAL--------RILTDKAKEFGAAALFNQI-L 125
T VK K++ ++L K++N TPP SAL +I DK N I L
Sbjct 432 NDTTSWVKPKRLNVVLDKLENATPPPPSSALANDTARTGKITKDKNNVLAPPTASNSIDL 491
Query 126 PLM 128
P++
Sbjct 492 PIL 494
> dre:554060 pcdh2g13; protocadherin 2 gamma 13
Length=963
Score = 28.9 bits (63), Expect = 4.8, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 22/35 (62%), Gaps = 3/35 (8%)
Query 68 FEEKSEDQMTQEEV---KEKKIQLLLLKIKNGTPP 99
FEEK + + +E+ KEK++ L+L + GTPP
Sbjct 186 FEEKYAELVLNKELDRRKEKEVNLILTAVDGGTPP 220
> xla:394391 map2k7, MKK7, map2k7-A; mitogen-activated protein
kinase kinase 7 (EC:2.7.12.2); K04431 mitogen-activated protein
kinase kinase 7 [EC:2.7.12.2]
Length=417
Score = 28.9 bits (63), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 19/71 (26%), Positives = 31/71 (43%), Gaps = 8/71 (11%)
Query 3 GPATPGVSSAATPLGVTPMYVMPEEGGISVKAVDPTGSLLDPQGLGEASGGVQMKAEDFH 62
GP + T V +Y + E+ G+ + V P+ LLD G Q+K DF
Sbjct 209 GPIPEAILGKMTVAIVNALYYLKEKHGVIHRDVKPSNILLDASG--------QIKLCDFG 260
Query 63 FFSKLFEEKSE 73
+L + K++
Sbjct 261 ISGRLVDSKAK 271
> dre:100170818 zgc:193751
Length=945
Score = 28.5 bits (62), Expect = 5.8, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 22/35 (62%), Gaps = 3/35 (8%)
Query 68 FEEKSEDQMTQEEV---KEKKIQLLLLKIKNGTPP 99
FEEK + + +E+ KEK++ L+L + GTPP
Sbjct 186 FEEKYAELVLNKELDREKEKEVTLILTAVDGGTPP 220
> dre:404618 adhfe1, MGC77479, zgc:77479; alcohol dehydrogenase,
iron containing, 1 (EC:1.1.1.1); K11173 hydroxyacid-oxoacid
transhydrogenase [EC:1.1.99.24]
Length=471
Score = 28.5 bits (62), Expect = 6.2, Method: Composition-based stats.
Identities = 13/41 (31%), Positives = 22/41 (53%), Gaps = 0/41 (0%)
Query 4 PATPGVSSAATPLGVTPMYVMPEEGGISVKAVDPTGSLLDP 44
P T G S T + + + M + GI+ +A+ PT ++DP
Sbjct 189 PTTAGTGSETTGVAIFDLEDMKAKTGIANRALKPTLGMVDP 229
> hsa:5609 MAP2K7, Jnkk2, MAPKK7, MKK7, PRKMK7; mitogen-activated
protein kinase kinase 7 (EC:2.7.12.2); K04431 mitogen-activated
protein kinase kinase 7 [EC:2.7.12.2]
Length=419
Score = 28.1 bits (61), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 19/71 (26%), Positives = 32/71 (45%), Gaps = 8/71 (11%)
Query 3 GPATPGVSSAATPLGVTPMYVMPEEGGISVKAVDPTGSLLDPQGLGEASGGVQMKAEDFH 62
GP + T V +Y + E+ G+ + V P+ LLD +G Q+K DF
Sbjct 212 GPIPERILGKMTVAIVKALYYLKEKHGVIHRDVKPSNILLDERG--------QIKLCDFG 263
Query 63 FFSKLFEEKSE 73
+L + K++
Sbjct 264 ISGRLVDSKAK 274
> bbo:BBOV_III000320 17.m10445; hypothetical protein
Length=512
Score = 28.1 bits (61), Expect = 8.9, Method: Composition-based stats.
Identities = 15/35 (42%), Positives = 17/35 (48%), Gaps = 0/35 (0%)
Query 31 SVKAVDPTGSLLDPQGLGEASGGVQMKAEDFHFFS 65
+V V TGS P G+ SG V K E FH S
Sbjct 318 TVTVVSNTGSAYCPTGMSVVSGFVMTKTEPFHIDS 352
> mmu:26400 Map2k7, 5930412N11Rik, Jnkk2, MKK7, Prkmk7, sek2;
mitogen-activated protein kinase kinase 7 (EC:2.7.12.2); K04431
mitogen-activated protein kinase kinase 7 [EC:2.7.12.2]
Length=468
Score = 27.7 bits (60), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 19/71 (26%), Positives = 32/71 (45%), Gaps = 8/71 (11%)
Query 3 GPATPGVSSAATPLGVTPMYVMPEEGGISVKAVDPTGSLLDPQGLGEASGGVQMKAEDFH 62
GP + T V +Y + E+ G+ + V P+ LLD +G Q+K DF
Sbjct 228 GPIPERILGKMTVAIVKALYYLKEKHGVIHRDVKPSNILLDERG--------QIKLCDFG 279
Query 63 FFSKLFEEKSE 73
+L + K++
Sbjct 280 ISGRLVDSKAK 290
Lambda K H
0.313 0.133 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2296762580
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40