bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0581_orf1
Length=336
Score E
Sequences producing significant alignments: (Bits) Value
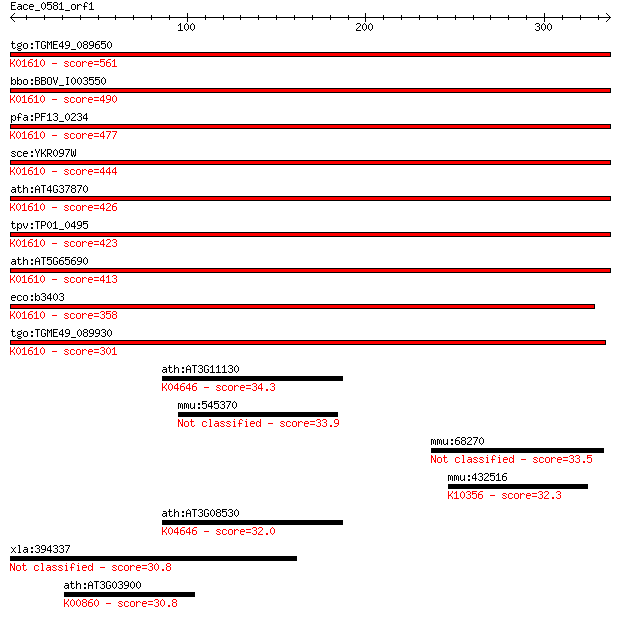
tgo:TGME49_089650 phosphoenolpyruvate carboxykinase (EC:4.1.1.... 561 1e-159
bbo:BBOV_I003550 19.m02385; phosphoenolpyruvate carboxykinase ... 490 3e-138
pfa:PF13_0234 PEPCK; phosphoenolpyruvate carboxykinase (EC:4.1... 477 3e-134
sce:YKR097W PCK1, JPM2, PPC1; Pck1p (EC:4.1.1.49); K01610 phos... 444 2e-124
ath:AT4G37870 PCK1; PCK1 (PHOSPHOENOLPYRUVATE CARBOXYKINASE 1)... 426 8e-119
tpv:TP01_0495 phosphoenolpyruvate carboxykinase; K01610 phosph... 423 5e-118
ath:AT5G65690 PCK2; PCK2 (PHOSPHOENOLPYRUVATE CARBOXYKINASE 2)... 413 5e-115
eco:b3403 pck, ECK3390, JW3366, pckA; phosphoenolpyruvate carb... 358 2e-98
tgo:TGME49_089930 phosphoenolpyruvate carboxykinase, putative ... 301 3e-81
ath:AT3G11130 clathrin heavy chain, putative; K04646 clathrin ... 34.3 0.74
mmu:545370 Hmcn1, EG545370, Gm201; hemicentin 1 33.9
mmu:68270 Lrrc50, 4930457P18Rik; leucine rich repeat containin... 33.5 1.3
mmu:432516 Myo1a, BBM-I, Myhl; myosin IA; K10356 myosin I 32.3 2.4
ath:AT3G08530 clathrin heavy chain, putative; K04646 clathrin ... 32.0 3.7
xla:394337 aifm3, MGC84340, nfrl-A; apoptosis-inducing factor,... 30.8 7.8
ath:AT3G03900 adenylylsulfate kinase, putative; K00860 adenyly... 30.8 8.3
> tgo:TGME49_089650 phosphoenolpyruvate carboxykinase (EC:4.1.1.49);
K01610 phosphoenolpyruvate carboxykinase (ATP) [EC:4.1.1.49]
Length=677
Score = 561 bits (1447), Expect = 1e-159, Method: Compositional matrix adjust.
Identities = 255/336 (75%), Positives = 296/336 (88%), Gaps = 0/336 (0%)
Query 1 YAGEMKKGILTLMMYLMPKQGRLPLHSSCNIGDKGDVTLFFGLSGTGKTTLSADPRRELI 60
YAGEMKKGILTLMMYLMPK+G+LPLHSSCN+G KGDVTLFFGLSGTGKTTLSADP R+LI
Sbjct 338 YAGEMKKGILTLMMYLMPKRGQLPLHSSCNVGPKGDVTLFFGLSGTGKTTLSADPSRQLI 397
Query 61 GDDEHVWTSKGVFNIEGGCYAKCKDLSREQEPEIYNAIRFGAVLENVVLEEEERNVDFSD 120
GDDEHVWT KGVFNIEGGCYAKCKDLS+EQEPEI+NAIRFG+VLENV+++ R VDF D
Sbjct 398 GDDEHVWTDKGVFNIEGGCYAKCKDLSKEQEPEIWNAIRFGSVLENVIIDGTTRVVDFRD 457
Query 121 STITENTRCAYPLHFIPNAKIPAAVNRHPSNIILLTCDAFGVLPPVWKLSASQVMYHFIS 180
+ITENTRCAYPL FIPNA +PA V+ HPSNIILLTCDAFGVLPP+ KL+ QVMYHFIS
Sbjct 458 VSITENTRCAYPLSFIPNALLPAKVDTHPSNIILLTCDAFGVLPPLSKLTPDQVMYHFIS 517
Query 181 GYTSKMAGTEIGILKPAATFSACYGSPFLAMHPMVYAEMLAAKLQQYNADAWLLNTGWVC 240
GYTSKMAGTEIGIL+P ATFSACYG+PFLAMHPMVYAEMLA KL+++NA AWLLNTGW+C
Sbjct 518 GYTSKMAGTEIGILEPTATFSACYGAPFLAMHPMVYAEMLAKKLKEHNAHAWLLNTGWIC 577
Query 241 GGYGAKEGRRISLQYTRAMVDAIHDGSLAKQQFEEMPIFKLRVPTAVPGVPSELLMPQRA 300
GGYG EGRRI L+YTR MVDAIH+G+ + +++ MP+F L VPTA+ GVPS+LL+PQ
Sbjct 578 GGYGMNEGRRIPLKYTRLMVDAIHNGAALQTEYQTMPVFNLAVPTAINGVPSDLLLPQTC 637
Query 301 WRDKKDLNRQLKKLAALFVENFVQYHDKATPEIIQA 336
W D+ L++Q++KLA+LFVENF Y D+ATP ++ A
Sbjct 638 WADQNKLDKQMRKLASLFVENFSNYTDRATPNVVAA 673
> bbo:BBOV_I003550 19.m02385; phosphoenolpyruvate carboxykinase
(EC:4.1.1.49); K01610 phosphoenolpyruvate carboxykinase (ATP)
[EC:4.1.1.49]
Length=589
Score = 490 bits (1261), Expect = 3e-138, Method: Compositional matrix adjust.
Identities = 226/336 (67%), Positives = 269/336 (80%), Gaps = 1/336 (0%)
Query 1 YAGEMKKGILTLMMYLMPKQGRLPLHSSCNIGDKGDVTLFFGLSGTGKTTLSADPRRELI 60
YAGEMKKGI TLMMYLMP +G LPLHSSCN+G+KGDVTLFFGLSGTGKTTLSADPRR LI
Sbjct 250 YAGEMKKGIFTLMMYLMPLRGLLPLHSSCNVGEKGDVTLFFGLSGTGKTTLSADPRRRLI 309
Query 61 GDDEHVWTSKGVFNIEGGCYAKCKDLSREQEPEIYNAIRFGAVLENVVLEEEERNVDFSD 120
GDDEHVWT GVFNIEGGCYAKCKDL RE+EPEI+NAI+FG+V+ENVV + +DF+D
Sbjct 310 GDDEHVWTESGVFNIEGGCYAKCKDLCREKEPEIFNAIKFGSVVENVVYDPATHEIDFTD 369
Query 121 STITENTRCAYPLHFIPNAKIPAAVNRHPSNIILLTCDAFGVLPPVWKLSASQVMYHFIS 180
+ITENTRCAYPL +IPN IPA V+ HPSNII LTCDAFGV+PPV KL+ Q MYHF+S
Sbjct 370 CSITENTRCAYPLEYIPNVMIPAKVDHHPSNIIFLTCDAFGVIPPVSKLTIEQAMYHFVS 429
Query 181 GYTSKMAGTEIGILKPAATFSACYGSPFLAMHPMVYAEMLAAKLQQYNADAWLLNTGWVC 240
GYTSKM GTE+G+ KP ATFSACY PFLAMHPMVYA++L KL+ ++ D WLLNTGW+
Sbjct 430 GYTSKMIGTEMGVTKPTATFSACYAEPFLAMHPMVYAKLLEDKLKAHSTDIWLLNTGWIQ 489
Query 241 GGYGAKEGRRISLQYTRAMVDAIHDGSLAKQQFEEMPIFKLRVPTAVPGVPSELLMPQRA 300
G +++GRRI L+YTRAMV+AIHDGSL K +F+ M +F L VPTA+ G+P+E+L +
Sbjct 490 GTIDSEKGRRIPLRYTRAMVNAIHDGSLKKAKFKPMDVFGLMVPTAIEGIPAEVLDQRLG 549
Query 301 WRDKKDLNRQLKKLAALFVENFVQYHDKATPEIIQA 336
W + QLK+LA F ENF QY +A I++
Sbjct 550 WGSAA-YDTQLKELANKFAENFKQYQSEANESILRG 584
> pfa:PF13_0234 PEPCK; phosphoenolpyruvate carboxykinase (EC:4.1.1.49);
K01610 phosphoenolpyruvate carboxykinase (ATP) [EC:4.1.1.49]
Length=583
Score = 477 bits (1227), Expect = 3e-134, Method: Compositional matrix adjust.
Identities = 218/336 (64%), Positives = 262/336 (77%), Gaps = 0/336 (0%)
Query 1 YAGEMKKGILTLMMYLMPKQGRLPLHSSCNIGDKGDVTLFFGLSGTGKTTLSADPRRELI 60
YAGEMKKGILTL MY MPK+G+LPLHSSCNIG K DVTLFFGLSGTGKTTLSAD R LI
Sbjct 244 YAGEMKKGILTLFMYKMPKEGKLPLHSSCNIGKKNDVTLFFGLSGTGKTTLSADANRYLI 303
Query 61 GDDEHVWTSKGVFNIEGGCYAKCKDLSREQEPEIYNAIRFGAVLENVVLEEEERNVDFSD 120
GDDEHVWT G+FNIEGGCYAKCK LS+ QEPEIY AI+FGA+LENVV++ R VD+++
Sbjct 304 GDDEHVWTDDGIFNIEGGCYAKCKGLSKRQEPEIYKAIKFGAILENVVMDPVTREVDYNN 363
Query 121 STITENTRCAYPLHFIPNAKIPAAVNRHPSNIILLTCDAFGVLPPVWKLSASQVMYHFIS 180
TITENTRCAYPL +I NAKIPA ++ HP NIILLTCDAFGV+PP+ KL Q+MYHF+S
Sbjct 364 CTITENTRCAYPLSYIENAKIPAYIHTHPQNIILLTCDAFGVIPPLCKLDVYQMMYHFVS 423
Query 181 GYTSKMAGTEIGILKPAATFSACYGSPFLAMHPMVYAEMLAAKLQQYNADAWLLNTGWVC 240
GYTSKMAGTE ILKP ATFS+CY +PFLA+HPM+YA+MLA K Q++ + WLLNTGW+
Sbjct 424 GYTSKMAGTEDNILKPTATFSSCYAAPFLALHPMIYAQMLADKYQKHKPNVWLLNTGWIY 483
Query 241 GGYGAKEGRRISLQYTRAMVDAIHDGSLAKQQFEEMPIFKLRVPTAVPGVPSELLMPQRA 300
G YG+ G RI L+YTR +VD IH+ L Q+++ PIF +P + G+P E++ P
Sbjct 484 GSYGSDNGIRIPLKYTRLLVDYIHENKLNNIQYKKTPIFNFNIPEHLEGIPDEVIDPLIG 543
Query 301 WRDKKDLNRQLKKLAALFVENFVQYHDKATPEIIQA 336
W+DK+D L+ LA F+ NF Y DKA PEI+
Sbjct 544 WKDKEDYLTNLQILAKEFINNFSLYLDKAGPEILSG 579
> sce:YKR097W PCK1, JPM2, PPC1; Pck1p (EC:4.1.1.49); K01610 phosphoenolpyruvate
carboxykinase (ATP) [EC:4.1.1.49]
Length=549
Score = 444 bits (1143), Expect = 2e-124, Method: Compositional matrix adjust.
Identities = 213/339 (62%), Positives = 260/339 (76%), Gaps = 4/339 (1%)
Query 1 YAGEMKKGILTLMMYLMP-KQGRLPLHSSCNIG-DKGDVTLFFGLSGTGKTTLSADPRRE 58
YAGEMKKGI T+M YLMP L LHSS N G GDVTLFFGLSGTGKTTLSADP R
Sbjct 207 YAGEMKKGIFTVMFYLMPVHHNVLTLHSSANQGIQNGDVTLFFGLSGTGKTTLSADPHRL 266
Query 59 LIGDDEHVWTSKGVFNIEGGCYAKCKDLSREQEPEIYNAIRFGAVLENVVLEEEERNVDF 118
LIGDDEH W+ GVFNIEGGCYAKC +LS E+EPEI++AI+FG+VLENV+ +E+ VD+
Sbjct 267 LIGDDEHCWSDHGVFNIEGGCYAKCINLSAEKEPEIFDAIKFGSVLENVIYDEKSHVVDY 326
Query 119 SDSTITENTRCAYPLHFIPNAKIPAAVNRHPSNIILLTCDAFGVLPPVWKLSASQVMYHF 178
DS+ITENTRCAYP+ +IP+AKIP + HP NIILLTCDA GVLPPV KL+ QVMYHF
Sbjct 327 DDSSITENTRCAYPIDYIPSAKIPCLADSHPKNIILLTCDASGVLPPVSKLTPEQVMYHF 386
Query 179 ISGYTSKMAGTEIGILKPAATFSACYGSPFLAMHPMVYAEMLAAKLQQYNADAWLLNTGW 238
ISGYTSKMAGTE G+ +P TFS+C+G PFLA+HP+ YA MLA K+ Q+ A+A+L+NTGW
Sbjct 387 ISGYTSKMAGTEQGVTEPEPTFSSCFGQPFLALHPIRYATMLATKMSQHKANAYLINTGW 446
Query 239 VCGGYGAKEGRRISLQYTRAMVDAIHDGSLAKQQFEEMPIFKLRVPTAVPGVPSELLMPQ 298
Y G+R L+YTRA++D+IHDGSLA + +E +PIF L+VPT V GVP+ELL P
Sbjct 447 TGSSY-VSGGKRCPLKYTRAILDSIHDGSLANETYETLPIFNLQVPTKVNGVPAELLNPA 505
Query 299 RAWRDKKDLNR-QLKKLAALFVENFVQYHDKATPEIIQA 336
+ W + R + LA LFV+NF Y D+ATP+++ A
Sbjct 506 KNWSQGESKYRGAVTNLANLFVQNFKIYQDRATPDVLAA 544
> ath:AT4G37870 PCK1; PCK1 (PHOSPHOENOLPYRUVATE CARBOXYKINASE
1); ATP binding / phosphoenolpyruvate carboxykinase (ATP)/ phosphoenolpyruvate
carboxykinase/ purine nucleotide binding
(EC:4.1.1.49); K01610 phosphoenolpyruvate carboxykinase (ATP)
[EC:4.1.1.49]
Length=671
Score = 426 bits (1094), Expect = 8e-119, Method: Compositional matrix adjust.
Identities = 205/343 (59%), Positives = 247/343 (72%), Gaps = 10/343 (2%)
Query 1 YAGEMKKGILTLMMYLMPKQGRLPLHSSCNIGDKGDVTLFFGLSGTGKTTLSADPRRELI 60
YAGEMKKG+ ++M YLMPK+ L LHS CN+G GDV LFFGLSGTGKTTLS D R LI
Sbjct 328 YAGEMKKGLFSVMHYLMPKRRILSLHSGCNMGKDGDVALFFGLSGTGKTTLSTDHNRYLI 387
Query 61 GDDEHVWTSKGVFNIEGGCYAKCKDLSREQEPEIYNAIRFGAVLENVVLEEEERNVDFSD 120
GDDEH WT GV NIEGGCYAKC DLSRE+EP+I+NAI+FG VLENVV +E R VD+SD
Sbjct 388 GDDEHCWTETGVSNIEGGCYAKCVDLSREKEPDIWNAIKFGTVLENVVFDEHTREVDYSD 447
Query 121 STITENTRCAYPLHFIPNAKIPAAVNRHPSNIILLTCDAFGVLPPVWKLSASQVMYHFIS 180
++TENTR AYP+ FIPNAKIP V HP+N+ILL CDAFGVLPPV KL+ +Q MYHFIS
Sbjct 448 KSVTENTRAAYPIEFIPNAKIP-CVGPHPTNVILLACDAFGVLPPVSKLNLAQTMYHFIS 506
Query 181 GYTSKMAGTEIGILKPAATFSACYGSPFLAMHPMVYAEMLAAKLQQYNADAWLLNTGWVC 240
GYT+ +AGTE GI +P ATFSAC+G+ F+ +HP YA MLA K++ A WL+NTGW
Sbjct 507 GYTALVAGTEDGIKEPTATFSACFGAAFIMLHPTKYAAMLAEKMKSQGATGWLVNTGWSG 566
Query 241 GGYGAKEGRRISLQYTRAMVDAIHDGSLAKQQFEEMPIFKLRVPTAVPGVPSELLMPQRA 300
G YG G RI L YTR ++DAIH GSL K +++ IF +PT + G+PSE+L P +
Sbjct 567 GSYGV--GNRIKLAYTRKIIDAIHSGSLLKANYKKTEIFGFEIPTEIEGIPSEILDPVNS 624
Query 301 WRDKKDLNRQLKKLAALFVENFVQYHD-------KATPEIIQA 336
W DKK L KL LF +NF + + K T EI+ A
Sbjct 625 WSDKKAHKDTLVKLGGLFKKNFEVFANHKIGVDGKLTEEILAA 667
> tpv:TP01_0495 phosphoenolpyruvate carboxykinase; K01610 phosphoenolpyruvate
carboxykinase (ATP) [EC:4.1.1.49]
Length=580
Score = 423 bits (1088), Expect = 5e-118, Method: Compositional matrix adjust.
Identities = 199/345 (57%), Positives = 255/345 (73%), Gaps = 10/345 (2%)
Query 1 YAGEMKKGILTLMMYLMPKQGRLPLHSSCNIGDKGDVTLFFGLSGTGKTTLSADPRRELI 60
YAGEMKKG+LTLMMYL+P++G LPLHSSCN+ +G+VTLFFGLSGTGKTTLS +P R+L+
Sbjct 229 YAGEMKKGVLTLMMYLLPQKGLLPLHSSCNVDSEGNVTLFFGLSGTGKTTLSTEPGRQLV 288
Query 61 GDDEHVWTSKGVFNIEGGCYAKCKDLSREQEPEIYNAIRFGAVLENVVLEEEERNVDFSD 120
GDDEHVWT +GVFN+EGGCYAKCKDLS E+EPEI+ A+RFG+VLENVVL + + VD++D
Sbjct 289 GDDEHVWTPEGVFNVEGGCYAKCKDLSAEREPEIFEAVRFGSVLENVVL-DSSKVVDYAD 347
Query 121 STITENTRCAYPLHFIPNAKIPAAVNRHPSNIILLTCDAFGVLPPVWKLSASQVMYHFIS 180
++TENTRCAYPL I N +PA V RHP+NII LTCDAFG LPPV L+ Q +YHF+S
Sbjct 348 VSVTENTRCAYPLRHIRNVCLPALVQRHPNNIIFLTCDAFGALPPVSLLNVHQAIYHFVS 407
Query 181 GYTSKMAGTEIGILKPAATFSACYGSPFLAMHPMVYAEMLAAKLQQYNAD-------AWL 233
GYT+KM GTEIG+ KP ATFSACY PFLA+ P+ YA +L KL + WL
Sbjct 408 GYTTKMVGTEIGVTKPTATFSACYAGPFLALSPLTYANLLYEKLTSTGSTNVKGSVRVWL 467
Query 234 LNTGWVCGGYGAKEGRRISLQYTRAMVDAIHDGSLAKQQ--FEEMPIFKLRVPTAVPGVP 291
LNTGW+ G + +G RI L+Y+R +VDAI+ G +++ Q FE P F VP V GVP
Sbjct 468 LNTGWIGGSSESDKGMRIPLRYSRRIVDAINRGEISESQDDFELFPYFGFLVPKEVTGVP 527
Query 292 SELLMPQRAWRDKKDLNRQLKKLAALFVENFVQYHDKATPEIIQA 336
E+L + +W D ++ Q+K +A F++NF QY +A +I++
Sbjct 528 KEVLRQELSWEDSQEHLDQVKLVAKKFIKNFNQYRAEANDDILKG 572
> ath:AT5G65690 PCK2; PCK2 (PHOSPHOENOLPYRUVATE CARBOXYKINASE
2); ATP binding / phosphoenolpyruvate carboxykinase (ATP)/ phosphoenolpyruvate
carboxykinase/ purine nucleotide binding
(EC:4.1.1.49); K01610 phosphoenolpyruvate carboxykinase (ATP)
[EC:4.1.1.49]
Length=670
Score = 413 bits (1061), Expect = 5e-115, Method: Compositional matrix adjust.
Identities = 200/343 (58%), Positives = 241/343 (70%), Gaps = 10/343 (2%)
Query 1 YAGEMKKGILTLMMYLMPKQGRLPLHSSCNIGDKGDVTLFFGLSGTGKTTLSADPRRELI 60
YAGEMKKG+ +M YLMPK+ L LHS CN+G GDV LFFGLSGTGKTTLS D R LI
Sbjct 327 YAGEMKKGLFGVMHYLMPKRKILSLHSGCNMGKDGDVALFFGLSGTGKTTLSTDHNRYLI 386
Query 61 GDDEHVWTSKGVFNIEGGCYAKCKDLSREQEPEIYNAIRFGAVLENVVLEEEERNVDFSD 120
GDDEH W+ GV NIEGGCYAKC DL+R++EP+I+NAI+FG VLENVV +E R VD++D
Sbjct 387 GDDEHCWSEAGVSNIEGGCYAKCIDLARDKEPDIWNAIKFGTVLENVVFDEHTREVDYTD 446
Query 121 STITENTRCAYPLHFIPNAKIPAAVNRHPSNIILLTCDAFGVLPPVWKLSASQVMYHFIS 180
++TENTR AYP+ +IPN+KIP V HP N+ILL CDAFGVLPP+ KL+ +Q MYHFIS
Sbjct 447 KSVTENTRAAYPIEYIPNSKIP-CVGPHPKNVILLACDAFGVLPPISKLNLAQTMYHFIS 505
Query 181 GYTSKMAGTEIGILKPAATFSACYGSPFLAMHPMVYAEMLAAKLQQYNADAWLLNTGWVC 240
GYT+ +AGTE G+ +P ATFSAC+G+ F+ +HP YA MLA K+Q A WL+NTGW
Sbjct 506 GYTALVAGTEEGVKEPRATFSACFGAAFIMLHPTKYAAMLAEKMQAQGATGWLVNTGWSG 565
Query 241 GGYGAKEGRRISLQYTRAMVDAIHDGSLAKQQFEEMPIFKLRVPTAVPGVPSELLMPQRA 300
G YG G RI L YTR ++DAIH GSL + + IF L +P V GVPSE+L P A
Sbjct 566 GSYGT--GSRIKLAYTRKIIDAIHSGSLLNASYRKTDIFGLEIPNEVEGVPSEILEPINA 623
Query 301 WRDKKDLNRQLKKLAALFVENFVQYHD-------KATPEIIQA 336
W DK L KLA LF NF + K T EI+ A
Sbjct 624 WPDKMAYEDTLLKLAGLFKSNFETFTSHKIGDDGKLTEEILAA 666
> eco:b3403 pck, ECK3390, JW3366, pckA; phosphoenolpyruvate carboxykinase
(EC:4.1.1.49); K01610 phosphoenolpyruvate carboxykinase
(ATP) [EC:4.1.1.49]
Length=540
Score = 358 bits (918), Expect = 2e-98, Method: Compositional matrix adjust.
Identities = 169/327 (51%), Positives = 229/327 (70%), Gaps = 7/327 (2%)
Query 1 YAGEMKKGILTLMMYLMPKQGRLPLHSSCNIGDKGDVTLFFGLSGTGKTTLSADPRRELI 60
Y GEMKKG+ ++M YL+P +G +H S N+G+KGDV +FFGLSGTGKTTLS DP+R LI
Sbjct 207 YGGEMKKGMFSMMNYLLPLKGIASMHCSANVGEKGDVAVFFGLSGTGKTTLSTDPKRRLI 266
Query 61 GDDEHVWTSKGVFNIEGGCYAKCKDLSREQEPEIYNAIRFGAVLENVVLEEEERNVDFSD 120
GDDEH W GVFN EGGCYAK LS+E EPEIYNAIR A+LENV + E+ +DF D
Sbjct 267 GDDEHGWDDDGVFNFEGGCYAKTIKLSKEAEPEIYNAIRRDALLENVTV-REDGTIDFDD 325
Query 121 STITENTRCAYPLHFIPNAKIPAAVNRHPSNIILLTCDAFGVLPPVWKLSASQVMYHFIS 180
+ TENTR +YP++ I N P + H + +I LT DAFGVLPPV +L+A Q YHF+S
Sbjct 326 GSKTENTRVSYPIYHIDNIVKPVSKAGHATKVIFLTADAFGVLPPVSRLTADQTQYHFLS 385
Query 181 GYTSKMAGTEIGILKPAATFSACYGSPFLAMHPMVYAEMLAAKLQQYNADAWLLNTGWVC 240
G+T+K+AGTE GI +P TFSAC+G+ FL++HP YAE+L ++Q A A+L+NTGW
Sbjct 386 GFTAKLAGTERGITEPTPTFSACFGAAFLSLHPTQYAEVLVKRMQAAGAQAYLVNTGW-- 443
Query 241 GGYGAKEGRRISLQYTRAMVDAIHDGSLAKQQFEEMPIFKLRVPTAVPGVPSELLMPQRA 300
G+RIS++ TRA++DAI +GSL + +P+F L +PT +PGV +++L P+
Sbjct 444 ----NGTGKRISIKDTRAIIDAILNGSLDNAETFTLPMFNLAIPTELPGVDTKILDPRNT 499
Query 301 WRDKKDLNRQLKKLAALFVENFVQYHD 327
+ + + + LA LF++NF +Y D
Sbjct 500 YASPEQWQEKAETLAKLFIDNFDKYTD 526
> tgo:TGME49_089930 phosphoenolpyruvate carboxykinase, putative
(EC:4.1.1.49); K01610 phosphoenolpyruvate carboxykinase (ATP)
[EC:4.1.1.49]
Length=610
Score = 301 bits (770), Expect = 3e-81, Method: Compositional matrix adjust.
Identities = 146/352 (41%), Positives = 223/352 (63%), Gaps = 19/352 (5%)
Query 1 YAGEMKKGILTLMMYLMPKQGRLPLHSSCNIGD-KGD--VTLFFGLSGTGKTTLSADPRR 57
YAGE+ KGI T M Y+MP +G LPLH+SC +G KGD +++ GL+ TGKT L A
Sbjct 241 YAGELHKGIFTYMNYVMPPKGVLPLHASCIVGSAKGDSDISVLLGLTATGKTALVATAEG 300
Query 58 ELIGDDEHVWTSKGVFNIEGGCYAKCKDLSREQEPEIYNAIRFGAVLENVVLEEEERNVD 117
+++ DDE +WT +GV + GGCY +CKD+ ++ NA+ +G+V+ENVVL+EE R V
Sbjct 301 KVLADDEVLWTPEGVSGVLGGCYVRCKDIDKDPCQTFVNAMVYGSVMENVVLDEETRRVY 360
Query 118 FSDSTITENTRCAYPLHFIPNA-KIPAAVNRHPSNIILLTCDAFGVLPPVWKLSASQVMY 176
F D+T+T+NTRC YPL F+ + K +V HP + I+L D FGV PPV +LS Q ++
Sbjct 361 FHDTTLTDNTRCTYPLTFLEHGMKGLPSVPLHPKHFIMLVNDTFGVFPPVARLSLPQALF 420
Query 177 HFISGYTSKMA-------GTEIGILKPAATFSACYGSPFLAMHPMVYAEMLAAKLQQYNA 229
+F+SG+T K GT + + + TFSAC G PFL +HP VY+++L K++++
Sbjct 421 YFLSGFTCKETTAEKRRDGTTLEMRRRIVTFSACSGCPFLPLHPTVYSDLLEKKIREHGT 480
Query 230 DAWLLNTGWVCG-GYGA---KEGRRISLQYTRAMVDAIHDGSLAKQQFEEMPIFKLRVPT 285
WL+NTGWV G YG G ++ ++ +R +V+AIHDG++++ F+ +P+F L +P
Sbjct 481 TVWLMNTGWVGGPAYGISFRSTGEKVPVEISRRIVNAIHDGTMSECTFKILPVFDLEIPV 540
Query 286 AVPGVPSELLMPQRAWR----DKKDLNRQLKKLAALFVENFVQYHDKATPEI 333
+P E L P +AW D + + + +A+LFV+NF Q+ + + E+
Sbjct 541 TFADIPEETLSPLQAWTARTGDSRAFENEARHVASLFVDNFKQFEGRVSSEV 592
> ath:AT3G11130 clathrin heavy chain, putative; K04646 clathrin
heavy chain
Length=1705
Score = 34.3 bits (77), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 30/106 (28%), Positives = 48/106 (45%), Gaps = 22/106 (20%)
Query 86 LSREQEPEIY-----NAIRFGAVLENVVLEEEERNVDFSDSTITENTRCAYPLHFIPNAK 140
LS ++PEI+ A + G + E +E R +F D+ T+N F+ AK
Sbjct 733 LSMSEDPEIHFKYIEAAAKTGQIKE---VERVTRESNFYDAEKTKN--------FLMEAK 781
Query 141 IPAAVNRHPSNIILLTCDAFGVLPPVWKLSASQVMYHFISGYTSKM 186
+P A P ++ CD FG +P + + M +I GY K+
Sbjct 782 LPDA---RP---LINVCDRFGFVPDLTHYLYTNNMLRYIEGYVQKV 821
> mmu:545370 Hmcn1, EG545370, Gm201; hemicentin 1
Length=5634
Score = 33.9 bits (76), Expect = 0.89, Method: Compositional matrix adjust.
Identities = 26/96 (27%), Positives = 49/96 (51%), Gaps = 7/96 (7%)
Query 95 YNAIRFGA-VLENVVLEEEERNVDFSDSTITENTR-CAYPLHFIPN-AKIPAAVNRHPSN 151
Y A +G +LENVVLE+ +++ E+TR +H +P ++P ++ +
Sbjct 4215 YTAQPYGELILENVVLEDSGTYTCVANNAAGEDTRIVTLAVHTLPTFTELPGDLSLNKGE 4274
Query 152 IILLTCDAFGVLPP--VWKLSASQVMYHF--ISGYT 183
+ L+C A G+ P W + + + HF I+G++
Sbjct 4275 QLRLSCKAVGIPLPKLTWTFNNNIIPAHFDSINGHS 4310
> mmu:68270 Lrrc50, 4930457P18Rik; leucine rich repeat containing
50
Length=634
Score = 33.5 bits (75), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 26/98 (26%), Positives = 47/98 (47%), Gaps = 4/98 (4%)
Query 237 GWVCGGYGAK--EGRRISLQYTRAMVDAIHDGSLAKQQFEEMPIFKLRVPTAVPGVPSEL 294
W GGY A+ E R+ + + + D++ ++ K++ EE + R T +P SE
Sbjct 287 AWARGGYAAEKEERRQWESREHKKITDSLEALAMIKRRAEERKKARDRGETPLPD--SEG 344
Query 295 LMPQRAWRDKKDLNRQLKKLAALFVENFVQYHDKATPE 332
+P ++K +++K LFVE + D+ PE
Sbjct 345 SIPTSPEAEEKQPMGEIQKKMELFVEESFEAKDELFPE 382
> mmu:432516 Myo1a, BBM-I, Myhl; myosin IA; K10356 myosin I
Length=1043
Score = 32.3 bits (72), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 20/90 (22%), Positives = 42/90 (46%), Gaps = 12/90 (13%)
Query 246 KEGRRISLQYTRAMVDAIHDGSLAKQQFEEMPIFKLRVPTAVPG---------VPSELLM 296
+E RR+ LQ ++ ++ G + +++M ++ + G + S +L+
Sbjct 690 EEQRRLRLQQLATLIQKVYRGWRCRTHYQQMRKSQILISAWFRGNKQKKHYGKIRSSVLL 749
Query 297 PQ---RAWRDKKDLNRQLKKLAALFVENFV 323
Q R WR +K+ + + AAL + NF+
Sbjct 750 IQAFVRGWRARKNYRKYFRSGAALTLANFI 779
> ath:AT3G08530 clathrin heavy chain, putative; K04646 clathrin
heavy chain
Length=1703
Score = 32.0 bits (71), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 29/106 (27%), Positives = 47/106 (44%), Gaps = 22/106 (20%)
Query 86 LSREQEPEIY-----NAIRFGAVLENVVLEEEERNVDFSDSTITENTRCAYPLHFIPNAK 140
LS ++PEI+ A + G + E +E R +F D+ T+N F+ AK
Sbjct 733 LSMSEDPEIHFKYIEAAAKTGQIKE---VERVTRESNFYDAEKTKN--------FLMEAK 781
Query 141 IPAAVNRHPSNIILLTCDAFGVLPPVWKLSASQVMYHFISGYTSKM 186
+P A P ++ CD F +P + + M +I GY K+
Sbjct 782 LPDA---RP---LINVCDRFSFVPDLTHYLYTNNMLRYIEGYVQKV 821
> xla:394337 aifm3, MGC84340, nfrl-A; apoptosis-inducing factor,
mitochondrion-associated, 3
Length=605
Score = 30.8 bits (68), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 38/160 (23%), Positives = 60/160 (37%), Gaps = 14/160 (8%)
Query 1 YAGEMKKGILTLMMYLMPKQGRLPLHSSCNIGDKGDVTLFFGLSGTGKTTLSADPRRELI 60
Y + KG+L+ + R P H +C GD+ F GL G K + + R I
Sbjct 112 YGAPLVKGVLS------KGRVRCPWHGACFNIATGDIEDFPGLDGLPKFQVKIEKERVYI 165
Query 61 GDDEHVWTSKGVFNIEGGCYAKCKDLSREQEPEIYNAIRFGAVLENVVLEEEERNVDFSD 120
+ S+ + AKC LS I N + GA +V E R FSD
Sbjct 166 RASKQALLSQRRTKV----MAKCIALS-NYSTAITNILIIGAGPAGLVCAETLRQEGFSD 220
Query 121 STITENTRCAYPLHFIPNAKIPAAVNRHPSNIILLTCDAF 160
+ + P +K+ +++ I L + + F
Sbjct 221 RIVMCTSEKNLPYD---RSKLSKSMDSQAEQIFLRSKEFF 257
> ath:AT3G03900 adenylylsulfate kinase, putative; K00860 adenylylsulfate
kinase [EC:2.7.1.25]
Length=208
Score = 30.8 bits (68), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 34/73 (46%), Gaps = 4/73 (5%)
Query 31 IGDKGDVTLFFGLSGTGKTTLSADPRRELIGDDEHVWTSKGVFNIEGGCYAKCKDLSREQ 90
+ KG V GLSG+GK+TL+ REL + + G N+ G KDL +
Sbjct 26 LNQKGCVVWITGLSGSGKSTLACSLSRELNNRGKLSYILDGD-NLRHGLN---KDLGFKA 81
Query 91 EPEIYNAIRFGAV 103
E + N R G V
Sbjct 82 EDRVENIRRVGEV 94
Lambda K H
0.320 0.136 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 14156784820
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40