bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0619_orf2
Length=210
Score E
Sequences producing significant alignments: (Bits) Value
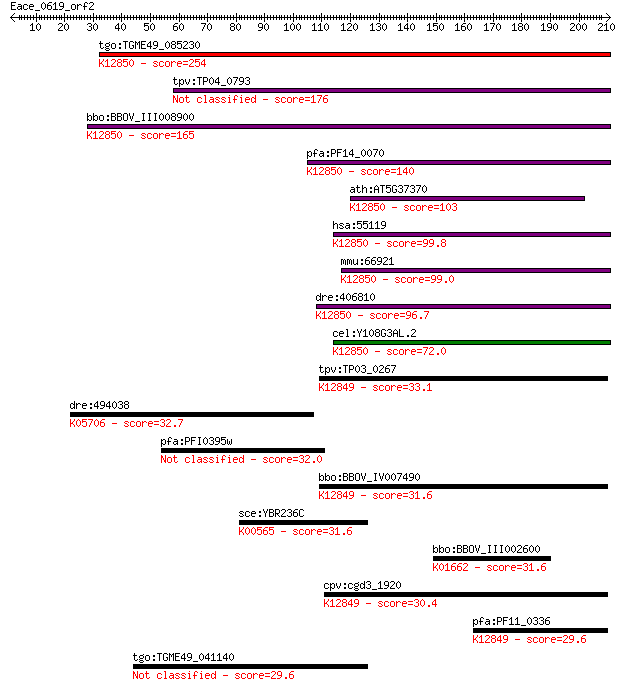
tgo:TGME49_085230 PRP38 family domain-containing protein ; K12... 254 2e-67
tpv:TP04_0793 hypothetical protein 176 5e-44
bbo:BBOV_III008900 17.m07776; PRP38 family protein; K12850 pre... 165 1e-40
pfa:PF14_0070 pre-mRNA splicing factor, putative; K12850 pre-m... 140 4e-33
ath:AT5G37370 ATSRL1; ATSRL1; binding; K12850 pre-mRNA-splicin... 103 3e-22
hsa:55119 PRPF38B, FLJ10330, MGC163218, MGC41809, NET1, RP11-2... 99.8 7e-21
mmu:66921 Prpf38b, 1110021E09Rik, AU018955; PRP38 pre-mRNA pro... 99.0 1e-20
dre:406810 prpf38b, wu:fc43b05, zgc:63496; PRP38 pre-mRNA proc... 96.7 5e-20
cel:Y108G3AL.2 hypothetical protein; K12850 pre-mRNA-splicing ... 72.0 2e-12
tpv:TP03_0267 hypothetical protein; K12849 pre-mRNA-splicing f... 33.1 0.70
dre:494038 snai2, cb147, cb355, id:ibd5003, sb:cb355, sb:cb569... 32.7 0.81
pfa:PFI0395w conserved Plasmodium protein, unknown function 32.0 1.7
bbo:BBOV_IV007490 23.m06033; pre-mRNA splicing factor; K12849 ... 31.6 1.9
sce:YBR236C ABD1; Abd1p (EC:2.1.1.56); K00565 mRNA (guanine-N7... 31.6 2.3
bbo:BBOV_III002600 17.m07248; 1-deoxy-D-xylulose-5-phosphate s... 31.6 2.3
cpv:cgd3_1920 possible PRP38 pre-mRNA splicing factor family m... 30.4 5.0
pfa:PF11_0336 pre-mRNA splicing factor, putative; K12849 pre-m... 29.6 7.2
tgo:TGME49_041140 DEAD/DEAH box helicase, putative (EC:6.1.1.11) 29.6 8.3
> tgo:TGME49_085230 PRP38 family domain-containing protein ; K12850
pre-mRNA-splicing factor 38B
Length=614
Score = 254 bits (648), Expect = 2e-67, Method: Compositional matrix adjust.
Identities = 121/183 (66%), Positives = 143/183 (78%), Gaps = 5/183 (2%)
Query 32 SAAAPVAVAPLAAAPATDVAAAENDKEDEEE--AIARCHLHTKPQLSCKFCRKYKSFTQQ 89
SA P VA PAT A + +ED + A+ RCHLHTKPQL CKFCRKYKSF QQ
Sbjct 60 SAGTPAVVA-APGPPATTAVGAVDAEEDSGDTWAVNRCHLHTKPQLHCKFCRKYKSFAQQ 118
Query 90 MGVLQQRAIA--ADEETQKKNMVQMTDSTTYNVNQLLRANILSSEYFKSLHQLKSFHEVV 147
+G+LQ+R + D++ K+NMV+MTDSTTYNVN LLR+NILSSEYFKSLH+LK+ EVV
Sbjct 119 LGILQKRELKQQEDDDGMKRNMVEMTDSTTYNVNALLRSNILSSEYFKSLHELKTVPEVV 178
Query 148 AEVAAYADHAEPYCSGSTRAPSTLFCCLYKLFTLKLTDKQMHMLLNHRESPYVRCTGFLY 207
E+ YA HAEPYCSGS+RAPSTLFCCLYKLFT+KLT KQ+ LL++ +SPYVRCTGFLY
Sbjct 179 DEITQYAQHAEPYCSGSSRAPSTLFCCLYKLFTMKLTTKQLEQLLDYSDSPYVRCTGFLY 238
Query 208 LRY 210
LRY
Sbjct 239 LRY 241
> tpv:TP04_0793 hypothetical protein
Length=239
Score = 176 bits (446), Expect = 5e-44, Method: Compositional matrix adjust.
Identities = 83/153 (54%), Positives = 115/153 (75%), Gaps = 2/153 (1%)
Query 58 EDEEEAIARCHLHTKPQLSCKFCRKYKSFTQQMGVLQQRAIAADEETQKKNMVQMTDSTT 117
++ E++ RCHLHTKP+L+CKFCRK+KS ++ + ++ + K N + MT+S T
Sbjct 77 QNSLESVQRCHLHTKPELNCKFCRKFKSSVHEISK-IAQKKSSSSKEDKANQIPMTNSVT 135
Query 118 YNVNQLLRANILSSEYFKSLHQLKSFHEVVAEVAAYADHAEPYCSGSTRAPSTLFCCLYK 177
YN+N LLR+NILSSEY+KSL +K+F++V E+ +A H+EPYCS +TRAPST+FCCLY+
Sbjct 136 YNMNDLLRSNILSSEYYKSL-SVKNFYQVFDELVQFATHSEPYCSTATRAPSTIFCCLYR 194
Query 178 LFTLKLTDKQMHMLLNHRESPYVRCTGFLYLRY 210
LKLT+KQM+ LL + +SPY RC GFLYLRY
Sbjct 195 FLVLKLTEKQMNFLLENVKSPYARCCGFLYLRY 227
> bbo:BBOV_III008900 17.m07776; PRP38 family protein; K12850 pre-mRNA-splicing
factor 38B
Length=483
Score = 165 bits (417), Expect = 1e-40, Method: Compositional matrix adjust.
Identities = 91/200 (45%), Positives = 130/200 (65%), Gaps = 19/200 (9%)
Query 28 TCVASAAAPVAVAPLAAAPAT--DVAAAE---------------NDKEDEEEAIARCHLH 70
T ++++ V + P+A T ++AAA ED E++ CH+H
Sbjct 74 TASSTSSVNVGIQPIATELGTSDNIAAASVPGHMELPPTSVSYTTHTEDLLESVQTCHMH 133
Query 71 TKPQLSCKFCRKYKSFTQQMGVLQQRAIAADEETQKKNMVQMTDSTTYNVNQLLRANILS 130
TKP L+CKFCRKYKS ++ L Q + + E T++ + + MT+S+TYN+N LL NIL+
Sbjct 134 TKPDLNCKFCRKYKSAVHELSRLSQ--LRSQEATERPDQLDMTNSSTYNMNTLLLNNILN 191
Query 131 SEYFKSLHQLKSFHEVVAEVAAYADHAEPYCSGSTRAPSTLFCCLYKLFTLKLTDKQMHM 190
S+Y+KSL + S H ++ E+A YADH EPYC +TR PSTLFCCL+KLFTL+LT++QM
Sbjct 192 SDYYKSLSTMTSHHSIIDELAQYADHVEPYCKTATRVPSTLFCCLHKLFTLRLTERQMED 251
Query 191 LLNHRESPYVRCTGFLYLRY 210
L++ +SPY RC GFLYLR+
Sbjct 252 LIDCTKSPYPRCCGFLYLRF 271
> pfa:PF14_0070 pre-mRNA splicing factor, putative; K12850 pre-mRNA-splicing
factor 38B
Length=690
Score = 140 bits (352), Expect = 4e-33, Method: Composition-based stats.
Identities = 69/106 (65%), Positives = 88/106 (83%), Gaps = 0/106 (0%)
Query 105 QKKNMVQMTDSTTYNVNQLLRANILSSEYFKSLHQLKSFHEVVAEVAAYADHAEPYCSGS 164
+KKN ++MT++TTYNVN LLR NILSSEYFKSL +K+F EVV E+ +YADH EPYC GS
Sbjct 168 EKKNCLEMTNTTTYNVNTLLRNNILSSEYFKSLIPIKTFKEVVDEIHSYADHVEPYCIGS 227
Query 165 TRAPSTLFCCLYKLFTLKLTDKQMHMLLNHRESPYVRCTGFLYLRY 210
RAPSTLFCCLYK FT++L++KQ+ L+ +++S Y+R GFLYLRY
Sbjct 228 NRAPSTLFCCLYKFFTMQLSEKQLKSLIENKDSCYIRACGFLYLRY 273
> ath:AT5G37370 ATSRL1; ATSRL1; binding; K12850 pre-mRNA-splicing
factor 38B
Length=111
Score = 103 bits (258), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 44/82 (53%), Positives = 60/82 (73%), Gaps = 0/82 (0%)
Query 120 VNQLLRANILSSEYFKSLHQLKSFHEVVAEVAAYADHAEPYCSGSTRAPSTLFCCLYKLF 179
+ ++L NILSS+YFK L+ LK++HEV+ E+ +H EP+ G+ R PST +C LYK F
Sbjct 15 LEKVLSMNILSSDYFKELYGLKTYHEVIDEIYNQVNHVEPWMGGNCRGPSTAYCLLYKFF 74
Query 180 TLKLTDKQMHMLLNHRESPYVR 201
T+KLT KQMH LL H +SPY+R
Sbjct 75 TMKLTVKQMHGLLKHTDSPYIR 96
> hsa:55119 PRPF38B, FLJ10330, MGC163218, MGC41809, NET1, RP11-293A10.1;
PRP38 pre-mRNA processing factor 38 (yeast) domain
containing B; K12850 pre-mRNA-splicing factor 38B
Length=546
Score = 99.8 bits (247), Expect = 7e-21, Method: Compositional matrix adjust.
Identities = 51/116 (43%), Positives = 68/116 (58%), Gaps = 19/116 (16%)
Query 114 DSTTYNVNQLLRANILSSEYFK-SLHQLKSFHEVVAEVAAYADHAEPYCSGSTRA----- 167
+ T N+N ++ NILSS YFK L++LK++HEVV E+ H EP+ GS +
Sbjct 52 NEKTMNLNPMILTNILSSPYFKVQLYELKTYHEVVDEIYFKVTHVEPWEKGSRKTAGQTG 111
Query 168 -------------PSTLFCCLYKLFTLKLTDKQMHMLLNHRESPYVRCTGFLYLRY 210
ST FC LYKLFTLKLT KQ+ L+ H +SPY+R GF+Y+RY
Sbjct 112 MCGGVRGVGTGGIVSTAFCLLYKLFTLKLTRKQVMGLITHTDSPYIRALGFMYIRY 167
> mmu:66921 Prpf38b, 1110021E09Rik, AU018955; PRP38 pre-mRNA processing
factor 38 (yeast) domain containing B; K12850 pre-mRNA-splicing
factor 38B
Length=542
Score = 99.0 bits (245), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 51/113 (45%), Positives = 67/113 (59%), Gaps = 19/113 (16%)
Query 117 TYNVNQLLRANILSSEYFK-SLHQLKSFHEVVAEVAAYADHAEPYCSGSTRA-------- 167
T N+N ++ NILSS YFK L++LK++HEVV E+ H EP+ GS +
Sbjct 56 TMNLNPMILTNILSSPYFKVQLYELKTYHEVVDEIYFKVTHVEPWEKGSRKTAGQTGMCG 115
Query 168 ----------PSTLFCCLYKLFTLKLTDKQMHMLLNHRESPYVRCTGFLYLRY 210
ST FC LYKLFTLKLT KQ+ L+ H +SPY+R GF+Y+RY
Sbjct 116 GVRGVGTGGIVSTAFCLLYKLFTLKLTRKQVMGLITHTDSPYIRALGFMYIRY 168
> dre:406810 prpf38b, wu:fc43b05, zgc:63496; PRP38 pre-mRNA processing
factor 38 (yeast) domain containing B; K12850 pre-mRNA-splicing
factor 38B
Length=501
Score = 96.7 bits (239), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 50/122 (40%), Positives = 70/122 (57%), Gaps = 19/122 (15%)
Query 108 NMVQMTDSTTYNVNQLLRANILSSEYFK-SLHQLKSFHEVVAEVAAYADHAEPYCSGSTR 166
N+ + T N+N ++ N+LSS YFK L++LK++HEVV E+ +H EP+ GS +
Sbjct 24 NLPLWGNEKTMNLNPMILTNVLSSPYFKVQLYELKTYHEVVDEIYFKVNHVEPWEKGSRK 83
Query 167 A------------------PSTLFCCLYKLFTLKLTDKQMHMLLNHRESPYVRCTGFLYL 208
ST FC LYKLFTLKLT KQ+ L+ H +SP +R GF+Y+
Sbjct 84 TAGQTGMCGGVRGVGTGGIVSTAFCLLYKLFTLKLTRKQVMGLITHSDSPDIRALGFMYI 143
Query 209 RY 210
RY
Sbjct 144 RY 145
> cel:Y108G3AL.2 hypothetical protein; K12850 pre-mRNA-splicing
factor 38B
Length=327
Score = 72.0 bits (175), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 37/116 (31%), Positives = 62/116 (53%), Gaps = 19/116 (16%)
Query 114 DSTTYNVNQLLRANILSSEYFKS-LHQLKSFHEVVAEVAAYADHAEPYCSGSTR------ 166
+ T N+N L+ NI S Y+K+ L ++ +F ++ ++ H EP+ G+ R
Sbjct 60 NKVTMNLNTLVLENIRESYYYKNNLVEIDNFQTLIEQIFYQVKHLEPWEKGTRRLQGMTG 119
Query 167 ------------APSTLFCCLYKLFTLKLTDKQMHMLLNHRESPYVRCTGFLYLRY 210
S+ +C LY+LF LK++ KQ+ +LN R+S Y+R GF+Y+RY
Sbjct 120 MCGGVRGVGAGGVVSSAYCLLYRLFNLKISRKQLISMLNSRQSVYIRGLGFMYIRY 175
> tpv:TP03_0267 hypothetical protein; K12849 pre-mRNA-splicing
factor 38A
Length=193
Score = 33.1 bits (74), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 25/108 (23%), Positives = 46/108 (42%), Gaps = 7/108 (6%)
Query 109 MVQMTDSTTYNVN----QLLRANILSSEYFKSLHQLKSFHEVVAEVAAYADHAEPYCSGS 164
M TD T + ++ Q L + IL + + S + +S + AE Y G+
Sbjct 1 MANRTDPTAHLIHGTNPQFLFSKILRDKVYNSFYWKESCFGLTAESLIDKAVQIKYVGGT 60
Query 165 ---TRAPSTLFCCLYKLFTLKLTDKQMHMLLNHRESPYVRCTGFLYLR 209
R PS C + K+ ++ + +H + + + Y+R G Y+R
Sbjct 61 FGGNRQPSPFICLVLKMLQIQPEMEIVHEYIKNEDYKYLRALGIYYMR 108
> dre:494038 snai2, cb147, cb355, id:ibd5003, sb:cb355, sb:cb569,
slug, wu:fb83e12, wu:fd19g11, zgc:92564; snail homolog 2
(Drosophila); K05706 snail 2
Length=257
Score = 32.7 bits (73), Expect = 0.81, Method: Compositional matrix adjust.
Identities = 19/93 (20%), Positives = 44/93 (47%), Gaps = 10/93 (10%)
Query 22 IAQRETTCVASAAAPVAVAPLAAAPAT--------DVAAAENDKEDEEEAIARCHLHTKP 73
I T+ + + P ++P++ P++ D + +E+ + DE+E I L
Sbjct 56 ITVWTTSNLPLSPLPHDLSPISGYPSSLSDTSSNKDHSGSESPRSDEDERIQSTKLSDAE 115
Query 74 QLSCKFCRKYKSFTQQMGVLQQRAIAADEETQK 106
+ C C KS++ G+++ + + D +++K
Sbjct 116 KFQCGLCN--KSYSTYSGLMKHKQLHCDAQSRK 146
> pfa:PFI0395w conserved Plasmodium protein, unknown function
Length=1516
Score = 32.0 bits (71), Expect = 1.7, Method: Composition-based stats.
Identities = 17/57 (29%), Positives = 28/57 (49%), Gaps = 0/57 (0%)
Query 54 ENDKEDEEEAIARCHLHTKPQLSCKFCRKYKSFTQQMGVLQQRAIAADEETQKKNMV 110
EN+K+D ++ + C C KY F QQ Q++I ++ QKKN++
Sbjct 829 ENEKKDFDKYYKIINNMINQNTVCCACAKYNYFNQQKYKQTQQSINYHQDIQKKNII 885
> bbo:BBOV_IV007490 23.m06033; pre-mRNA splicing factor; K12849
pre-mRNA-splicing factor 38A
Length=202
Score = 31.6 bits (70), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 27/108 (25%), Positives = 45/108 (41%), Gaps = 7/108 (6%)
Query 109 MVQMTDSTTYNVN----QLLRANILSSEYFKSLHQLKSFHEVVAEVAAYADHAEPYCSGS 164
M TD T V+ Q L + IL + + S++ +S + AE Y G+
Sbjct 1 MANRTDPTAVMVHGTNPQNLFSKILRDKVYNSMYWKESCFGLTAESIIDKAIDLQYIGGT 60
Query 165 ---TRAPSTLFCCLYKLFTLKLTDKQMHMLLNHRESPYVRCTGFLYLR 209
R PS C + KL ++ + + + + E Y+R G Y+R
Sbjct 61 FGGNRQPSPFLCLVLKLLQIQPEIEIIQEYIRNEEFKYLRALGIYYMR 108
> sce:YBR236C ABD1; Abd1p (EC:2.1.1.56); K00565 mRNA (guanine-N7-)-methyltransferase
[EC:2.1.1.56]
Length=436
Score = 31.6 bits (70), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 30/45 (66%), Gaps = 0/45 (0%)
Query 81 RKYKSFTQQMGVLQQRAIAADEETQKKNMVQMTDSTTYNVNQLLR 125
R+++ + Q+ + +QRA EE K++ ++MT + + NV+Q++R
Sbjct 79 RRHERYDQEERLRKQRAQKLREEQLKRHEIEMTANRSINVDQIVR 123
> bbo:BBOV_III002600 17.m07248; 1-deoxy-D-xylulose-5-phosphate
synthase family protein (EC:2.2.1.7); K01662 1-deoxy-D-xylulose-5-phosphate
synthase [EC:2.2.1.7]
Length=686
Score = 31.6 bits (70), Expect = 2.3, Method: Composition-based stats.
Identities = 12/41 (29%), Positives = 22/41 (53%), Gaps = 0/41 (0%)
Query 149 EVAAYADHAEPYCSGSTRAPSTLFCCLYKLFTLKLTDKQMH 189
+V HA + +G+T + + FCC+Y F + D+ +H
Sbjct 412 DVGIAEQHAVTFAAGTTISGAKPFCCIYSTFMQRALDQVIH 452
> cpv:cgd3_1920 possible PRP38 pre-mRNA splicing factor family
member ; K12849 pre-mRNA-splicing factor 38A
Length=261
Score = 30.4 bits (67), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 25/100 (25%), Positives = 45/100 (45%), Gaps = 4/100 (4%)
Query 111 QMTDSTTYNVNQLLRANILSSEYFKSL-HQLKSFHEVVAEVAAYADHAEPYCSGSTRAPS 169
Q + ++V+ +LR + SS Y+K L S E + + A D+ G R +
Sbjct 5 QNSHHKLFSVSSILRDRVFSSIYWKGECFALDS--ETILDKAVLLDYIGT-TYGGDRKAT 61
Query 170 TLFCCLYKLFTLKLTDKQMHMLLNHRESPYVRCTGFLYLR 209
C L KL ++ + + + +N+ Y+ G +YLR
Sbjct 62 PFLCLLVKLLQIRPSTEIVLEYINNPRFKYLTALGIVYLR 101
> pfa:PF11_0336 pre-mRNA splicing factor, putative; K12849 pre-mRNA-splicing
factor 38A
Length=386
Score = 29.6 bits (65), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 12/47 (25%), Positives = 22/47 (46%), Gaps = 0/47 (0%)
Query 163 GSTRAPSTLFCCLYKLFTLKLTDKQMHMLLNHRESPYVRCTGFLYLR 209
G R P+ C + KL ++ ++ + + + Y+R G YLR
Sbjct 62 GGNRKPTRFLCLILKLLQIQPDKDIIYEYIKNEDFVYLRALGIFYLR 108
> tgo:TGME49_041140 DEAD/DEAH box helicase, putative (EC:6.1.1.11)
Length=1454
Score = 29.6 bits (65), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 19/82 (23%), Positives = 39/82 (47%), Gaps = 13/82 (15%)
Query 44 AAPATDVAAAENDKEDEEEAIARCHLHTKPQLSCKFCRKYKSFTQQMGVLQQRAIAADEE 103
AA A ++ +A++D+EDEE+A + K T+ + LQ R + A E+
Sbjct 906 AADAGEIESAKDDEEDEEKAFVSAE-------------ERKEMTRVLKRLQSRGLVACED 952
Query 104 TQKKNMVQMTDSTTYNVNQLLR 125
+ + + D+ + +L++
Sbjct 953 AMEGEIGKAVDTENESGKRLMK 974
Lambda K H
0.322 0.131 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6559193640
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40