bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0664_orf1
Length=129
Score E
Sequences producing significant alignments: (Bits) Value
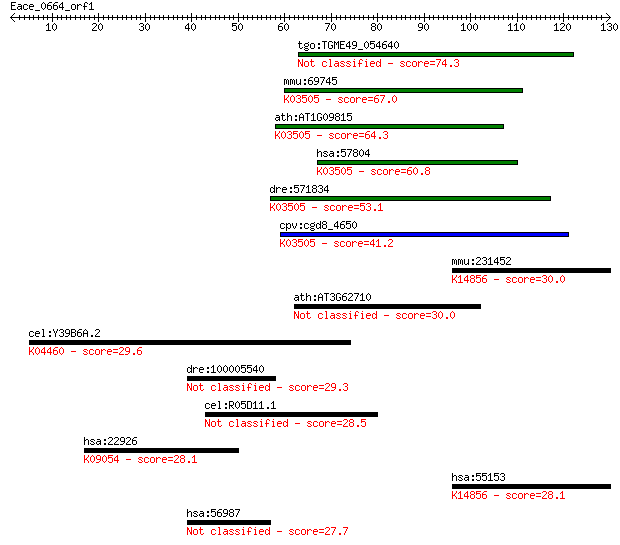
tgo:TGME49_054640 DNA polymerase delta subunit 4, putative 74.3 9e-14
mmu:69745 Pold4, 2410012M21Rik, AI463381, AW060307, Polds, p12... 67.0 1e-11
ath:AT1G09815 POLD4; POLD4 (POLYMERASE DELTA 4); DNA-directed ... 64.3 1e-10
hsa:57804 POLD4, POLDS, p12; polymerase (DNA-directed), delta ... 60.8 9e-10
dre:571834 im:7142218; si:dkey-28b4.7; K03505 DNA polymerase d... 53.1 2e-07
cpv:cgd8_4650 hypothetical protein ; K03505 DNA polymerase del... 41.2 8e-04
mmu:231452 Sdad1, 4931421J16, AA591032, AW538460; SDA1 domain ... 30.0 1.7
ath:AT3G62710 glycosyl hydrolase family 3 protein 30.0 2.0
cel:Y39B6A.2 pph-5; Protein PHosphatase family member (pph-5);... 29.6 2.8
dre:100005540 HMG-BOX transcription factor BBX-like 29.3 3.4
cel:R05D11.1 daf-8; abnormal DAuer Formation family member (da... 28.5 5.7
hsa:22926 ATF6, ATF6A; activating transcription factor 6; K090... 28.1 6.3
hsa:55153 SDAD1, DKFZp686E22207, FLJ10498; SDA1 domain contain... 28.1 7.4
hsa:56987 BBX, HBP2, HSPC339, MDS001; bobby sox homolog (Droso... 27.7 9.7
> tgo:TGME49_054640 DNA polymerase delta subunit 4, putative
Length=313
Score = 74.3 bits (181), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 33/59 (55%), Positives = 42/59 (71%), Gaps = 1/59 (1%)
Query 63 EQLLRRFDMEAMYGPCVGLTREERWKRAAEMGLDPPAEVMQALQRHPKSTLSVFDQRMS 121
E +L+RFDM YGPC GL+R ERW RA E+G+ PP EV QA+Q + S+FDQR+S
Sbjct 245 EPMLQRFDMAVQYGPCCGLSRSERWYRADELGMKPPLEVKQAIQ-FTRGDASIFDQRLS 302
> mmu:69745 Pold4, 2410012M21Rik, AI463381, AW060307, Polds, p12;
polymerase (DNA-directed), delta 4; K03505 DNA polymerase
delta subunit 4
Length=107
Score = 67.0 bits (162), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 30/52 (57%), Positives = 39/52 (75%), Gaps = 1/52 (1%)
Query 60 DETE-QLLRRFDMEAMYGPCVGLTREERWKRAAEMGLDPPAEVMQALQRHPK 110
+ETE +LLR+FD+ YGPC G+TR +RW RA +MGL PP EV Q L+ HP+
Sbjct 42 EETELELLRQFDLAWQYGPCTGITRLQRWSRAEQMGLKPPLEVYQVLKAHPE 93
> ath:AT1G09815 POLD4; POLD4 (POLYMERASE DELTA 4); DNA-directed
DNA polymerase; K03505 DNA polymerase delta subunit 4
Length=124
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 23/49 (46%), Positives = 38/49 (77%), Gaps = 0/49 (0%)
Query 58 EADETEQLLRRFDMEAMYGPCVGLTREERWKRAAEMGLDPPAEVMQALQ 106
+ D+ E++LR+FDM YGPC+G+TR +RW+RA +G++PP E+ + L+
Sbjct 62 DYDKEEEMLRQFDMNITYGPCLGMTRLDRWERAVRLGMNPPNEIEKLLK 110
> hsa:57804 POLD4, POLDS, p12; polymerase (DNA-directed), delta
4; K03505 DNA polymerase delta subunit 4
Length=107
Score = 60.8 bits (146), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 25/43 (58%), Positives = 32/43 (74%), Gaps = 0/43 (0%)
Query 67 RRFDMEAMYGPCVGLTREERWKRAAEMGLDPPAEVMQALQRHP 109
R+FD+ YGPC G+TR +RW RA +MGL+PP EV Q L+ HP
Sbjct 50 RQFDLAWQYGPCTGITRLQRWCRAKQMGLEPPPEVWQVLKTHP 92
> dre:571834 im:7142218; si:dkey-28b4.7; K03505 DNA polymerase
delta subunit 4
Length=117
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 24/63 (38%), Positives = 41/63 (65%), Gaps = 3/63 (4%)
Query 57 AEADETEQLLRRFDMEAMYGPCVGLTREERWKRAAEMGLDPPAEVMQALQRH---PKSTL 113
+E ++ Q L+ FD++ +GPC G++R +RW+RAA GL+PP E+ L + P+ T
Sbjct 50 SEREKDIQELKNFDLDWKFGPCTGISRLQRWERAALHGLNPPQEIKDILLKEDTDPEYTQ 109
Query 114 SVF 116
S++
Sbjct 110 SLW 112
> cpv:cgd8_4650 hypothetical protein ; K03505 DNA polymerase delta
subunit 4
Length=128
Score = 41.2 bits (95), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 19/70 (27%), Positives = 39/70 (55%), Gaps = 8/70 (11%)
Query 59 ADETEQLL---RRFDMEAMYGPCVGLTREERWKRAAEMGLDPPAEVMQALQRH-----PK 110
A++ EQLL FD++ +GPC ++R +R +RA + G+ + + + +H +
Sbjct 54 ANDNEQLLNTLHSFDLDHTFGPCSEISRSKRLERAKKFGIAIDLNLEKMINKHCGDVNSE 113
Query 111 STLSVFDQRM 120
++S+ D+ M
Sbjct 114 MSVSIIDKLM 123
> mmu:231452 Sdad1, 4931421J16, AA591032, AW538460; SDA1 domain
containing 1; K14856 protein SDA1
Length=687
Score = 30.0 bits (66), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 16/36 (44%), Positives = 23/36 (63%), Gaps = 4/36 (11%)
Query 96 DPPAEVMQALQR--HPKSTLSVFDQRMSPDKPSAAL 129
DPPA V + LQ+ H KS + +F ++ P+KPS L
Sbjct 23 DPPAYVEEFLQQYNHYKSNMEIF--KLQPNKPSKEL 56
> ath:AT3G62710 glycosyl hydrolase family 3 protein
Length=650
Score = 30.0 bits (66), Expect = 2.0, Method: Composition-based stats.
Identities = 14/40 (35%), Positives = 20/40 (50%), Gaps = 0/40 (0%)
Query 62 TEQLLRRFDMEAMYGPCVGLTREERWKRAAEMGLDPPAEV 101
T Q +R + + PCV + R+ RW R E + PA V
Sbjct 171 TAQEVRATGVAQAFAPCVAVCRDPRWGRCYESYSEDPAVV 210
> cel:Y39B6A.2 pph-5; Protein PHosphatase family member (pph-5);
K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=496
Score = 29.6 bits (65), Expect = 2.8, Method: Composition-based stats.
Identities = 26/73 (35%), Positives = 34/73 (46%), Gaps = 5/73 (6%)
Query 5 ATAATDAPTAAPVNPSK----YRSPERFAAPGFTEKELKDTADAVVKGNPRLKWCPAEAD 60
+A DA A ++PS YR A G +K L D AVVK P K A+ D
Sbjct 79 GSALEDADNAIAIDPSYVKGFYRRATANMALGRFKKALTDY-QAVVKVCPNDKDARAKFD 137
Query 61 ETEQLLRRFDMEA 73
E +++RR EA
Sbjct 138 ECSKIVRRQKFEA 150
> dre:100005540 HMG-BOX transcription factor BBX-like
Length=967
Score = 29.3 bits (64), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 11/19 (57%), Positives = 13/19 (68%), Gaps = 0/19 (0%)
Query 39 KDTADAVVKGNPRLKWCPA 57
K+ DA +K NP KWCPA
Sbjct 138 KEYKDAFMKANPGYKWCPA 156
> cel:R05D11.1 daf-8; abnormal DAuer Formation family member (daf-8)
Length=546
Score = 28.5 bits (62), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 15/37 (40%), Positives = 21/37 (56%), Gaps = 6/37 (16%)
Query 43 DAVVKGNPRLKWCPAEADETEQLLRRFDMEAMYGPCV 79
DAV+KG+P+ + CPA E+L+ F M CV
Sbjct 59 DAVIKGDPKTRCCPA---HNEKLIGNFGRAIM---CV 89
> hsa:22926 ATF6, ATF6A; activating transcription factor 6; K09054
activating transcription factor 6
Length=670
Score = 28.1 bits (61), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 12/33 (36%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 17 VNPSKYRSPERFAAPGFTEKELKDTADAVVKGN 49
+NPS + +R GF+ KE +DT+D +++ N
Sbjct 405 MNPSVSPANQRRHLLGFSAKEAQDTSDGIIQKN 437
> hsa:55153 SDAD1, DKFZp686E22207, FLJ10498; SDA1 domain containing
1; K14856 protein SDA1
Length=687
Score = 28.1 bits (61), Expect = 7.4, Method: Composition-based stats.
Identities = 15/36 (41%), Positives = 23/36 (63%), Gaps = 4/36 (11%)
Query 96 DPPAEVMQALQR--HPKSTLSVFDQRMSPDKPSAAL 129
DPPA + + LQ+ H KS + +F ++ P+KPS L
Sbjct 23 DPPAYIEEFLQQYNHYKSNVEIF--KLQPNKPSKEL 56
> hsa:56987 BBX, HBP2, HSPC339, MDS001; bobby sox homolog (Drosophila)
Length=941
Score = 27.7 bits (60), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 10/18 (55%), Positives = 12/18 (66%), Gaps = 0/18 (0%)
Query 39 KDTADAVVKGNPRLKWCP 56
K+ DA +K NP KWCP
Sbjct 134 KEYKDAFMKANPGYKWCP 151
Lambda K H
0.314 0.127 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2044474180
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40