bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0684_orf1
Length=153
Score E
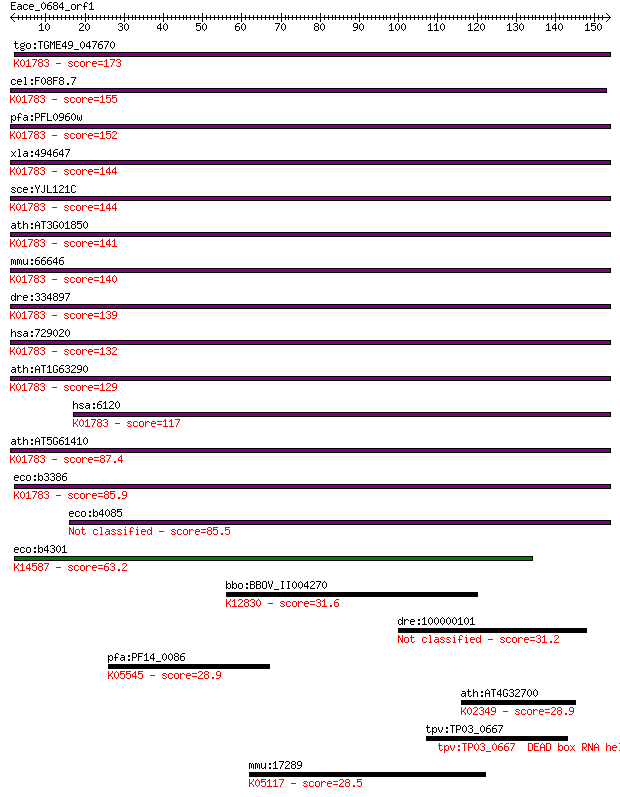
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_047670 ribulose-phosphate 3-epimerase (EC:5.1.3.1);... 173 2e-43
cel:F08F8.7 hypothetical protein; K01783 ribulose-phosphate 3-... 155 5e-38
pfa:PFL0960w D-ribulose-5-phosphate 3-epimerase, putative (EC:... 152 5e-37
xla:494647 rpe, rpe2-1; ribulose-5-phosphate-3-epimerase (EC:5... 144 8e-35
sce:YJL121C RPE1, EPI1, POS18; D-ribulose-5-phosphate 3-epimer... 144 1e-34
ath:AT3G01850 ribulose-phosphate 3-epimerase, cytosolic, putat... 141 7e-34
mmu:66646 Rpe, 2810429B02Rik, 5730518J08Rik; ribulose-5-phosph... 140 2e-33
dre:334897 rpe, ik:tdsubc_2c8, wu:fa07h08, wu:fb93d11, wu:fc20... 139 4e-33
hsa:729020 rcRPE; K01783 ribulose-phosphate 3-epimerase [EC:5.... 132 6e-31
ath:AT1G63290 ribulose-phosphate 3-epimerase, cytosolic, putat... 129 2e-30
hsa:6120 RPE, MGC2636, RPE2-1; ribulose-5-phosphate-3-epimeras... 117 2e-26
ath:AT5G61410 RPE; RPE; catalytic/ ribulose-phosphate 3-epimer... 87.4 2e-17
eco:b3386 rpe, dod, ECK3373, JW3349, yhfD; D-ribulose-5-phosph... 85.9 5e-17
eco:b4085 alsE, ECK4078, JW4046, yjcU; allulose-6-phosphate 3-... 85.5 6e-17
eco:b4301 sgcE, ECK4290, JW4263, yjhK; KpLE2 phage-like elemen... 63.2 3e-10
bbo:BBOV_II004270 18.m06355; splicing factor 3b, subunit 3, 13... 31.6 0.90
dre:100000101 kazald3, im:7144240; Kazal-type serine peptidase... 31.2 1.5
pfa:PF14_0086 tRNA-dihydrouridine synthase, putative; K05545 t... 28.9 6.8
ath:AT4G32700 DNA-directed DNA polymerase family protein; K023... 28.9 7.1
tpv:TP03_0667 DEAD box RNA helicase (EC:3.6.1.-); K01529 [EC:... 28.9 7.1
mmu:17289 Mertk, Eyk, Mer, Nyk; c-mer proto-oncogene tyrosine ... 28.5 8.9
> tgo:TGME49_047670 ribulose-phosphate 3-epimerase (EC:5.1.3.1);
K01783 ribulose-phosphate 3-epimerase [EC:5.1.3.1]
Length=230
Score = 173 bits (438), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 82/152 (53%), Positives = 108/152 (71%), Gaps = 0/152 (0%)
Query 2 VVRCLRRHIPSTFFDIHLMVNNPENLVEELNSSDASQITFHLESVDNNEEAAAALCSKIR 61
VV+ LR H+ S FFD+HLMV+ PE ++ + A+ ITFH ESV + + AA L +I+
Sbjct 55 VVKALRGHLKSAFFDVHLMVSEPEKWIQPFADAGANSITFHWESVGGDLQRAAELAKRIQ 114
Query 62 SKGIKAGIAIKPNTSIQLVKHLLTNNLIDNLLIMTVEPGFGGQTFMNNQMSKVKEARHLN 121
++GIKAG+AIKP T + + L + D LL+MTVEPGFGGQ FM + + KV+ AR L
Sbjct 115 ARGIKAGLAIKPATKFEDLGEALAGDNFDMLLVMTVEPGFGGQKFMADMLQKVRTARSLF 174
Query 122 PNIDIQVDGGLNEETVKIAAESGANVIVAGTS 153
P ++IQVDGGL+ ETVK AA +GANVIVAGTS
Sbjct 175 PKLNIQVDGGLDGETVKPAASAGANVIVAGTS 206
> cel:F08F8.7 hypothetical protein; K01783 ribulose-phosphate
3-epimerase [EC:5.1.3.1]
Length=227
Score = 155 bits (392), Expect = 5e-38, Method: Compositional matrix adjust.
Identities = 79/154 (51%), Positives = 107/154 (69%), Gaps = 6/154 (3%)
Query 1 PVVRCLRRHIPS-TFFDIHLMVNNPENLVEELNSSDASQITFHLESVDNNEEAAAALCSK 59
PVV LR+ + + FFD+HLMV+NP VE + + ASQ TFH E+VD + A + L K
Sbjct 52 PVVESLRKSLGAEPFFDVHLMVSNPGQWVEPMAKAGASQFTFHYEAVDGDV-AVSELIEK 110
Query 60 IRSKGIKAGIAIKPNTSIQ-LVKHLLTNNLIDNLLIMTVEPGFGGQTFMNNQMSKVKEAR 118
IR G+K G+++KP TS++ ++KH N +DN LIMTVEPGFGGQ FM N M KV+ R
Sbjct 111 IRKSGMKVGLSVKPGTSVEHILKH---ANHLDNALIMTVEPGFGGQKFMENMMEKVRTIR 167
Query 119 HLNPNIDIQVDGGLNEETVKIAAESGANVIVAGT 152
PN+ IQVDGG+ E ++I+A++GAN IV+GT
Sbjct 168 SKYPNLTIQVDGGVTPENIEISAQAGANAIVSGT 201
> pfa:PFL0960w D-ribulose-5-phosphate 3-epimerase, putative (EC:5.1.3.1);
K01783 ribulose-phosphate 3-epimerase [EC:5.1.3.1]
Length=227
Score = 152 bits (383), Expect = 5e-37, Method: Compositional matrix adjust.
Identities = 75/153 (49%), Positives = 104/153 (67%), Gaps = 2/153 (1%)
Query 1 PVVRCLRRHIPSTFFDIHLMVNNPENLVEELNSSDASQITFHLESVDNNEEAAAALCSKI 60
PV+ L+++ S FFD+HLMV PE V L +S+ Q+TFH E+++ + E L +I
Sbjct 53 PVINNLKKYTKSIFFDVHLMVEYPEKYVPLLKTSN--QLTFHFEALNEDTERCIQLAKEI 110
Query 61 RSKGIKAGIAIKPNTSIQLVKHLLTNNLIDNLLIMTVEPGFGGQTFMNNQMSKVKEARHL 120
R + GI+IKP T +Q + +L NLI+ +L+MTVEPGFGGQ+FM++ M KV R
Sbjct 111 RDNNLWCGISIKPKTDVQKLVPILDTNLINTVLVMTVEPGFGGQSFMHDMMGKVSFLRKK 170
Query 121 NPNIDIQVDGGLNEETVKIAAESGANVIVAGTS 153
N++IQVDGGLN ET +I+A GAN+IVAGTS
Sbjct 171 YKNLNIQVDGGLNIETTEISASHGANIIVAGTS 203
> xla:494647 rpe, rpe2-1; ribulose-5-phosphate-3-epimerase (EC:5.1.3.1);
K01783 ribulose-phosphate 3-epimerase [EC:5.1.3.1]
Length=228
Score = 144 bits (364), Expect = 8e-35, Method: Compositional matrix adjust.
Identities = 72/154 (46%), Positives = 102/154 (66%), Gaps = 7/154 (4%)
Query 1 PVVRCLRRHIPS-TFFDIHLMVNNPENLVEELNSSDASQITFHLESVDNNEEAAAALCSK 59
PVV CLR+ + S FFD+H+MV PE V+ + ++ A+Q TFHLE+ +N AL
Sbjct 52 PVVECLRKQLGSNPFFDMHMMVAQPEQWVKPMATAGANQYTFHLEATNNT----GALIKD 107
Query 60 IRSKGIKAGIAIKPNTSIQLVKHLLTNNLIDNLLIMTVEPGFGGQTFMNNQMSKVKEARH 119
IR G+K G+AIKPNT+++ + N ID L+MTVEPGFGGQ FM + M KV R
Sbjct 108 IRESGMKVGLAIKPNTTVEYLAPWA--NQIDMALVMTVEPGFGGQKFMEDMMPKVHWLRS 165
Query 120 LNPNIDIQVDGGLNEETVKIAAESGANVIVAGTS 153
P++DI+VDGG+ + + AE+GAN+IV+G++
Sbjct 166 QFPSLDIEVDGGVGPDNIHRCAEAGANMIVSGSA 199
> sce:YJL121C RPE1, EPI1, POS18; D-ribulose-5-phosphate 3-epimerase,
catalyzes a reaction in the non-oxidative part of the
pentose-phosphate pathway; mutants are sensitive to oxidative
stress (EC:5.1.3.1); K01783 ribulose-phosphate 3-epimerase
[EC:5.1.3.1]
Length=238
Score = 144 bits (362), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 77/165 (46%), Positives = 101/165 (61%), Gaps = 18/165 (10%)
Query 1 PVVRCLRRHIP------------STFFDIHLMVNNPENLVEELNSSDASQITFHLESVDN 48
P+V LRR +P + FFD H+MV NPE V++ A Q TFH E+ +
Sbjct 51 PIVTSLRRSVPRPGDASNTEKKPTAFFDCHMMVENPEKWVDDFAKCGADQFTFHYEATQD 110
Query 49 NEEAAAALCSKIRSKGIKAGIAIKPNTSIQLVKHLLTNNLIDNLLIMTVEPGFGGQTFMN 108
L I+SKGIKA AIKP TS+ ++ L + +D L+MTVEPGFGGQ FM
Sbjct 111 ----PLHLVKLIKSKGIKAACAIKPGTSVDVLFELAPH--LDMALVMTVEPGFGGQKFME 164
Query 109 NQMSKVKEARHLNPNIDIQVDGGLNEETVKIAAESGANVIVAGTS 153
+ M KV+ R P+++IQVDGGL +ET+ AA++GANVIVAGTS
Sbjct 165 DMMPKVETLRAKFPHLNIQVDGGLGKETIPKAAKAGANVIVAGTS 209
> ath:AT3G01850 ribulose-phosphate 3-epimerase, cytosolic, putative
/ pentose-5-phosphate 3-epimerase, putative (EC:5.1.3.1);
K01783 ribulose-phosphate 3-epimerase [EC:5.1.3.1]
Length=225
Score = 141 bits (356), Expect = 7e-34, Method: Compositional matrix adjust.
Identities = 69/154 (44%), Positives = 98/154 (63%), Gaps = 5/154 (3%)
Query 1 PVVRCLRRHIPSTFFDIHLMVNNPENLVEELNSSDASQITFHLESVDNNEEAAAALCSKI 60
PV+ LR+H + + D HLMV NP + V ++ + AS TFH+E +N + L KI
Sbjct 51 PVIESLRKHT-NAYLDCHLMVTNPMDYVAQMAKAGASGFTFHVEVAQDNWQQ---LVEKI 106
Query 61 RSKGIKAGIAIKPNTSIQLVKHLLTNNL-IDNLLIMTVEPGFGGQTFMNNQMSKVKEARH 119
+S G++ G+A+KP T ++ V L+ ++ +L+MTVEPGFGGQ FM + M KV+ R
Sbjct 107 KSTGMRPGVALKPGTPVEQVYPLVEGTTPVEMVLVMTVEPGFGGQKFMPDMMDKVRALRQ 166
Query 120 LNPNIDIQVDGGLNEETVKIAAESGANVIVAGTS 153
P +DIQVDGGL T+ AA +GAN IVAG+S
Sbjct 167 KYPTLDIQVDGGLGPSTIDTAAAAGANCIVAGSS 200
> mmu:66646 Rpe, 2810429B02Rik, 5730518J08Rik; ribulose-5-phosphate-3-epimerase
(EC:5.1.3.1); K01783 ribulose-phosphate 3-epimerase
[EC:5.1.3.1]
Length=228
Score = 140 bits (352), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 71/154 (46%), Positives = 100/154 (64%), Gaps = 7/154 (4%)
Query 1 PVVRCLRRHI-PSTFFDIHLMVNNPENLVEELNSSDASQITFHLESVDNNEEAAAALCSK 59
PVV LR+ + FFD+H+MV+ PE V+ + + A+Q TFHLE+ +N AL
Sbjct 52 PVVESLRKQLGQDPFFDMHMMVSRPEQWVKPMAVAGANQYTFHLEATEN----PGALIKD 107
Query 60 IRSKGIKAGIAIKPNTSIQLVKHLLTNNLIDNLLIMTVEPGFGGQTFMNNQMSKVKEARH 119
IR G+K G+AIKP T+++ + N ID L+MTVEPGFGGQ FM + M KV R
Sbjct 108 IRENGMKVGLAIKPGTTVEYLAPW--ANQIDMALVMTVEPGFGGQKFMEDMMPKVHWLRT 165
Query 120 LNPNIDIQVDGGLNEETVKIAAESGANVIVAGTS 153
P +DI+VDGG+ +TV+ AE+GAN+IV+G++
Sbjct 166 QFPTLDIEVDGGVGPDTVQKCAEAGANMIVSGSA 199
> dre:334897 rpe, ik:tdsubc_2c8, wu:fa07h08, wu:fb93d11, wu:fc20b11,
xx:tdsubc_2c8; ribulose-5-phosphate-3-epimerase (EC:5.1.3.1);
K01783 ribulose-phosphate 3-epimerase [EC:5.1.3.1]
Length=228
Score = 139 bits (350), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 69/154 (44%), Positives = 100/154 (64%), Gaps = 7/154 (4%)
Query 1 PVVRCLRRHI-PSTFFDIHLMVNNPENLVEELNSSDASQITFHLESVDNNEEAAAALCSK 59
P+V CLR I P FFD+H+MV+ PE V+ + ++ A+Q TFHLE+ N L +
Sbjct 52 PMVECLRSCIGPDPFFDMHMMVSRPEQWVKPMAAAGANQYTFHLEATSN----PGNLIKE 107
Query 60 IRSKGIKAGIAIKPNTSIQLVKHLLTNNLIDNLLIMTVEPGFGGQTFMNNQMSKVKEARH 119
IR G+K G+AIKP T+++ + ID L+MTVEPGFGGQ FM + M KV R
Sbjct 108 IRESGMKVGLAIKPGTTVEELAPWAGQ--IDMALVMTVEPGFGGQKFMEDMMPKVSWLRG 165
Query 120 LNPNIDIQVDGGLNEETVKIAAESGANVIVAGTS 153
P++DI+VDGG+ +++ AE+GAN+IV+G++
Sbjct 166 QFPSLDIEVDGGVGPDSIHRCAEAGANMIVSGSA 199
> hsa:729020 rcRPE; K01783 ribulose-phosphate 3-epimerase [EC:5.1.3.1]
Length=228
Score = 132 bits (331), Expect = 6e-31, Method: Compositional matrix adjust.
Identities = 68/154 (44%), Positives = 97/154 (62%), Gaps = 7/154 (4%)
Query 1 PVVRCLRRHI-PSTFFDIHLMVNNPENLVEELNSSDASQITFHLESVDNNEEAAAALCSK 59
PVV LR+ + FFD+H+MV+ PE V+ + ++A+Q TFHLE+ +N L
Sbjct 52 PVVESLRKQLGQDPFFDMHMMVSKPEQWVKPMAVAEANQYTFHLEATEN----PGTLIKD 107
Query 60 IRSKGIKAGIAIKPNTSIQLVKHLLTNNLIDNLLIMTVEPGFGGQTFMNNQMSKVKEARH 119
IR G+K G+AIKP TS++ + N ID L+MTVEPGFG Q FM + M KV R
Sbjct 108 IRENGMKVGLAIKPGTSVEYLAPWA--NQIDMALVMTVEPGFGEQKFMEDMMPKVHWLRT 165
Query 120 LNPNIDIQVDGGLNEETVKIAAESGANVIVAGTS 153
P++DI+ DGG+ +TV AE+GAN+ V+G++
Sbjct 166 QFPSLDIEGDGGVGSDTVHKCAEAGANMTVSGSA 199
> ath:AT1G63290 ribulose-phosphate 3-epimerase, cytosolic, putative
/ pentose-5-phosphate 3-epimerase, putative (EC:5.1.3.1);
K01783 ribulose-phosphate 3-epimerase [EC:5.1.3.1]
Length=227
Score = 129 bits (325), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 68/154 (44%), Positives = 100/154 (64%), Gaps = 5/154 (3%)
Query 1 PVVRCLRRHIPSTFFDIHLMVNNPENLVEELNSSDASQITFHLESVDNNEEAAAALCSKI 60
PV+ LR+H + + D HLMV NP + V+++ + AS TFH+E N + L KI
Sbjct 53 PVIESLRKHT-NAYLDCHLMVTNPMDYVDQMAKAGASGFTFHVEVAQENWQE---LVKKI 108
Query 61 RSKGIKAGIAIKPNTSIQLVKHLLTN-NLIDNLLIMTVEPGFGGQTFMNNQMSKVKEARH 119
++ G++ G+A+KP T ++ V L+ N ++ +L+MTVEPGFGGQ FM + M KV+ R+
Sbjct 109 KAAGMRPGVALKPGTPVEQVYPLVEGTNPVEMVLVMTVEPGFGGQKFMPSMMDKVRALRN 168
Query 120 LNPNIDIQVDGGLNEETVKIAAESGANVIVAGTS 153
P +DI+VDGGL T+ AA +GAN IVAG+S
Sbjct 169 KYPTLDIEVDGGLGPSTIDAAAAAGANCIVAGSS 202
> hsa:6120 RPE, MGC2636, RPE2-1; ribulose-5-phosphate-3-epimerase
(EC:5.1.3.1); K01783 ribulose-phosphate 3-epimerase [EC:5.1.3.1]
Length=178
Score = 117 bits (292), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 64/155 (41%), Positives = 91/155 (58%), Gaps = 24/155 (15%)
Query 17 IHLMVNNPENLVEELNSSDASQITFHLESVDNNEEAAAALCSKIRSKGIKA--------- 67
+H+MV+ PE V+ + + A+Q TFHLE+ +N AL IR G+K+
Sbjct 1 MHMMVSKPEQWVKPMAVAGANQYTFHLEATEN----PGALIKDIRENGMKSCSVTQAEVQ 56
Query 68 ---------GIAIKPNTSIQLVKHLLTNNLIDNLLIMTVEPGFGGQTFMNNQMSKVKEAR 118
G+AIKP TS++ + N ID L+MTVEPGFGGQ FM + M KV R
Sbjct 57 WHSQGPLQVGLAIKPGTSVEYLAPWA--NQIDMALVMTVEPGFGGQKFMEDMMPKVHWLR 114
Query 119 HLNPNIDIQVDGGLNEETVKIAAESGANVIVAGTS 153
P++DI+VDGG+ +TV AE+GAN+IV+G++
Sbjct 115 TQFPSLDIEVDGGVGPDTVHKCAEAGANMIVSGSA 149
> ath:AT5G61410 RPE; RPE; catalytic/ ribulose-phosphate 3-epimerase
(EC:5.1.3.1); K01783 ribulose-phosphate 3-epimerase [EC:5.1.3.1]
Length=281
Score = 87.4 bits (215), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 51/160 (31%), Positives = 90/160 (56%), Gaps = 14/160 (8%)
Query 1 PVVRCLRRHIPSTFFDIHLMVNNPENLVEELNSSDASQITFHLESVDNNEEAAAAL---C 57
P+V R + D+HLM+ PE V + + A ++ H E +++ L
Sbjct 104 PLVVDALRPVTDLPLDVHLMIVEPEQRVPDFIKAGADIVSVHCE-----QQSTIHLHRTV 158
Query 58 SKIRSKGIKAGIAIKPNTSIQLVKHLLTNNLIDNLLIMTVEPGFGGQTFMNNQMSKVKEA 117
++I+S G KAG+ + P T + ++++L +++D +LIM+V PGFGGQ+F+ +Q+ K+ +
Sbjct 159 NQIKSLGAKAGVVLNPGTPLSAIEYVL--DMVDLVLIMSVNPGFGGQSFIESQVKKISDL 216
Query 118 RHL----NPNIDIQVDGGLNEETVKIAAESGANVIVAGTS 153
R + N I+VDGG+ E+GAN +VAG++
Sbjct 217 RKMCAEKGVNPWIEVDGGVTPANAYKVIEAGANALVAGSA 256
> eco:b3386 rpe, dod, ECK3373, JW3349, yhfD; D-ribulose-5-phosphate
3-epimerase (EC:5.1.3.1); K01783 ribulose-phosphate 3-epimerase
[EC:5.1.3.1]
Length=225
Score = 85.9 bits (211), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 48/156 (30%), Positives = 88/156 (56%), Gaps = 10/156 (6%)
Query 2 VVRCLRRHIPSTFFDIHLMVNNPENLVEELNSSDASQITFHLESVDNNEEAAAALCSKIR 61
V++ LR + + D+HLMV + +V + ++ AS ITFH E+ ++ + I+
Sbjct 52 VLKSLRNYGITAPIDVHLMVKPVDRIVPDFAAAGASIITFHPEASEHVDRT----LQLIK 107
Query 62 SKGIKAGIAIKPNTSIQLVKHLLTNNLIDNLLIMTVEPGFGGQTFMNNQMSKVKEARHL- 120
G KAG+ P T + + +++ +D +L+M+V PGFGGQ+F+ + K++E R
Sbjct 108 ENGCKAGLVFNPATPLSYLDYVMDK--LDVILLMSVNPGFGGQSFIPQTLDKLREVRRRI 165
Query 121 ---NPNIDIQVDGGLNEETVKIAAESGANVIVAGTS 153
+I ++VDGG+ + A +GA++ VAG++
Sbjct 166 DESGFDIRLEVDGGVKVNNIGEIAAAGADMFVAGSA 201
> eco:b4085 alsE, ECK4078, JW4046, yjcU; allulose-6-phosphate
3-epimerase
Length=231
Score = 85.5 bits (210), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 45/142 (31%), Positives = 76/142 (53%), Gaps = 9/142 (6%)
Query 16 DIHLMVNNPENLVEELNSSDASQITFHLESVDNNEEAAAALCSKIRSKGIKAGIAIKPNT 75
D HLMV P++ + +L + A IT H E+++ A L +IR +K G+ + P T
Sbjct 61 DCHLMVTRPQDYIAQLARAGADFITLHPETINGQ---AFRLIDEIRRHDMKVGLILNPET 117
Query 76 SIQLVKHLLTNNLIDNLLIMTVEPGFGGQTFMNNQMSKVKEARHLNPN----IDIQVDGG 131
++ +K+ + D + +MTV+PGF GQ F+ + K+ E + +I+VDG
Sbjct 118 PVEAMKYYIHKA--DKITVMTVDPGFAGQPFIPEMLDKLAELKAWREREGLEYEIEVDGS 175
Query 132 LNEETVKIAAESGANVIVAGTS 153
N+ T + +GA+V + GTS
Sbjct 176 CNQATYEKLMAAGADVFIVGTS 197
> eco:b4301 sgcE, ECK4290, JW4263, yjhK; KpLE2 phage-like element;
predicted epimerase; K14587 protein sgcE [EC:5.1.3.-]
Length=210
Score = 63.2 bits (152), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 35/132 (26%), Positives = 60/132 (45%), Gaps = 7/132 (5%)
Query 2 VVRCLRRHIPSTFFDIHLMVNNPENLVEELNSSDASQITFHLESVDNNEEAAAALCSKIR 61
V+ + R P H M+ P+ L + I H E++D E ++IR
Sbjct 49 TVQAVARQTPHPL-SFHFMLARPQRWFNALAEIRPAWIFVHAETLDYPSET----LTEIR 103
Query 62 SKGIKAGIAIKPNTSIQLVKHLLTNNLIDNLLIMTVEPGFGGQTFMNNQMSKVKEARHLN 121
G +AG+ P T I ++L + +D +++MT EP GQ F+ + K+++ R
Sbjct 104 HTGARAGLVFNPATPIDAWRYLASE--LDGVMVMTSEPDGQGQRFIPSMCEKIQKVRTAF 161
Query 122 PNIDIQVDGGLN 133
P + DGG+
Sbjct 162 PQTECWADGGIT 173
> bbo:BBOV_II004270 18.m06355; splicing factor 3b, subunit 3,
130kD; K12830 splicing factor 3B subunit 3
Length=1169
Score = 31.6 bits (70), Expect = 0.90, Method: Composition-based stats.
Identities = 23/70 (32%), Positives = 36/70 (51%), Gaps = 7/70 (10%)
Query 56 LCSKIRSKGIKAGIAIKP--NTSIQLVKHLLTNNLIDNLLIMTVEPGFGGQTFM----NN 109
LCS I +G A +A KP N ++QLV L + + I +L ++ V+ G G Q
Sbjct 372 LCSSIHPQGKDAIVAFKPRVNQNLQLVDELSSLSAITDLKVIDVQ-GLGQQQIFLGCGKG 430
Query 110 QMSKVKEARH 119
+ S ++ RH
Sbjct 431 ERSTLRVLRH 440
> dre:100000101 kazald3, im:7144240; Kazal-type serine peptidase
inhibitor domain 3
Length=296
Score = 31.2 bits (69), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 17/48 (35%), Positives = 26/48 (54%), Gaps = 2/48 (4%)
Query 100 GFGGQTFMNNQMSKVKEARHLNPNIDIQVDGGLNEETVKIAAESGANV 147
G GQT+MN + K KEA +L P +++ E +K+A + NV
Sbjct 135 GSDGQTYMN--VCKYKEAAYLKPGLNVSDGPCRTEPIIKVAPHNLVNV 180
> pfa:PF14_0086 tRNA-dihydrouridine synthase, putative; K05545
tRNA-dihydrouridine synthase 4 [EC:1.-.-.-]
Length=340
Score = 28.9 bits (63), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 12/41 (29%), Positives = 24/41 (58%), Gaps = 0/41 (0%)
Query 26 NLVEELNSSDASQITFHLESVDNNEEAAAALCSKIRSKGIK 66
NL+ ++ ++ IT + +DN+ + LC ++S+GIK
Sbjct 138 NLISDITNNCVIPITCKIRKIDNDYQKTLNLCYDLQSRGIK 178
> ath:AT4G32700 DNA-directed DNA polymerase family protein; K02349
DNA polymerase theta subunit [EC:2.7.7.7]
Length=2154
Score = 28.9 bits (63), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 14/29 (48%), Positives = 17/29 (58%), Gaps = 0/29 (0%)
Query 116 EARHLNPNIDIQVDGGLNEETVKIAAESG 144
E RH NPNID +V L+ E + A SG
Sbjct 1637 EERHSNPNIDKEVKKRLSPEAAEAANRSG 1665
> tpv:TP03_0667 DEAD box RNA helicase (EC:3.6.1.-); K01529 [EC:3.6.1.-]
Length=2359
Score = 28.9 bits (63), Expect = 7.1, Method: Composition-based stats.
Identities = 13/40 (32%), Positives = 25/40 (62%), Gaps = 4/40 (10%)
Query 107 MNNQMSKVKEARHLN----PNIDIQVDGGLNEETVKIAAE 142
+NNQ++++ H+N +++ V+ G+NEETV+ E
Sbjct 1523 LNNQITQINRVNHVNHIVDKDVEKDVEHGVNEETVESVRE 1562
> mmu:17289 Mertk, Eyk, Mer, Nyk; c-mer proto-oncogene tyrosine
kinase (EC:2.7.10.1); K05117 c-mer proto-oncogene tyrosine
kinase [EC:2.7.10.1]
Length=994
Score = 28.5 bits (62), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 34/60 (56%), Gaps = 4/60 (6%)
Query 62 SKGIKAGIAIKPNTSIQLVKHLLTNNLIDNLLIMTVEPGFGGQTFMNNQMSKVKEARHLN 121
SKG+ I + P+ ++ H+L N+ ++L+ V PGF G + + N +VKEA L+
Sbjct 268 SKGVHINIKVIPSPPTEV--HIL-NSTAHSILVSWV-PGFDGYSPLQNCSIQVKEADRLS 323
Lambda K H
0.315 0.131 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3256415000
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40