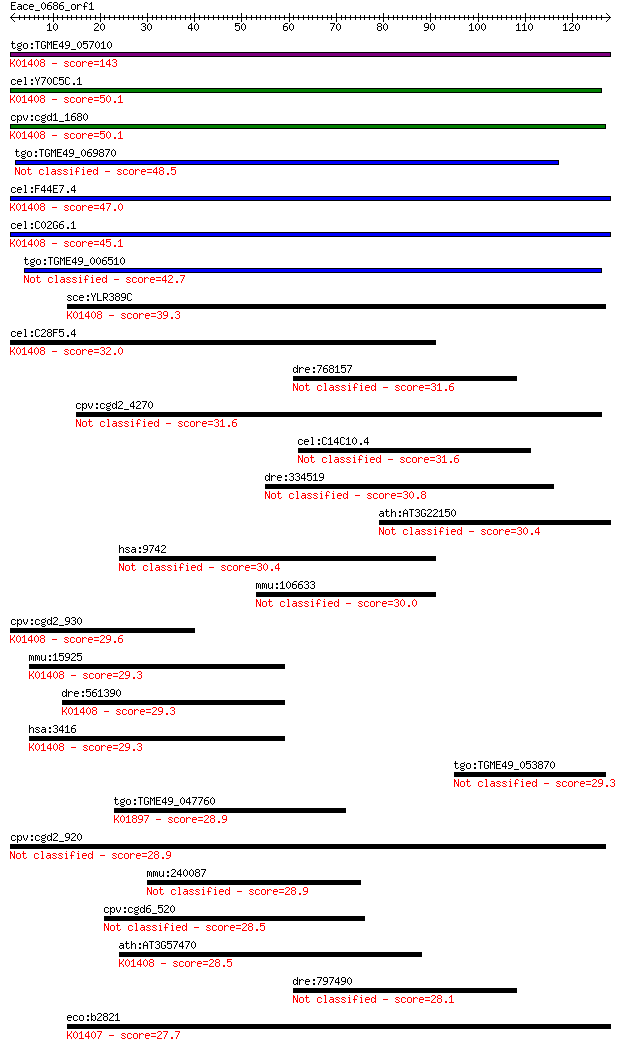
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_0686_orf1
Length=127
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_057010 insulysin, putative (EC:3.4.24.56); K01408 i... 143 2e-34
cel:Y70C5C.1 hypothetical protein; K01408 insulysin [EC:3.4.24... 50.1 2e-06
cpv:cgd1_1680 insulinase like protease, signal peptide ; K0140... 50.1 2e-06
tgo:TGME49_069870 hypothetical protein 48.5 5e-06
cel:F44E7.4 hypothetical protein; K01408 insulysin [EC:3.4.24.56] 47.0 1e-05
cel:C02G6.1 hypothetical protein; K01408 insulysin [EC:3.4.24.56] 45.1 6e-05
tgo:TGME49_006510 peptidase M16 domain containing protein (EC:... 42.7 3e-04
sce:YLR389C STE23; Metalloprotease involved, with homolog Axl1... 39.3 0.003
cel:C28F5.4 hypothetical protein; K01408 insulysin [EC:3.4.24.56] 32.0 0.43
dre:768157 cplx4a, CPLX4, si:ch211-233a1.5, zgc:153429; comple... 31.6 0.59
cpv:cgd2_4270 secreted insulinase-like peptidase 31.6 0.62
cel:C14C10.4 hypothetical protein 31.6 0.69
dre:334519 rnf17, fj98c04, im:7147535, wu:fj98c04; ring finger... 30.8 1.2
ath:AT3G22150 pentatricopeptide (PPR) repeat-containing protein 30.4 1.3
hsa:9742 IFT140, DKFZp564L232, FLJ10306, FLJ30571, KIAA0590, W... 30.4 1.4
mmu:106633 Ift140, AI661311, MGC170632, Tce5, Wdtc2, mKIAA0590... 30.0 1.7
cpv:cgd2_930 peptidase'insulinase-like peptidase' ; K01408 ins... 29.6 2.4
mmu:15925 Ide, 1300012G03Rik, 4833415K22Rik, AA675336, AI50753... 29.3 2.9
dre:561390 ide, MGC162603, zgc:162603; insulin-degrading enzym... 29.3 3.4
hsa:3416 IDE, FLJ35968, INSULYSIN; insulin-degrading enzyme (E... 29.3 3.6
tgo:TGME49_053870 hypothetical protein 29.3 3.6
tgo:TGME49_047760 long chain acyl-CoA synthetase, putative (EC... 28.9 3.9
cpv:cgd2_920 peptidase'insulinase-like peptidase' 28.9 4.0
mmu:240087 Mdc1, 6820401C03, AA413496, Nfbd1, mKIAA0170; media... 28.9 4.5
cpv:cgd6_520 Ser/Thr protein kinase 28.5 5.1
ath:AT3G57470 peptidase M16 family protein / insulinase family... 28.5 5.1
dre:797490 cplx4b, si:ch211-261c8.4; complexin 4b 28.1 7.3
eco:b2821 ptrA, ECK2817, JW2789, ptr; protease III (EC:3.4.24.... 27.7 9.0
> tgo:TGME49_057010 insulysin, putative (EC:3.4.24.56); K01408
insulysin [EC:3.4.24.56]
Length=953
Score = 143 bits (360), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 65/127 (51%), Positives = 90/127 (70%), Gaps = 0/127 (0%)
Query 1 LGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTH 60
LG+PSIE+RVNLAVL Q LNRR++D LRTE QLGYI GA+ +S L+C +EG++ H
Sbjct 778 LGVPSIEERVNLAVLGQMLNRRLFDRLRTEEQLGYIVGARSYIDSSVESLRCVLEGSRKH 837
Query 61 PDEVVKMIDEELTKAKEYLANMPDAEMARWKEAAHAKLTKMEANFSEDFKKSAEEIFSHS 120
PDE+ +ID+EL K ++L ++ D E+ WKE+A A+L K F E+F +S +I +H
Sbjct 838 PDEIADLIDKELWKMNDHLQSISDGELDHWKESARAELEKPTETFYEEFGRSWGQIANHG 897
Query 121 NCFTKRD 127
+CF KRD
Sbjct 898 HCFNKRD 904
> cel:Y70C5C.1 hypothetical protein; K01408 insulysin [EC:3.4.24.56]
Length=985
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 33/125 (26%), Positives = 56/125 (44%), Gaps = 1/125 (0%)
Query 1 LGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTH 60
+G+ + D + ++ + +D+LRT+ LGYI + T LQ V+G K+
Sbjct 764 IGVQNTYDNAVIGLIKNLITEPAFDTLRTKESLGYIVWTRTHFNCGTVALQILVQGPKS- 822
Query 61 PDEVVKMIDEELTKAKEYLANMPDAEMARWKEAAHAKLTKMEANFSEDFKKSAEEIFSHS 120
D V++ I+ L ++ + MP E A+L + S FKK +EI
Sbjct 823 VDHVLERIEAFLESVRKEIVEMPQEEFENRVSGLIAQLEEKPKTLSCRFKKFWDEIECRQ 882
Query 121 NCFTK 125
FT+
Sbjct 883 YNFTR 887
> cpv:cgd1_1680 insulinase like protease, signal peptide ; K01408
insulysin [EC:3.4.24.56]
Length=1033
Score = 50.1 bits (118), Expect = 2e-06, Method: Composition-based stats.
Identities = 40/127 (31%), Positives = 59/127 (46%), Gaps = 18/127 (14%)
Query 1 LGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTH 60
G+PS E++++L L + IYD+LRT QLGYI A +ST LL VEG +
Sbjct 829 FGVPSFEEKLHLMALQPIIQGYIYDNLRTNKQLGYIIFANIVPISSTRLLVVGVEGDNNN 888
Query 61 PDEVVKMIDEELTKAKEYLANMPDAEMARWKEAAHAKLTKMEANFSEDFKKS-AEEIFSH 119
E + E + + Y E + KL ME++ ED K + +E S
Sbjct 889 SVEKI----ESIIRNTLY-------------EFSTRKLGNMESHMFEDIKSALIQEAKSI 931
Query 120 SNCFTKR 126
N F ++
Sbjct 932 GNSFNQK 938
> tgo:TGME49_069870 hypothetical protein
Length=413
Score = 48.5 bits (114), Expect = 5e-06, Method: Composition-based stats.
Identities = 30/120 (25%), Positives = 59/120 (49%), Gaps = 13/120 (10%)
Query 2 GIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGA---K 58
+P+I DR L ++++++++R ++ LRTE QLGY+ S+ + F+
Sbjct 215 ALPNIHDRAVLYMVSRWMSQRFFNKLRTEQQLGYLTAMHSSRLEDRFYYRFFITSTYDPA 274
Query 59 THPDEVVKMIDEELTK--AKEYLANMPDAEMARWKEAAHAKLTKMEANFSEDFKKSAEEI 116
D +V+ I+ E +K +E A + A + WK+ N E+F+K+ ++
Sbjct 275 EVADRIVEFINAERSKIPTQEEFATLKQAAIDVWKQKPK--------NIFEEFRKNRRQV 326
> cel:F44E7.4 hypothetical protein; K01408 insulysin [EC:3.4.24.56]
Length=1051
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 31/127 (24%), Positives = 55/127 (43%), Gaps = 1/127 (0%)
Query 1 LGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTH 60
+G+ + D + ++ Q + +++LRT LGYI T L V+G K+
Sbjct 823 IGVQNTYDNAVVGLIDQLIREPAFNTLRTNEALGYIVWTGSRLNCGTVALNVIVQGPKS- 881
Query 61 PDEVVKMIDEELTKAKEYLANMPDAEMARWKEAAHAKLTKMEANFSEDFKKSAEEIFSHS 120
D V++ I+ L ++ +A MP E A+L + S F++ EI
Sbjct 882 VDHVLERIEVFLESVRKEIAEMPQEEFDNQVSGMIARLEEKPKTLSSRFRRFWNEIECRQ 941
Query 121 NCFTKRD 127
F +R+
Sbjct 942 YNFARRE 948
> cel:C02G6.1 hypothetical protein; K01408 insulysin [EC:3.4.24.56]
Length=980
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 31/127 (24%), Positives = 56/127 (44%), Gaps = 1/127 (0%)
Query 1 LGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTH 60
+G+ + D + ++ Q + ++D+LRT LGYI L FV+G K+
Sbjct 753 IGVQNKYDNAVVGLIDQLIKEPVFDTLRTNEALGYIVWTGCRFNCGAVALNIFVQGPKS- 811
Query 61 PDEVVKMIDEELTKAKEYLANMPDAEMARWKEAAHAKLTKMEANFSEDFKKSAEEIFSHS 120
D V++ I+ L ++ + MP E + A+L + S FK+ +I
Sbjct 812 VDYVLERIEVFLESVRKEIIEMPQDEFEKKVAGMIARLEEKPKTLSNRFKRFWYQIECRQ 871
Query 121 NCFTKRD 127
F +R+
Sbjct 872 YDFARRE 878
> tgo:TGME49_006510 peptidase M16 domain containing protein (EC:4.1.1.70
3.4.24.13 3.4.24.56 3.4.21.10 3.4.24.35 3.2.1.91)
Length=2435
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 27/124 (21%), Positives = 62/124 (50%), Gaps = 2/124 (1%)
Query 4 PSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTHPDE 63
P + + V +++ + ++ +D++RT GY+A A + L V+G++ PDE
Sbjct 1203 PDMMEMVVYSLIGEVISSPFFDTIRTHWMDGYVAAAAVREVPPAMTLATIVQGSQRKPDE 1262
Query 64 VVKMIDEELTKAKEYLAN--MPDAEMARWKEAAHAKLTKMEANFSEDFKKSAEEIFSHSN 121
+ + + L + +E + + +A + R + + +K + +FS+ F + +I S +
Sbjct 1263 LERHVCAFLAEMEENIGSSMTTEAFLERLRWLSSSKFHRSATSFSDYFGEVTSQIASRNF 1322
Query 122 CFTK 125
CF +
Sbjct 1323 CFIR 1326
> sce:YLR389C STE23; Metalloprotease involved, with homolog Axl1p,
in N-terminal processing of pro-A-factor to the mature
form; member of the insulin-degrading enzyme family (EC:3.4.24.-);
K01408 insulysin [EC:3.4.24.56]
Length=1027
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 27/114 (23%), Positives = 51/114 (44%), Gaps = 1/114 (0%)
Query 13 AVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTHPDEVVKMIDEEL 72
+ Q ++ +D+LRT+ QLGY+ + TA ++ ++ T P + I+
Sbjct 818 GLFAQLIHEPCFDTLRTKEQLGYVVFSSSLNNHGTANIRILIQSEHTTP-YLEWRINNFY 876
Query 73 TKAKEYLANMPDAEMARWKEAAHAKLTKMEANFSEDFKKSAEEIFSHSNCFTKR 126
+ L +MP+ + + KEA L + N +E+ + I+ FT R
Sbjct 877 ETFGQVLRDMPEEDFEKHKEALCNSLLQKFKNMAEESARYTAAIYLGDYNFTHR 930
> cel:C28F5.4 hypothetical protein; K01408 insulysin [EC:3.4.24.56]
Length=856
Score = 32.0 bits (71), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 23/92 (25%), Positives = 39/92 (42%), Gaps = 3/92 (3%)
Query 1 LGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTH 60
+G+ S + +L + + Y LRT LGY + L V+G ++
Sbjct 695 IGVQSTYNNSVNKLLNELIKNPAYTILRTNEALGYNVSTESRLNDGNVYLHVIVQGPES- 753
Query 61 PDEVVKMIDEELTKAKEYLANMP--DAEMARW 90
D V++ I+ L A+E + MP D + W
Sbjct 754 ADHVLERIEVFLESAREEIVAMPQEDFDYQVW 785
> dre:768157 cplx4a, CPLX4, si:ch211-233a1.5, zgc:153429; complexin
4a
Length=159
Score = 31.6 bits (70), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 18/53 (33%), Positives = 33/53 (62%), Gaps = 6/53 (11%)
Query 61 PDEVVKMIDEELTKAKE------YLANMPDAEMARWKEAAHAKLTKMEANFSE 107
P+E++KM+DE+ T+ +E + N+ + +M + KE A A LT+M++ E
Sbjct 102 PEELLKMVDEDATEEEEKDSIMGQIQNLQNMDMDQIKEKASATLTEMKSKAEE 154
> cpv:cgd2_4270 secreted insulinase-like peptidase
Length=1257
Score = 31.6 bits (70), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 31/118 (26%), Positives = 52/118 (44%), Gaps = 8/118 (6%)
Query 15 LTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTHPDEVVKMIDEELTK 74
+ + LN YD+LRTE Q GY+A A L V+ A+ + +V + L K
Sbjct 948 IGEILNSPFYDTLRTEWQDGYVAFATTKYETPIISLIGAVQSAEKLSETLVCHMFSALKK 1007
Query 75 ----AKEYLANMPDAEM---ARWKEAAHAKLTKMEANFSEDFKKSAEEIFSHSNCFTK 125
+E L + +E RW + K+++ F++ + + + SH CF K
Sbjct 1008 VSKDVEEDLKEISKSEFEDKIRWFGLSKYSSQKLDS-FTKYIEHFGKLVVSHELCFEK 1064
> cel:C14C10.4 hypothetical protein
Length=1058
Score = 31.6 bits (70), Expect = 0.69, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 29/60 (48%), Gaps = 11/60 (18%)
Query 62 DEVVKMIDEELTKAKEYLANMPD-----------AEMARWKEAAHAKLTKMEANFSEDFK 110
D V++ +EELT A+ L + P+ A M RW+ +L K +NFS D K
Sbjct 650 DPRVQLNNEELTDARNVLKSSPEKAELLGKLRKSATMQRWRATDVNELLKELSNFSNDVK 709
> dre:334519 rnf17, fj98c04, im:7147535, wu:fj98c04; ring finger
protein 17
Length=1485
Score = 30.8 bits (68), Expect = 1.2, Method: Composition-based stats.
Identities = 21/61 (34%), Positives = 30/61 (49%), Gaps = 2/61 (3%)
Query 55 EGAKTHPDEVVKMIDEELTKAKEYLANMPDAEMARWKEAAHAKLTKMEANFSEDFKKSAE 114
E A+ PD VVK++DE LTKA E L N D H +L K + + ++ E
Sbjct 114 ENAEGKPD-VVKVLDEALTKATENL-NQLDKLHQTLANGVHGQLRKERSRIIREIDEAVE 171
Query 115 E 115
+
Sbjct 172 K 172
> ath:AT3G22150 pentatricopeptide (PPR) repeat-containing protein
Length=820
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 23/49 (46%), Gaps = 1/49 (2%)
Query 79 LANMPDAEMARWKEAAHAKLTKMEANFSEDFKKSAEEIFSHSNCFTKRD 127
L+NM AE +WK + E ++ +S EI + NCF RD
Sbjct 733 LSNMY-AEEQKWKSVDKVRRGMREKGLKKEVGRSGIEIAGYVNCFVSRD 780
> hsa:9742 IFT140, DKFZp564L232, FLJ10306, FLJ30571, KIAA0590,
WDTC2, c305C8.4, c380F5.1, gs114; intraflagellar transport
140 homolog (Chlamydomonas)
Length=1462
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 32/67 (47%), Gaps = 1/67 (1%)
Query 24 YDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTHPDEVVKMIDEELTKAKEYLANMP 83
+D + + AG E+ A + L + E + TH EV +M+ E+L + Y+ M
Sbjct 895 HDRVHLRSTYHRYAGHLEASADCSRALS-YYEKSDTHRFEVPRMLSEDLPSLELYVNKMK 953
Query 84 DAEMARW 90
D + RW
Sbjct 954 DKTLWRW 960
> mmu:106633 Ift140, AI661311, MGC170632, Tce5, Wdtc2, mKIAA0590;
intraflagellar transport 140 homolog (Chlamydomonas)
Length=1464
Score = 30.0 bits (66), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 13/38 (34%), Positives = 21/38 (55%), Gaps = 0/38 (0%)
Query 53 FVEGAKTHPDEVVKMIDEELTKAKEYLANMPDAEMARW 90
+ E + TH EV +M+ E+L + Y+ M D + RW
Sbjct 924 YYEKSDTHRFEVPRMLSEDLQSLELYINRMKDKTLWRW 961
> cpv:cgd2_930 peptidase'insulinase-like peptidase' ; K01408 insulysin
[EC:3.4.24.56]
Length=1013
Score = 29.6 bits (65), Expect = 2.4, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 24/39 (61%), Gaps = 0/39 (0%)
Query 1 LGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGA 39
+G ++ + V L +L+++L+ Y LRT QLGYI A
Sbjct 812 MGEYNLRNYVLLELLSKYLDSNCYLELRTNQQLGYIVHA 850
> mmu:15925 Ide, 1300012G03Rik, 4833415K22Rik, AA675336, AI507533;
insulin degrading enzyme (EC:3.4.24.56); K01408 insulysin
[EC:3.4.24.56]
Length=1019
Score = 29.3 bits (64), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 14/54 (25%), Positives = 28/54 (51%), Gaps = 0/54 (0%)
Query 5 SIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAK 58
S + + L + Q ++ +++LRT+ QLGYI + +A L+ ++ K
Sbjct 801 STSENMFLELFCQIISEPCFNTLRTKEQLGYIVFSGPRRANGIQGLRFIIQSEK 854
> dre:561390 ide, MGC162603, zgc:162603; insulin-degrading enzyme
(EC:3.4.24.56); K01408 insulysin [EC:3.4.24.56]
Length=978
Score = 29.3 bits (64), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 13/47 (27%), Positives = 25/47 (53%), Gaps = 0/47 (0%)
Query 12 LAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAK 58
L + Q ++ +++LRT+ QLGYI + +A L+ ++ K
Sbjct 767 LELFCQIISEPCFNTLRTKEQLGYIVFSGPRRANGVQGLRFIIQSEK 813
> hsa:3416 IDE, FLJ35968, INSULYSIN; insulin-degrading enzyme
(EC:3.4.24.56); K01408 insulysin [EC:3.4.24.56]
Length=464
Score = 29.3 bits (64), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 14/54 (25%), Positives = 28/54 (51%), Gaps = 0/54 (0%)
Query 5 SIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAK 58
S + + L + Q ++ +++LRT+ QLGYI + +A L+ ++ K
Sbjct 246 STSENMFLELFCQIISEPCFNTLRTKEQLGYIVFSGPRRANGIQGLRFIIQSEK 299
> tgo:TGME49_053870 hypothetical protein
Length=4787
Score = 29.3 bits (64), Expect = 3.6, Method: Composition-based stats.
Identities = 15/34 (44%), Positives = 19/34 (55%), Gaps = 2/34 (5%)
Query 95 HAKLTKMEA--NFSEDFKKSAEEIFSHSNCFTKR 126
+ +L K+E FS DF KSA I H+ C KR
Sbjct 2244 YTRLGKLERLLRFSADFTKSAHHILLHAACLRKR 2277
> tgo:TGME49_047760 long chain acyl-CoA synthetase, putative (EC:6.2.1.3);
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=793
Score = 28.9 bits (63), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 21/49 (42%), Positives = 26/49 (53%), Gaps = 10/49 (20%)
Query 23 IYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTHPDEVVKMIDEE 71
+YD+L EA L YI G T L FVEGAK HP + ++ EE
Sbjct 266 LYDTLGHEALL-YIIGL-------TKLQVLFVEGAKLHP--ALNLVTEE 304
> cpv:cgd2_920 peptidase'insulinase-like peptidase'
Length=1028
Score = 28.9 bits (63), Expect = 4.0, Method: Composition-based stats.
Identities = 25/128 (19%), Positives = 61/128 (47%), Gaps = 2/128 (1%)
Query 1 LGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAK-T 59
LG ++ +V ++ F++ + LRT QL Y+ A + ++ ++ +++ ++ T
Sbjct 817 LGEYNLRKQVLCDLILPFVSSEAFADLRTNQQLAYVVRATQIFSSPAIIIGYYLQSSEYT 876
Query 60 HPDEVVKMIDEELTKAKEYLANMPDAEM-ARWKEAAHAKLTKMEANFSEDFKKSAEEIFS 118
+ + ++++ + K K L + + EM + K++ L+ + +++K EI
Sbjct 877 NALTLERLLEFHINKTKVELKSKLNKEMFIKLKDSTIQTLSSNPKSIFDEYKTYLHEINE 936
Query 119 HSNCFTKR 126
S F R
Sbjct 937 RSYLFDIR 944
> mmu:240087 Mdc1, 6820401C03, AA413496, Nfbd1, mKIAA0170; mediator
of DNA damage checkpoint 1
Length=1708
Score = 28.9 bits (63), Expect = 4.5, Method: Composition-based stats.
Identities = 20/50 (40%), Positives = 25/50 (50%), Gaps = 6/50 (12%)
Query 30 EAQLGYIAGAKESQAASTALL-----QCFVEGAKTHPDEVVKMIDEELTK 74
EA +G AKE A L QCFVEG HP E V+ ++ E T+
Sbjct 631 EAHVGRTKSAKECCDAEPEDLCLPATQCFVEGESQHP-EAVQSLENEPTQ 679
> cpv:cgd6_520 Ser/Thr protein kinase
Length=1103
Score = 28.5 bits (62), Expect = 5.1, Method: Composition-based stats.
Identities = 18/55 (32%), Positives = 25/55 (45%), Gaps = 1/55 (1%)
Query 21 RRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTHPDEVVKMIDEELTKA 75
RR Y T YIA K + +T LL CF G HP E++ + + + K
Sbjct 235 RRKYYVKVTPINTKYIAIVK-TITNNTPLLLCFEHGKDRHPKEIINLKNSNIYKG 288
> ath:AT3G57470 peptidase M16 family protein / insulinase family
protein; K01408 insulysin [EC:3.4.24.56]
Length=891
Score = 28.5 bits (62), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 16/64 (25%), Positives = 26/64 (40%), Gaps = 0/64 (0%)
Query 24 YDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTHPDEVVKMIDEELTKAKEYLANMP 83
+ LRT QLGYI S + +Q ++ + P + ++ L + NM
Sbjct 711 FHQLRTIEQLGYITSLSLSNDSGVYGVQFIIQSSVKGPGHIDSRVESLLKDLESKFYNMS 770
Query 84 DAEM 87
D E
Sbjct 771 DEEF 774
> dre:797490 cplx4b, si:ch211-261c8.4; complexin 4b
Length=126
Score = 28.1 bits (61), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 30/53 (56%), Gaps = 6/53 (11%)
Query 61 PDEVVKMIDEELTKAKE------YLANMPDAEMARWKEAAHAKLTKMEANFSE 107
P+E+VKM+D E + +E + N+ + ++ + KE A A L +M++ E
Sbjct 69 PEELVKMVDGEAPQEEENPSIMAQMQNLQNMDVGQLKEKAQATLVEMKSKAEE 121
> eco:b2821 ptrA, ECK2817, JW2789, ptr; protease III (EC:3.4.24.55);
K01407 protease III [EC:3.4.24.55]
Length=962
Score = 27.7 bits (60), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 24/115 (20%), Positives = 45/115 (39%), Gaps = 0/115 (0%)
Query 13 AVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTHPDEVVKMIDEEL 72
++L Q + Y+ LRTE QLGY A + ++ P + +
Sbjct 777 SLLGQIVQPWFYNQLRTEEQLGYAVFAFPMSVGRQWGMGFLLQSNDKQPSFLWERYKAFF 836
Query 73 TKAKEYLANMPDAEMARWKEAAHAKLTKMEANFSEDFKKSAEEIFSHSNCFTKRD 127
A+ L M E A+ ++A ++ + E+ K +++ + F RD
Sbjct 837 PTAEAKLRAMKPDEFAQIQQAVITQMLQAPQTLGEEASKLSKDFDRGNMRFDSRD 891
Lambda K H
0.315 0.128 0.355
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2054672932
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40